Introduction
Coronary artery disease (CAD) is a major cause of
morbidity and mortality worldwide, with particularly high levels in
India. The predisposing risk factors identified at present
(diabetes, hypertension, obesity and smoking) do not give a good
prediction of the risk of an individual developing CAD. Complex
diseases such as CAD are caused by a combination of environmental
and genetic factors, and altered molecular mechanisms. Network
biology-based approaches have been previously used as powerful
tools in studying and understanding the physical and functional
interactions between these factors (1). Furthermore, these methods are being
applied in the development of novel approaches for disease
classification and the improved understanding of pathophenotypes
(2).
Several studies suggesting that infections may
predispose to CAD (3–8) are based on the evidence that
infectious agents reside in the wall of atherosclerotic vessels
(5), and seroepidemiological
studies demonstrating an association between pathogen-specific
immunoglobulin G antibodies and atherosclerosis. However, a
comprehensive network analysis of infection and CAD has not been
performed to understand the molecular associations between these
two pathologies, specifically in humans.
A previous study by Gulbahce et al (9) provided several novel insights into
viruses and diseases by constructing a viral disease network.
Subsequently, numerous studies aiming to uncover the novel disease
associations, in order to understand associations between clinical
presentation and molecular networks, have been conducted (10–16).
The present study aimed to use complex clinical phenotype
information and molecular networks to elucidate the functional
associations between infection, inflammation and CAD. Integration
of discrete data sets from high throughput technologies with
clinical phenotype information can potentially lead to the
identification of the functional networks that respond to
environmental and genetic factors.
Tools are often used with networks to graphically
represent the nodes and edges, thus identifying the associations,
interactions, co-expression, coregulation and modulations in normal
and disease conditions. The addition of gene ontologies to these
networks can provide a higher level of information of the
alterations in biological processes/functions in diseases, thus may
aid in the elucidation of causal associations between certain
factors and disease. A similar study completed using
macrophage-enriched metabolic networks in mice that were also
conserved in humans identified potential causative mechanisms for
several metabolic diseases (17).
The current study identified that infections may trigger the
networks of mechanisms including xenobiotic responses, cell surface
anchoring and inflammation in myocardial infarction (MI).
Furthermore, the analysis conducted additionally identified a
simple and cost effective potential biomarker for identifying
people at high risk of CAD and MI.
Materials and methods
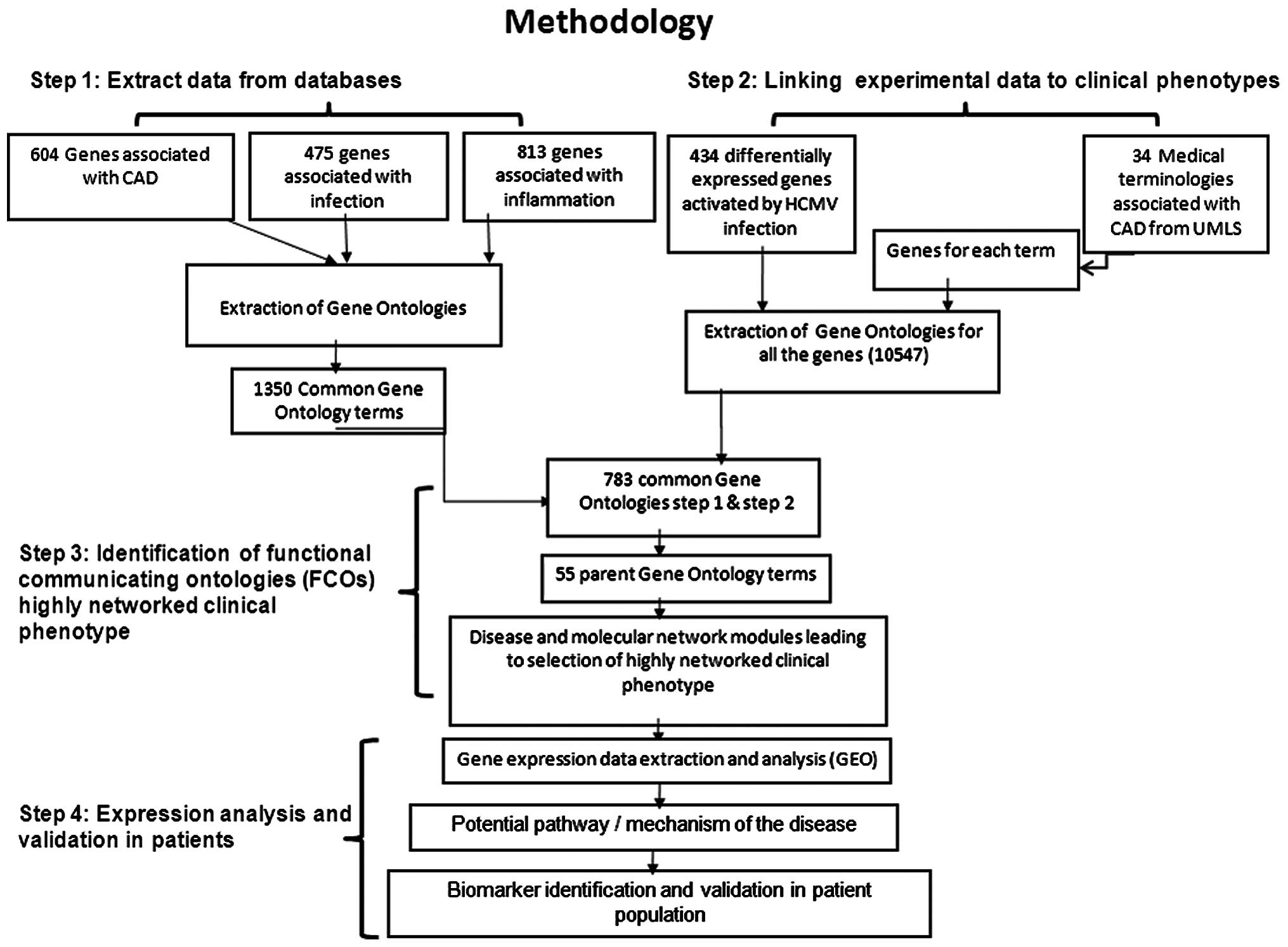
The methodology adapted as presented in Fig. 1 was divided into four steps.
