Introduction
Normal bone is maintained in a dynamic state of
equilibrium between osseous absorption and reconstruction. The
process of bone remodeling is associated with the coordinated
regulation of bone-resorbing osteoclasts and bone-forming
osteoblasts (1). Weight-bearing
mechanical loading is essential for maintaining the balance of bone
metabolism. Substantial studies have confirmed that physiological
cyclic loading effectively promotes bone mass and maintains the
optimization of skeletal structures, and the lack of mechanical
stimuli to the skeleton leads to lower bone formation and inferior
bone quality (2). Numerous
experiments have demonstrated that bone cells (including
osteoblasts and osteocytes) secrete various essential cytokines
(including Ca2+, prostaglandin E2 and nitric
oxide) in response to various physiological mechanical stimuli,
including fluid shear stress (3),
compressive force and cyclic stretch (4–6).
However, how bone cells sense the external mechanical signals and,
thus, transduce the them into intracellular biochemical signals
remains unclear, which is regarded as an important question for
deciphering the mechanisms of bone mechanotransduction and
adaptation.
The cytoskeleton is a network of protein filaments
that modulate cellular shape, motility and mechanical properties
(7). Cytoskeletal deformation is
believed to be one of the earliest cellular events in response to
mechanical stimuli, and is involved in regulating transmembrane
signal transduction (6). The
cytoskeleton is composed of three types of protein filaments,
actin, microtubules and intermediate filaments. The actin-based
cytoskeleton has previously been demonstrated to be important for
the biochemical response of bone cells to mechanical loading
(8). A previous study demonstrated
that the deformed actin cytoskeletal network provided enhanced
docking and activation sites for kinases (9). Disruption of the actin cytoskeleton
impaired the ability of bone cells to respond to fluid shear
stress, and enhanced actin polymerization increases osteogenic
differentiation (10). However,
the key signaling pathways that mediate cyclic strain-induced
osteoblastic cytoskeletal deformation and rearrangement have not
been fully elucidated, which is critical for understanding the
mechanism of how the bone cell cytoskeleton responds to external
mechanical signals.
Osteoblast cytoskeletal reconstruction depends on
the regulation of mechanosensitive proteins. Cofilin is an
actin-binding protein that is essential for the depolymerization of
actin filaments (11,12). When an actin filament is activated,
cofilin can sever the actin filament, and in turn increase the
polymerization of muscle actin monomers (13). A previous study demonstrated that
Rho small GTPase, and its downstream effector molecules, regulate
the cyclic strain-induced migration of vascular smooth muscle cells
(9). This mechanism is notable
because Rho is a major organizer of the cytoskeleton (14) and can regulate the formation of
actin stress fibers by activating Rho-associated coiled-coil
containing protein kinase 1 (ROCK), which in turn phosphorylates
cofilin. LIM domain kinase (LIMK) can regulate actin dynamics
through the phosphorylation of cofilin (15). Thus, Rho regulates cofilin via ROCK
and LIMK, and this signal transduction pathway modulates actin
assembly in various cell types in response to numerous
extracellular stimuli. Cofilin binds to actin monomers and
polymers, and promotes the disassembly of actin filaments (16). These observations led to the
hypothesis that the cofilin signaling pathway may be important for
osteoblastic mechanosensing leading to the cytoskeletal
rearrangement of osteoblasts.
Thus, in the present study, the role of cofilin in
regulating cyclic stretch-induced osteogenesis was investigated in
human osteoblast-like MG-63 cells, which are derived from
osteosar-comas and exhibit various osteoblast-like characteristics
(17). MG-63 cells were subjected
to physiological cyclic strain stimulation (12% elongation), and
the expression levels of cofilin and osteogenesis-associated genes
were quantified with reverse transcription-quantitative polymerase
chain reaction (RT-qPCR), immunofluorescence staining and western
blotting analysis. Additionally, RNA interference (RNAi) was
performed to knockdown cofilin expression, and the expression
levels of osteogenesis-associated genes were evaluated.
Materials and methods
Cell culture
Human osteoblast-like MG-63 cells (RIKEN BioResource
Center, Tsukuba, Japan) were maintained in a logarithmic growth
phase in a hybrid modified Eagle's medium (Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 10
µg/ml insulin-transferring-selenium (Gibco; Thermo Fisher
Scientific, Inc.). Cells were cultured at 37°C in a humidified
atmosphere of 5% CO2. Cells were initially seeded at
6×104 cells/well density in 6-well plates. The medium
was replaced every 2 days.