Step 1: Extraction of knowledge base
The human gene sets (flat files) were collected
using the search terms, ''infection'' and ''infammation'' from the
UniProt database, which resulted in 475 and 814 genes (search
conducted on Oct 31, 2013). For CAD, all 604 genes listed in the
CAD Gene Database (http://www.bioguo.org/CADgene/) (18) were considered. Gene ontolgies (GO)
for all the genes were extracted from the UniProt flat files. In
order to understand common molecular mechanisms and functions, GO
terms of the three gene sets were matched. A unique list of GO
terms was used for each gene set in each of the steps.
Step 2: Linking the experimental data to
clinical phenotypes
In order to understand the role and molecular
connections of infection, specifically of cytomegalovirus (CMV),
434 genes that were demonstrated to be differentially expressed
upon infection of the human foreskin fibroblast cell line (CC-2509)
with CMV (19) were investigated.
Furthermore, 34 clinical terms associated with CAD extracted from
the Unified Medical Language System (http://www.nlm.nih.gov/research/umls/) (Table I) were used as search terms for
extracting corresponding genes from the UniProt database. Out of 34
clinical phenotypes, 23 resulted in 3,489 genes. The GO terms of
all of these genes were combined and a unique list of 10,547 terms
was generated. These terms associate CMV infection-mediated gene
expression with clinical phenotypes.
 | Table IThe 34 medical terms associated with
coronary artery disease extracted from the Unified Medical Language
System. |
Table I
The 34 medical terms associated with
coronary artery disease extracted from the Unified Medical Language
System.
| Medical term |
|---|
| 1. Tobacco
abusea |
| 2. Angina |
| 3. Exercise stress
test abnormal |
| 4. Chest pain |
| 5. Shortness of
breath |
| 6. Obesity |
| 7. Coronary artery
disease risk high |
| 8. Hypertension |
| 9. Coronary artery
bypass grafta |
| 10 Organic heart
disease |
| 11. Heart |
| 12. Angina
stable |
| 13. Epigastric
paina |
| 14. Myocardial
infarction acute |
| 15. Angina
unstable |
| 16. Orthopneaa |
| 17. Coronary
atherosclerotic heart disease |
| 18. Cardiac
arrest |
| 19. Diabetic
retinopathy |
| 20. Lower extremity
edemaa |
| 21. Congestive heart
failure |
| 22. Myocardial
infarction |
| 23. Cardiac
catheterizationa |
| 24.
Electrocardiogram abnormala |
| 25. Percutaneous
transluminal coronary angioplasty planneda |
| 26. Pyuriaa |
| 27.
Hypercholesterolemia |
| 28. Diabetes
mellitus |
| 29. Atrial
fibrillation new onset |
| 30. Ischemic heart
disease silenta |
| 31. Weight
loss |
| 32. Diabetes
mellitus insulin dependent |
| 33. Dyspnea
paroxysmal nocturnala |
| 34.
Hyperlipidemia |
Step 3: Identification of functional
communicator ontologies (FCOs) and the highly networked clinical
phenotype
In order to identify the FCOs which link different
clinical phenotypes, 783 common GO terms from step 1 and step 2
were used. Due to the complexity of representing and evaluating a
large network, parent ontologies termed as FCOs were used in the
final construction of network using Cytoscape 3.0.2 software
(http://www.cytoscape.org/). Network
topology statistics such as node degree distribution and the
average short path were used as parameters for selection of highly
networked clinical phenotypes and FCOs.
Step 4: Expression analysis and
validation in patients Identification of differentially expressed
genes in patients
Based on the network parameters described in step 3,
MI was identified to be a highly networked clinical phenotype.
Therefore, the data set GSE48060 previously published by Suresh
et al (20) was selected
for further gene expression analysis. The raw microarray data was
downloaded from the Gene Expression Ominibus (http://www.ncbi.nlm.nih.gov/geo/) and was
analyzed using GeneSpring software, version 12.5 (Agilent
Technologies, Inc., Santa Clara, CA, USA). The GO terms of the
differentially expressed genes were matched to 783 GO terms in
order to elucidate the potential mechanism of CMV infection leading
to CAD. The gene(s) containing the matched GO terms were used for
experimental validation in the selected patient population.
Selection of patient population for
validation of potential biomarker
For validation of potential biomarker(s) patients
enrolled in the Indian Atherosclerosis Research Study (IARS) were
selected. The IARS is a prospective cohort study designed to
investigate and understand the molecular basis of CAD in the Indian
population. The design and details of data collection and the
procedures used were described previously (21). The study was designed in accordance
with the principles and the guidelines of World Medical Association
Declaration of Helsinki and the Indian Council of Medical Research
and approved by the Thrombosis Research Institute Ethics committee.
Signed informed consent was obtained from all the participants who
were enrolled from Narayana Institute of Cardiac Sciences
(Bangalore, India) and the Asian Heart Institute and Research
Centre (Mumbai, India). Between 2004 and 2006, 2,332 subjects were
enrolled of which 772 (33%) were patients with CAD as diagnosed by
an angiogram. Out of these, 200 suffered from MI. These patients
with MI were age and gender matched to 200 unaffected subjects (as
indicated by a normal electrocardiogram) and the serum samples of
all these subjects were used for biomarker assays. All blood
samples were collected subsequent to overnight (12–14 h) fasting
and the plasma and serum aliquots were stored at −80°C. Detailed
information on the medical history, demographics and relevant
information including a 3-generation pedigree was collected and
recorded in a printed questionnaire subsequent to a personal
interview. Diagnosis of diabetes and hypertension was based on
medical reports while body mass index was calculated.
Telephone assessment of cardiac health
status
Periodic telephone follow-up of all study
participants was undertaken to obtain an update on the cardiac
health status, with five rounds of follow-up completed. Follow-up
was performed once every 18 months on average from the time of
recruitment. Endpoints included non-fatal events (redo percutaneous
transluminal coronary angioplasty/redo coronary artery bypass
graft/heart attack/stroke) and fatal events (fatal heart
attack/fatal stroke/sudden death/cardiac arrest). This information
was verified through hospital records wherever available.
γ-glutamyl transferase-5 (GGT-5) assays
and statistical analysis
The plasma GGT-5 concentration was measured using
Flex Reagent Cartridges (Siemens Healthcare GmbH, Erlangen,
Germany) in Dimension Xpand Plus Integrated Chemistry System
(Siemens Healthcare GmbH). The inter assay coefficient of variation
was 5.35%.
Statistical analysis was conducted using SPSS,
version 17.0 (SPSS, Inc., Chicago, IL, USA). Results were presented
as the mean ± standard error for the continuous variables.
Student's independent t-test was used to test for mean differences
in quantitative variables between the MI affected and unaffected
subjects, while the χ2 test was used for testing the
association of qualitative (discrete) variables with CAD.
Hypertension, diabetes and smoking were considered as the
conventional risk factors (CRFs). To assess the association of
individual biomarkers and combinations, logistical regression
analysis was performed. To assess the accuracy of discrimination by
different models of GGT-5, receiver operating characteristics or C
statistics analysis was performed. The statistical significance
between the area under the curve (AUCs) was calculated using the
DeLong method (22) using the R
statistical program, version 3.2.1 (https://cran.r-project.org/). P<0.05 was considered
to indicate a statistically significant difference. For the same
set of subjects, CMV experimental data (200 controls and 200
CAD-affected subjects) were used from a previous publication by our
group (8).
Results
Functions as molecular communicators
between different clinical phenotypes
As presented in Table
II, 4,277 GO terms were identified for the CAD genes followed
by 2,556 for infection and 4,525 for inflammation. These search
terms resulted in 1,350 common GO terms in step 1. In step 2, 2,263
unique GO terms were identified for the differentially expressed
genes upon CMV infection and 8,284 for 23 clinical phenotypes.
Furthermore 783 GO terms matched between step 1 and step 2 were
identified, which represent potential links between diseases and
molecular functions.
 | Table IINumber of genes, their ontologies and
identified FCOs linking molecular data and clinical phenotypes. |
Table II
Number of genes, their ontologies and
identified FCOs linking molecular data and clinical phenotypes.
| Category | No. of genes | No. of gene
ontology terms | No. of common
ontologies | No. of parent
FCOs |
|---|
| Coronary artery
disease | 604 | 4277 | 783 | 55 |
| Infection | 475 | 2556 | | |
| Inflammation | 814 | 4525 | | |
|
Cytomegalovirus | 433 | 2263 | | |
| UMLS (medical
ontologies) | 3489 | 8284 | | |
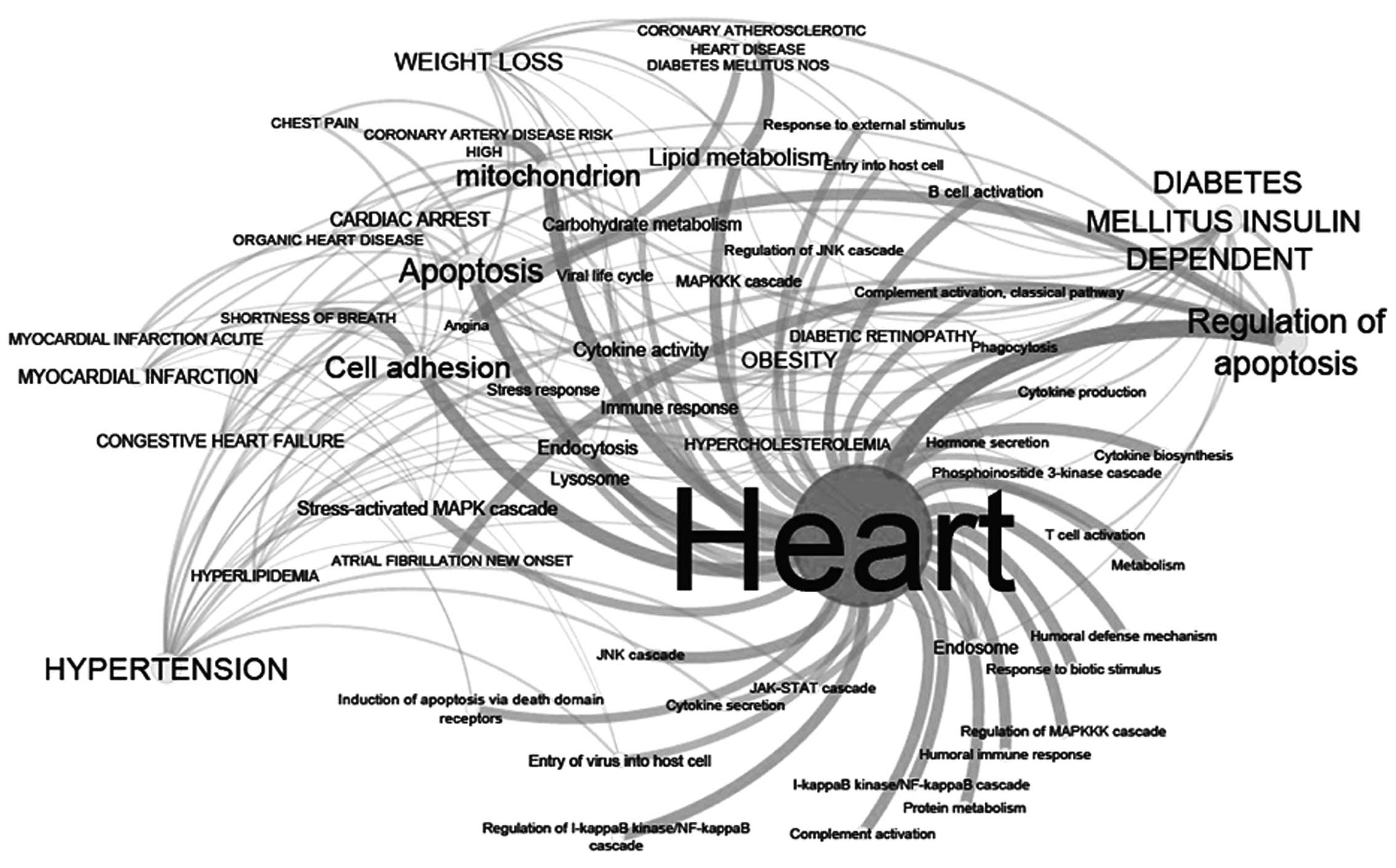
Network of clinical and molecular
ontologies
In order to identify the molecular functions
networking with 23 clinical phenotypes, 55 parent ontologies of 783
GO terms were extracted, which were termed FCOs. As presented in
Fig. 2, different FCOs and
clinical phenotypes are associated with various degrees of
connectivity. Using node degree (ND) and average short path (ASP)
analysis (Table III), it was
identified that FCOs such as apoptosis (ND:14 and ASP:1.8) and cell
adhesion (ND:12, ASP:1.9) were two important molecular functions.