Mechanical stretch stimulation
MG-63 cells were plated in collagen II coated 6-well
BioFlex culture plates with elastic bottom (Flexcell International
Corporation, Burlington, NC, USA) and incubated for 24 h. Cells
were subjected to cyclic stretch with 12% elongation at 0.1 Hz (5
sec stretch/5 sec relaxation) using FX-4000 Tension System
(Flexcell International Corporation) and harvested for biochemical
assays at 1, 4, 8, 12 and 24 h after the application of mechanical
stretch. The control groups were seeded on the same plates and
maintained in the same experimental condition without applying
mechanical stretch.
Total RNA extraction and RT-qPCR
Following mechanical stretching, total RNA was
extracted using TRIzol (Gibco; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions, and quantified with a
NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Inc.).
First-strand cDNA synthesis was performed using the High Capacity
cDNA Archive kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The cDNA reactions
were incubated at 42°C for 60 min and the reaction was terminated
by heating to 70°C for 5 min. The sequences of the primers are as
follows: i) Alkaline phosphatase (ALP), sense
5′-GGACCATTCCCACGTCTTCAC-3′, anti-sense 5′-CCTTGTAGCCAGGCCCATTG-3′;
ii) collagen-1 (COL1), sense 5′-CCAGAAGAACTGGTACATCAGCAA-3′,
antisense 5′-CGCCATACTCGAACTGGAATC-3′; iii) runt-related
transcription factor-2 (Runx2), sense 5′-GGCATGTCCCTCGGTATG-3′,
antisense 5′-CGGAAGCATTCTGGAAGGA-3′; iv) osteocalcin (OCN), sense
5′-GGCAGCGAGGTAGTGAAG-3′, antisense 5′-CGTAGAAGCGCCGATAGG-3′; v)
cofilin, sense 5′-CACCTTTGTCAAGATGCT-3′, antisense
5′-GGAGCTGGCATAAATCAT-3′; and vi) glyceraldehyde 3-phosphate
dehydrogenase (GAPDH), 5′-GTCATCCCAGAGCTGAAC-3′, antisense
5′-TCAGTGTAGCCCAAGATG-3′. The sequences of all primers were
designed by Takara Biotechnology Co., Ltd. (Dalian, China). qPCR
was performed on 1 µl cDNA in a 20 µl reaction with
SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.) using the
Bio-Rad CFX96 real-time PCR detection system (Bio-Rad Laboratories,
Inc. Hercules, CA, USA). GAPDH was used as an internal control for
normalization. The relative quantity of mRNA was calculated using
the 2−ΔΔCq method (18). Triplicate RT-qPCR reactions were
preformed using three separate samples.
Protein extraction and western blot
analysis
Following cyclic stretch loading, cells were washed
once with ice-cold phosphate-buffered saline immediately and lysed
in 500 µl sodium dodecyl sulfate (SDS) buffer. Samples were
then boiled at 95°C for 5 min. The protein concentration was
determined by bicinchoninic acid assay (Thermo Fisher Scientific,
Inc.). Subsequently, 10 µl protein aliquots were subjected
to electrophoretic separation by Tris-glycine SDS-polyacrylamide
gel (Thermo Fisher Scientific, Inc.) electrophoresis (10% resolving
gel) and transferred to polyvinylidene fluoride membranes (Bio-Rad
Laboratories, Inc.). The membranes were blocked in Tris-buffered
saline (0.5% Tween-20) containing 5% bovine serum albumin for 2 h
at 4°C. The membranes were incubated with monoclonal rabbit
anti-cofilin (1:700; cat. no. 5175; Cell Signaling Technology,
Inc., Danvers, MA, USA), monoclonal rabbit anti-phospho-cofilin
(1:500; cat. no. 3313; Cell Signaling Technology, Inc.) and
monoclonal rabbit anti-GAPDH (1:1,000; cat. no. 2118; Cell
Signaling Technology, Inc.) antibodies overnight at 4°C, and then
incubated with horseradish peroxidase-conjugated goat anti-rabbit
secondary antibodies (1:1,000; cat. no. AP307P; EMD Millipore,
Billerica, MA, USA) for 1 h at room temperature. Images for western
blot analysis were visualized by an enhanced chemiluminescence
system (GE ImageQuant 350, GE Healthcare Life Sciences).