These functions were highly networked with different clinical
phenotypes, followed by mitochondria, regulation of apoptosis,
cytokine activity, lipid metabolism, viral entry and immune
response. These functions are in concordance with viral infections.
Of the 23 clinical phenotypes, MI (ND:15, ASP:2.5) and cardiac
arrest (ND:10, ASP:2.3) were observed to be highly networked with
several FCOs (Table IV). MI was
considered for further analysis as it was highly connected with
important FCOs including viral infections, cell adhesion, entry of
virus into host cell, apoptosis, regulation of apoptosis, stress
response and immune response (Table
IV).
 | Table IIIHighly connected functional
communicator ontologies based on node degree distribution and
average short path in the network. |
Table III
Highly connected functional
communicator ontologies based on node degree distribution and
average short path in the network.
| Gene ontology
term | Average short
path | Node degree |
|---|
| Apoptosis | 1.86440678 | 14 |
| Cell adhesion | 1.93220339 | 12 |
| Mitochondrion | 1.96610169 | 11 |
| Regulation of
apoptosis | 2 | 10 |
| Cytokine activity,
endocytosis | 2.06779661 | 8 |
| Lipid metabolism,
stress-activated MAPK cascade, endosome, carbohydrate metabolism,
immune response | 2.10169492 | 7 |
| Lysosome | 2.13559322 | 6 |
| Viral life cycle,
stress response, MAPKKK cascade | 2.16949153 | 5 |
| Entry of virus into
host cell | 2.20338983 | 4 |
| Response to
external stimulus, B cell activation, induction of apoptosis via
death domain receptors, entry into host cell, phosphoinositide
3-kinase cascade | 2.23728814 | 3 |
| Regulation of c-Jun
N-terminal kinase cascade | 2.20338983 | 4 |
| Complement
activation-classical pathway, hormone secretion, phagocytosis,
cytokine production, T cell activation, cytokine secretion, Janus
kinase-signal transducer and activator of transcription
cascade | 2.27118644 | 2 |
| Complement
activation, cytokine biosynthesis, humoral defense mechanism &
immune response, protein metabolism, regulation of I-κB
kinase/NF-κB cascade, regulation of MAPKKK cascade, response to
biotic stimulus | 2.30508475 | 1 |
 | Table IVTop 5 highly networked clinical
terminologies based on node degree distribution. |
Table IV
Top 5 highly networked clinical
terminologies based on node degree distribution.
| Name | Average short
path | Node degree
(functional communicator ontologies) |
|---|
| MI including acute
MI | 2.52542373 | 14 (entry of virus
into host cell, endosome, apoptosis, regulation of apoptosis,
stress-activated MAPK cascade, stress response, immune response,
immunity, cell adhesion, endocytosis, apoptosis via death domain
receptors, inflammation, regulation of infammation, external side
of membrane) |
| Cardiac arrest | 2.37288136 | 10 (viral life
cycle, cell adhesion, stress response, apoptosis, regulation of
apoptosis, cytokine activity, immune response, mitochondrion,
carbohydrate metabolism, stress-activated MAPK cascade) |
| Congestive heart
failure | 2.47457627 | 7 (cell adhesion,
apoptosis, regulation of apoptosis, apoptosis by death domain
receptors, mitochondrion, stress-activated MAPK cascade,
lysosome) |
| Chest pain | 2.6779661 | 3 (cell adhesion,
apoptosis, mitochondrion) |
| Angina | 2.98305085 | 1 (regulation of
apoptosis) |
The network of clinical terms and FCOs also suggest
that there are common mechanisms and sub-phenotype specific
molecular signals which may serve an important role in disease
onset and progression. As presented in Table IV, the carbohydrate metabolism
appears to be a specific molecular function associated with cardiac
arrest and lysosome function for congestive heart failure.
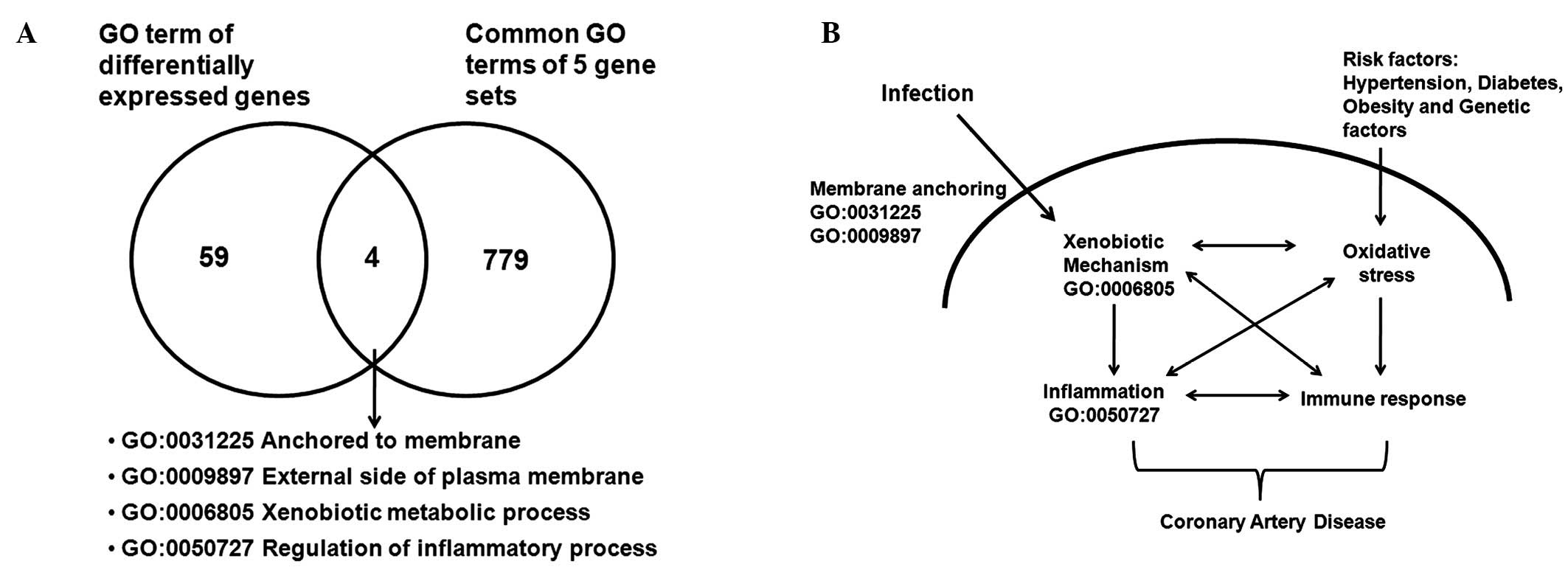
Fusion of global gene expression and FCOs
for mechanism and biomarker discovery
From the diseasome-function network (Fig. 2) for further understanding of the
potential mechanism of infection in CAD, the gene expression data
and FCOs were brought together. The global gene expression data
analysis of the MI dataset GSE48060 (19) indicated 243 differentially
expressed probe sets, of which 170 genes contained GO, gene symbol
and Entrez gene ID information. The 63 GO terms of these
differentially expressed genes were matched with 783 GO terms from
step 3. As presented in the Fig.
3A, the four matching GO terms were: Anchored to membrane,
external side of plasma membrane, xenobiotic metabolic process and
regulation of inflammatory process. These molecular functions and
processes suggest that persistent infection(s) by CMV may lead to
increased inflammation and activation of xenobiotic mechanisms
(Fig. 3B). Genes associated with
these four GO terms and were further analyzed, and it was
identified that the γ-glutamyl transferase family of genes (GGT5,
GGTL3 and GGT4) were involved in all 4 of the functions, and
additional genes such as TNFAIP3 interacting protein 1 and
complement receptor 1 were associated with a minumum of 2.
Therefore, for further analysis GGT5 was considered as a potential
biomarker for assessment in patient subjects.
Biomarker validation
Patient baseline characteristics
As presented in Table
V, mean differences of body mass index, waist:hip ratio, waist
circumference and triglycerides were not identified to be
significantly different between MI-affected and -unaffected
subjects, whereas the mean of high density lipoprotein (HDL) was
observed to be significantly higher in unaffected subjects
(P=0.011). The frequency of diabetes, hypertension, smoking and
usage of prescription medication including statins, beta blockers,
fibrates, calcium channel blockers, angiotensin-converting enzyme
inhibitors, hypoglycemic agents, nitrates and anti-platelet agents
was significantly increased in MI-affected subjects. In addition,
total cholesterol and low density lipoprotein (LDL) were observed
to be at low levels in affected subjects, which may be due to the
greater usage of statins in this group. Additional biomarkers
including GGT-5, CMV and CMV-NA titers were also observed to be
significantly increased in patients with MI in comparison with
unaffected individuals.
 | Table VBase line characteristics of patient
group selected for study. |
Table V
Base line characteristics of patient
group selected for study.