Semi-quantitative analysis was performed using the QuantityOne
software (version 4.6.3; Bio-Rad Laboratories, Inc.). GAPDH was
used as an internal control for normalization.
Immunofluorescence staining
Following stimulation with cyclic stretch, cells in
BioFlex culture plates were fixed with 4% formaldehyde for 30 min
at room temperature. The elastic bottom of each plate was cut into
8–10 pieces, which were then blocked in 2% goat serum (Beyotime
Institute of Biotechnology, Haimen, China) in phosphate-buffered
saline at 37°C for 1 h. Following blocking, the cells were
incubated with monoclonal rabbit anti-active cofilin primary
antibody (1:100; cat. no. 5175; Cell Signaling Technology, Inc.)
for 1 h. Cells were then incubated with fluorescein
isothiocyanate-conjugated goat anti-rabbit IgG secondary antibody
(1:500; cat. no. 65–6111; Thermo Fisher Scientific, Inc.) for 1 h
and counterstained with 4′,6-diamidino-2-phe-nylindole for 5 min at
room temperature. Cells were then visualized with an FV1200 laser
scanning confocal microscope (Olympus Corporation, Tokyo, Japan).
Six fields of view were randomly selected and the number of cells
exhibiting nuclear translocation of cofilin were counted and
normalized to the total number of cells in each field.
Cofilin RNAi knockdown
Different cofilin short hairpin (sh) RNA fragments
(GeneCopoeia, Inc., Rockville, MD, USA) were designed to achieve a
low cofilin protein expression model. The shRNA sequences used are
as follows: No. 1, GGACAAGAAGAACATCATC; no. 2, CCTACGCCACCTTTGTCAA;
no. 3, TGTGCGGCTCCTACTAAAC; no. 4, ATGCTGCCAACTTCTAACC; and no. 5,
blank shRNA sequence (control cells). Cells were transfected with
shRNA and pMIR-REPORT luciferase reporter vector (Promega
Corporation, Madison, WI, USA) mixture using Lipofectamine 2000
liposome transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Briefly, 50–100 nM shRNA was mixed with the luciferase reporter
vector and transfection reagent, and the mix was added to MG-63
cells (50–60% confluence). After transfection for 24–36 h, cells
were harvested to determine the cofilin mRNA and protein expression
levels via RT-qPCR and western blot analysis, respectively.
Statistical analysis
All data presented are expressed as the mean ±
standard deviation. Statistical analyses were performed using SPSS
software, version 16.0 (SPSS, Inc., Chicago, IL, USA). One-way
analysis of variance with Tukey's post hoc analysis was used to
determine the differences between groups. P<0.05 was considered
to indicate a statistically significant difference.
Results
Cyclic stretch increases the expression
of genes associated with osteoblastic activities in MG-63
cells
As demonstrated in Fig.
1, cyclic stretch for 1, 4, 8 and 12 h significantly increased
ALP mRNA expression compared with control (P<0.05). OCN and
Runx2 mRNA expression levels were also significantly increased
following 1, 4, 8 and 12 h cyclic stretch stimulation compared with
control (P<0.05). Furthermore, stretch for 1, 4, 8 and 12 h also
significantly increased the mRNA expression levels of COL-1
compared with control (P<0.05).