| Variables | Unaffected
(n=200) | Affected (MI,
n=200) | P-value |
|---|
| Agea | 51.88±0.74 | 51.90±0.74 | 0.98 |
| Gender (Male) | 157 (78.5%) | 157 (78.5%) | 0.548 |
| Gender
(Female) | 43 (21.5%) | 43 (21.5%) | |
| Body mass index
kg/m2 | 25.21±0.284 | 25.69±0.280 | 0.22 |
| Waist:hip
ratioa | 0.93±0.005 | 0.94±0.005 | 0.28 |
| Waist
circumferencea (cm) | 90.61±0.73 | 90.35±0.85 | 0.81 |
| Total
cholesterola (mg/dl) | 177.41±2.83 | 150.49±2.64 | 1.54a10−11 |
|
Triglyceridesa (mg/dl) | 160.71±7.716 | 159.44±5.10 | 0.89 |
| High-density
lipoproteina (mg/dl) | 37.58±0.66 | 35.29±0.60 | 0.011 |
| Low-density
lipoproteina (mg/dl) | 110.47±2.53 | 83.30±2.35 | 3.85a10−14 |
| Smoking, n (%) | 58 (29.0%) | 94 (47.0%) | 1.49a10−4 |
| Alcohol
consumption, n (%) | 42 (21.0%) | 27 (13.5%) | 0.032 |
| Hypertension, n
(%) | 50 (25.0%) | 82 (41.0%) | 4.72a10−4 |
| Diabetes mellitus,
n (%) | 48 (24.0%) | 88 (44.0%) | 1.76a10−5 |
| Statin, n (%) | 8 (4.0%) | 137 (68.5%) | 1.88a10−46 |
| Beta blocker, n
(%) | 26 (13.0%) | 124 (62.0%) | 2.68a10−25 |
| Fibrate, n (%) | 1 (0.5%) | 8 (4.0%) | 0.018 |
| Calcium channel
blocker, n (%) | 20 (10.0%) | 51 (25.5%) | 1.40a10−17 |
| ACE inhibitor, n
(%) | 15 (7.5%) | 87 (43.5%) | 1.40a10−17 |
| Hypoglycemic
agents, n (%) | 38 (19.0%) | 63 (31.5%) | 0.003 |
| Nitrate, n (%) | 1 (0.5%) | 86 (43.0%) | 7.09a10−30 |
| Antiplatelet, n
(%) | 7 (3.5%) | 177 (88.5%) | 1.24a10−76 |
| Biomarkers |
| GGTa U/l | 32.05±0.97 | 36.05±1.25 | 0.011 |
| CMVa | 9.69±0.514 | 12.55±0.66 | 0.001 |
| CMV-NA
titresb | 4.67±0.052 | 4.92±0.074 | 0.008 |
Association of GGT-5 in patients with
MI
In the logistical regression analysis (Table VI) for the biomarkers (GGT-5, CMV
and CMV-NA titers), it was identified that GGT-5 alone was
associated with an odds ratio of 1.947 (95% CI: 1.14–3.31,
P=0.014), which was greater than for the other markers. The odds
ratios for the combination of GGT-5 + CMV-NA titers was 1.872 (95%
CI, 1.03–3.41; P=0.041), and GGT-5 + CMV was 2.226 (95% CI,
1.26–3.92; P=0.006). When all three biomarkers were added into the
model the odds ratio was 2.133 (95% CI, 1.12–4.03; P=0.020) which
further increased to 2.338 (95% CI, 1.17–4.64; P=0.015) upon
addition of CRFs (waist circumference, hypertension, diabetes and
smoking) and lipids (HDL and LDL). In the final model, as GGT-5 is
known to be associated with alcohol consumption, this was adjusted
for and it was identified that the odds ratio increased to 2.561
(95% CI, 1.27–5.15; P=0.009). The C-statistics analysis (AUC)
indicated that CRFs alone had a AUC of 0.53 (95% CI, 0.47–0.58)
that increased in the final model to 0.711 (95% CI, 0.64–0.77).
This increase was significant as measured using the DeLong method
with P=0.005.
 | Table VILogistical regression analysis for
the association between GGT-5 and myocardial infarction in
combination with CMV infection markers. |
Table VI
Logistical regression analysis for
the association between GGT-5 and myocardial infarction in
combination with CMV infection markers.
| Model | Odds ratio (95%
CI) | AUC (95% CI) |
|---|
| CRFs | – | 0.530
(0.47–0.58)d |
| CMV | 1.31
(1.04–1.63)c | 0.593
(0.53–0.65)b |
| CMV-NA titres | 1.457
(1.10–1.92)b | 0.587
(0.52–0.65)b |
| GGT | 1.947
(1.14–3.310)c | 0.570
(0.51–0.62)c |
| GGT + CMV-NA
titres | 1.872
(1.03–3.41)c | 0.604
(0.54–0.67)a |
| GGT + CMV | 2.226
(1.26–3.92)b | 0.620
(0.56–0.67)a |
| GGT + CMV-NA titres
+ CMV | 2.133
(1.12–4.03)c | 0.633
(0.57–0.69)a |
| GGT + CMV-NA titres
+ CMV + CRFs + lipids | 2.338
(1.17–4.64)c | 0.704
(0.64–0.76)a |
| GGT + CMV-NA titres
+ CMV + CRFs + lipids + alcohol consumption | 2.561
(1.27–5.15)b | 0.711
(0.64–0.77)a |
Discussion
In order to prevent, diagnose and treat disease,
there is a requirement to integrate data from different sources
(biological experiments and clinopathological information), which
will be stored in different formats depending on the technologies
used. This integration of data will provide an improved
understanding of disease-disease associations, gene prioritization,
function prediction and significant comorbidity effects in
associated diseases (10–17). The current study developed, to the
best of our knowledge for the first time, a novel translational
informatics approach in combining molecular and clinical
information for translational research. One of the key aspects of
CAD research has been the identification of potential biomarkers
for risk prediction, with infection having been identified as an
important factor (1–7, 2 3–2 9). However, the molecular and
functional associations between the infection (by CMV) and CAD
remain to be fully understood. The current study used data stored
in molecular and clinical databases. The 55 FCOs (Table II) associated with 23 clinical
phenotypes indicates the system-level molecular associations
between diseases and molecular functions (Fig. 2 and Table III). This network may provide a
platform for exploring the associations between phenotype and
disease from a genetic to a functional level. The network
statistics analysis suggested that important FCOs such as
apoptosis, cell adhesion and lipid metabolism serve important roles
in CAD (Table III). These
functions may represent the associations between pathogen-induced
injury and CAD onset or progression. Cell adhesion of virus and
entry into endothelial cells may be the first steps towards vessel
wall injury (30). Furthermore, it
has been suggested that the inflammatory response to injury,
dysregulation of lipid metabolism and the presence of activated
macrophages and T-cells may lead to the initiation of
atherosclerotic lesions. At the diseasome level, the highly
networked disease MI was observed to network with important FCOs
including entry of virus into host cell, apoptosis, stress response
and immune response, which are also associated with infections
(Table IV). A previous study
suggested that infections may serve an important role in MI
(31), which may potentially be
due to modulation of the above pathways (Fig. 3 and Table V). This approach identified
molecular functions associated with clinical phenotypes, and those
associated with unique functions including carbohydrate metabolism
specifically associated with cardiac arrest (Table IV). Furthermore, by combining 783
common GO terms between different mechanisms with the
differentially expressed gene profiles of patients with MI, 4
important molecular functions were indicated, leading to the
identification of GGT-5 as a potential biomarker (Tables V and VI). GGT-5 has been previously
demonstrated to be an important marker for the onset of metabolic
syndrome and predicts incidence of cardiovascular disease (32). In the current study, GGT-5 was
demonstrated to be involved in the four identified GO terms
including xenobiotic response, and it is suggested that in addition
to its previously established role in oxidative stress and
metabolic syndrome (33), GGT-5
may serve an important role in CAD predisposition (Fig. 3B).
Previous studies have demonstrated the association
of infections and CAD, and additionally localization of infectious
agents in the plaque area (1–7,23–29),
suggesting that these infections may contribute to increases in
inflammation in the plaque area. It has been previously suggested
that indirect upregulation of inflammatory molecules may serve a
role in CMV infection (3).
However, in the current study (Fig. 3A
and B), it was observed that active infection by CMV may
trigger viral entry pathways including cell adhesion/plasma
membrane adhesion stimulating xenobiotic the mechanism, an increase
apoptosis, inflammation and modulation of immunity. This may
explain failure of antibiotic trials for CAD (33,34),
as infection may lead to modulation of the xenobiotic mechanism
(Fig. 3B), resulting in
ineffectiveness of the drugs. Infection has also been demonstrated
to accelerate atherosclerosis in hyperlipidemic animal models
(25–29), suggesting that infections in
addition to risk factors including obesity, diabetes and
hypertension may increase the oxidative stress which is known to
increase inflammation. Therefore, infection is able to stimulate a
cascade of molecular events (Fig.
3B) leading to imbalanced inflammatory and immune responses.
Persistent infections which trigger long term alterations in the
abovementioned molecular mechanisms are able to potentiate disease
progression and development from lesion generation to plaque
formation, which may explain the presence of infectious agents in
plaques. The methods used in the current study for fusion of
clinical and molecular data may aid in the advancement of patient
care and further research.
In conclusion, an integrative translational
informatics approach was established in the current study, which
may aid in the understanding of the molecular mechanisms in CAD,
and additionally identify potential biomarkers for use in risk
stratification. However, it is necessary to perform biomarker
assays in a larger patient population to validate the observations
of the current study.
Acknowledgments
The current study was supported by the Department of
Biotechnology, Ministry of Science and Technology, Government of
India (grant no. BT/01/CDE/08/07), the Tata Social Welfare Trust,
India (grant no. TSWT/IG/SNB/JP/Sdm) and the Bharati Foundation,
India (grant no. 005/2012–2013). Sponsors had no role in the
design, conduct, sample collection, analysis and interpretation of
the data or in the preparation, review or approval of the
manuscript. The authors would like to thank all investigators,
staff and administrative teams and participants of IARS at Narayana
Institute of Cardiac Sciences, Bangalore (India) and the Asian
Heart Institute, Mumbai (India) for their contributions. The
authors would also like to thank the patients and their family
members for participating in the study.
Abbreviations:
|
CAD
|
coronary artery disease
|
|
MI
|
myocardial infarction
|
|
CMV
|
cytomegalovirus
|
|
GO
|
gene ontology
|
|
FCOs
|
functional communicator ontologies
|
|
GGT-5
|
γ-glutamyl transferase 5
|
|
ASP
|
average short path
|
|
ND
|
node degree
|
References
|
1
|
Cho DY, Kim YA and Przytycka TM: Chapter
5: Network biology approach to complex diseases. PLOS Comput Biol.
8:e10028202012. View Article : Google Scholar
|
|
2
|
Loscallzo J, Kohanne I and Barabasi AL:
Human disease classification in the post genomic era: A complex
systems approach to human pathobiology. Mol Sys Biol.
3:1242007.
|
|
3
|
Epstein SE, Zhu J, Najafi AH and Burnett
MS: Insights into the role of infection in atherogenesis and in
plaque rupture. Circulation. 119:3133–3141. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Epstein SE, Zhou YF and Zhu J: Infection
and atherosclerosis: Emerging mechanistic paradigms. Circulation.
100:e20–e28. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosenfield ME and Campbell LA:
Pathogenesis and atherosclerosis: Update on potential contribution
of multiple infectious organisms to pathogenesis of
atherosclerosis. Thromb Haemost. 106:858–867. 2011. View Article : Google Scholar
|
|
6
|
Shah PK: Link between infection and
atherosclerosis: Who are the culprits: Virises, bacteria, both, or
neither? Circulation. 103:5–6. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stassen FR, Vainas T and Bruggeman CA:
Infection and atherosclerosis. An alternative view on an outdated
hypothesis. Pharmacol Rep. 60:85–92. 2008.PubMed/NCBI
|
|
8
|
Mundkur LA, Rao VS, Hebbagudi S, Shanker
J, Shivanandan H, Nagaraj RK and Kakkar VV: Pathogen burden,
cytomegalovirus infection and inflammatory markers in the risk of
premature coronary artery disease in individuals of Indian origin.
Exp Clin Cardiol. 17:63–68. 2012.PubMed/NCBI
|
|
9
|
Gulbahce N, Yan H, Dricot A, Padi M,
Byrdsong D, Franchi R, Lee DS, Rozenblatt-Rosen O, Mar JC,
Calderwood MA, et al: Viral perturbations of host networks reflect
disease etiology. PLoS Comput Biol. 8:e10025312012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee DS, Park J, Kay KA, Christakis NA,
Oltvai ZN and Barabási AL: The implications of human metabolic
network topology for disease comorbidity. Proc Natl Acad Sci USA.