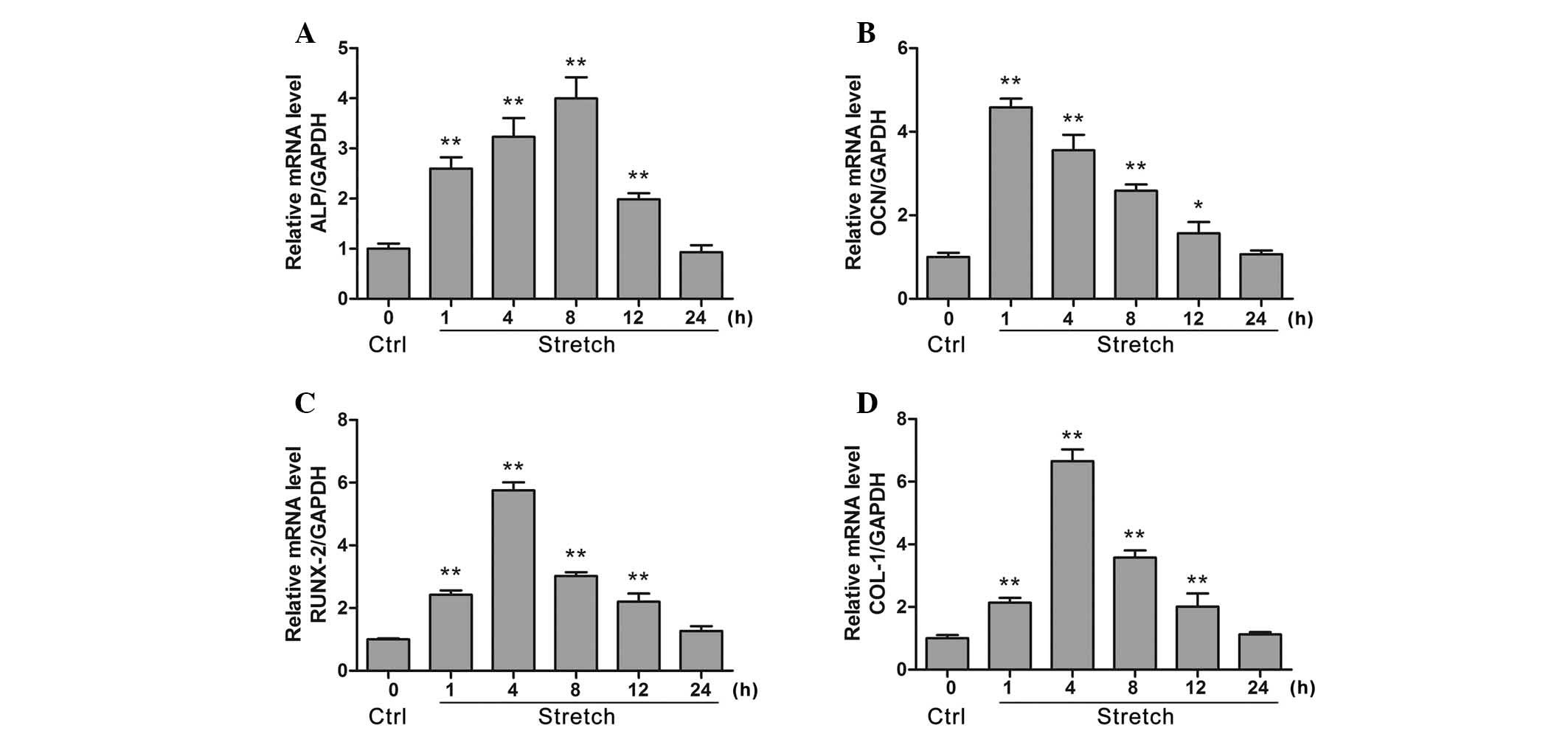 | Figure 1Evaluation for the mRNA expression
levels of osteogenesis-associated genes ALP, OCN, Runx2 and COL-1
in MG-63 cells under cyclic stretch (12% elongation, 0.1 Hz) via
reverse transcription-quantitative polymerase chain reaction
analysis. The effect of cyclic mechanical stretch on (A) ALP, (B)
OCN, (C) Runx2 and (D) COL-1 mRNA levels were measured at 1, 4, 8,
12 and 24 h. Data are presented as the mean ± standard deviation,
n=6. *P<0.05, **P<0.01 vs. control.