105:9880–9885. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goh KI, Cusick ME, Val le D, Childs B,
Vidal M and Barabási AL: The human disease network. Proc Natl Acad
Sci USA. 104:8685–8690. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Linghu B, Snitkin ES, Hu Z, Xia Y and
Delisi C: Genome-wide proirotization of disease genes and
identification of disease-disease associations from an integrated
human functional linkage network. Genome Biol. 10:R912009.
View Article : Google Scholar
|
|
13
|
Janić C and Pržulj N: Biological function
through network topology: A survey of the human diseasome. Brief
Funct Genomics. 11:522–532. 2012. View Article : Google Scholar
|
|
14
|
Emmert-Streib F, Tripathi S, de Matos
Simoes R, Hawwa AF and Dehmer M: The human disease network.
Opportunities for classification, diagnosis and prediction of
disorders and disease genes. Systems Biomedicine. 1:20–28. 2013.
View Article : Google Scholar
|
|
15
|
Piro RM and Di Cunto F: Computational
approached to disease-gene prediction: Rationale, classification
and successes. FEBS J. 279:678–696. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu S, Falck T, Daemen A, Tranchevent LC,
Suykens JA, De Moor B and Moreau Y: L2-norm multiple
kernel learning and its application to biomedical data fusion. BMC
Bioinformatics. 11:3092010. View Article : Google Scholar
|
|
17
|
Chen Y, Zhu J, Lum PY, Yang X, Pinto S,
MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al:
Variations in DNA molecular networks that cause disease. Nature.
452:429–435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu H, Liu W, Liao Y, Cheng L, Liu Q, Ren
X, Shi L, Tu X, Wang QK and Guo AY: CADgene: A comprehensive
database for coronary artery disease genes. Nucleic Acids Res.
39(Database issue): D991–D996. 2011. View Article : Google Scholar :
|
|
19
|
Wang A, Ren L and Li H: A systemic network
triggered by human cytomegalovirus entry. Adv Virol.
2011:2620802011. View Article : Google Scholar
|
|
20
|
Suresh R, Li X, Chiriac A, Goel K, Terzic
A, Perez-Terzic C and Nelson TJ: Transcriptome from circulating
cells suggests dysregulated pathways associated with long-term
recurrent events following frst-time myocardial infarction. J Mol
Cell Cardiol. 74:13–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shanker J, Mitra A, Rao VS, Mundkur L,
Dhanalakshmi B, Hebbagodi S and Kakkar VV: Rationale, design &
preliminary findings of the Indian atherosclerosis research study.
Indian Heart J. 62:286–295. 2010.
|
|
22
|
DeLong ER, DeLong DM and Clarke-Perason
DL: Comparing the areas under two or more correlated receiver
operating characteristics curves: A nonparametric approach.
Biometrics. 44:837–845. 1988. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ji YN, An L, Zhan P and Chen XH:
Cytomegalovirus infection and coronary heart disease risk: A
meta-analysis. Mol Biol Rep. 39:6537–6546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Span AH, Van Boven CP and Bruggeman CA:
The effect of cytomegalovirus infection on the adherence of
polymorphonuclear leucocytes to endothelial cells. Eur J Clin
Invest. 19:542–548. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Naghavi M, Wyde P, Litovsky S, Madjid M,
Akhtar A, Naguib S, Siadaty MS, Sanati S and Casscells W: Influenza
infection exerts prominent inflammatory and thrombotic effects on
the atherosclerotic plaques of apolipoprotein E-deficient mice.
Circulation. 107:762–768. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hsich E, Zhou YF, Paigen B, Johnson TM,
Burnett MS and Epstein SE: Cytomegalovirus infection increases
development of atherosclerosis in apolipoprotein-E knockout mice.
Atherosclerosis. 156:23–28. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Burnett MS, Gaydos CA, Madico GE, Glad SM,
Paigen B, Quinn TC and Epstein SE: Atherosclerosis in apoE knockout
mice infected with multiple pathogens. J Infect Dis. 183:226–231.
2001. View
Article : Google Scholar
|
|
28
|
Ezzahiri R, Neilssen-Vrancken HJ, Kurvers
HA, Stassen FR, Vliegen I, Grauls GE, van Pul MM, Kitslaar PJ and
Bruggeman CA: Chlamydophila pneumonia (Chlamidia pneumoniae)
accelerates the formation of complex atherosclerotic lesions in apo
E3-Leiden mice. Cardiovasc Res. 56:269–276. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ezzahiri R, Stassen FR, Kurvers HA, van
Pul MM, Kitslaar PJ and Bruggemann CA: Chlamydia pneumoniae
infection induces an uns- atherosclerotic plaque phenotype in
LDL-receptor, ApoE double knockout mice. Eur J Vasc Endovasc Surg.
26:88–95. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Janeway CA Jr, Travers P, Walport M and
Shlomchik MJ: The front line of host defense. Immunobiology: The
Immune System in Health and Disease. 5th edition. Garland Science;
New York: pp. 295–340. 2001
|
|
31
|
Smeeth L, Thomas SL, Hall AJ, Hubbard R,
Farrington P and Vallance P: Risk of myocardial infarction and
stroke after acute infection or vaccination. N Engl J Med.
351:2611–2618. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anderson JL and Muhlestein JB: Antiobiotic
trials for coronary heart disease. Tex Heart Inst J. 31:33–38.
2004.
|
|
33
|
Dhingra R, Gona P, Wang TJ, Fox CS,
D'Agostino RB Sr and Vasan RS: Serum gamma glutamyl transferase and
risk of heart failure in the community. Atheroscler Thromb Vasc
Biol. 30:1855–1860. 2010. View Article : Google Scholar
|
|
34
|
Gryaston JT: Antiobiotic treatment of
atherosclerotic cardiovascular disease. Circulation. 107:1228–1230.
2003. View Article : Google Scholar
|