ALP, alkaline phosphatase; OCN, osteocalcin; Runx2, runt related
transcription factor 2; COL-1, collagen 1; GAPDH, glyceraldehyde
3-phosphate dehydrogenase; Ctrl, control. |
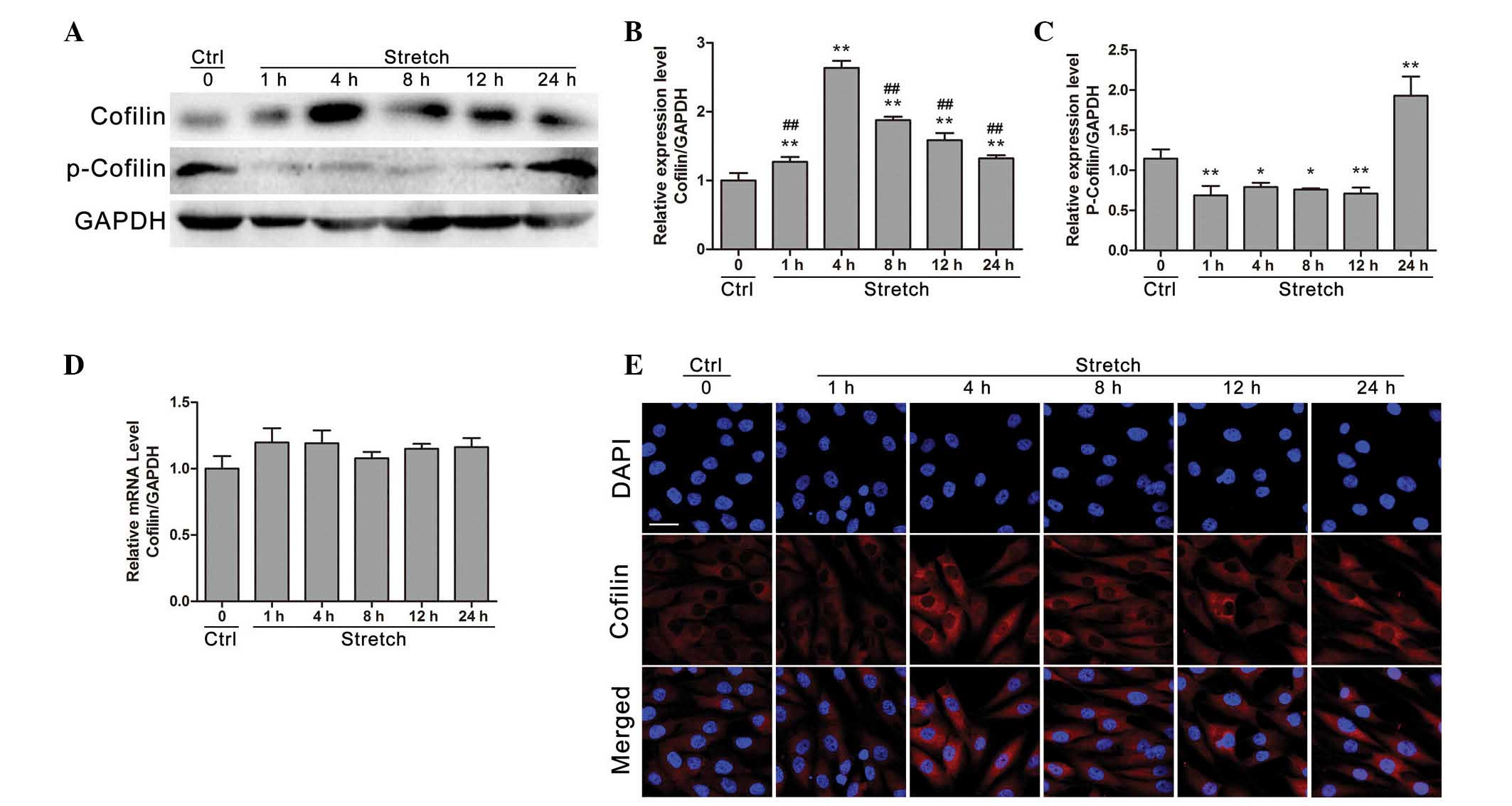
Cyclic stretch regulates the gene and
protein expression of cofilin in osteoblast-like MG-63 cells
Western blot analysis (Fig. 2A) demonstrated that cofilin protein
expression was increased within the first 4 h of mechanical strain
application compared with control (P<0.01). Following 8, 12 or
24 h of stretch stimulation, cofilin protein expression was
decreased compared with the 4 h stimulation group (P<0.05),
whereas it remained significantly higher compared with the control
group (P<0.01; Fig. 2B).
Furthermore, following cyclic stretch for 1, 4, 8 and 12 h the
phosphorylation levels of cofilin were significantly decreased
compared with control (P<0.05), whereas cofilin phosphorylation
was significantly increased at 24 h stretch compared with control
(P<0.01; Fig. 2C). However, as
presented in Fig. 2D, cofilin mRNA
expression levels demonstrated no significant change compared with
control following cyclic stretch stimulation for 1, 4, 8, 12 or 24
h (P>0.05). Additionally, the regulatory effects of cyclic
stretch on cofilin protein expression were further confirmed by
immunofluorescence analysis, which demonstrated that cofilin
protein expression was increased under 4 h cyclic stretch compared
with control.
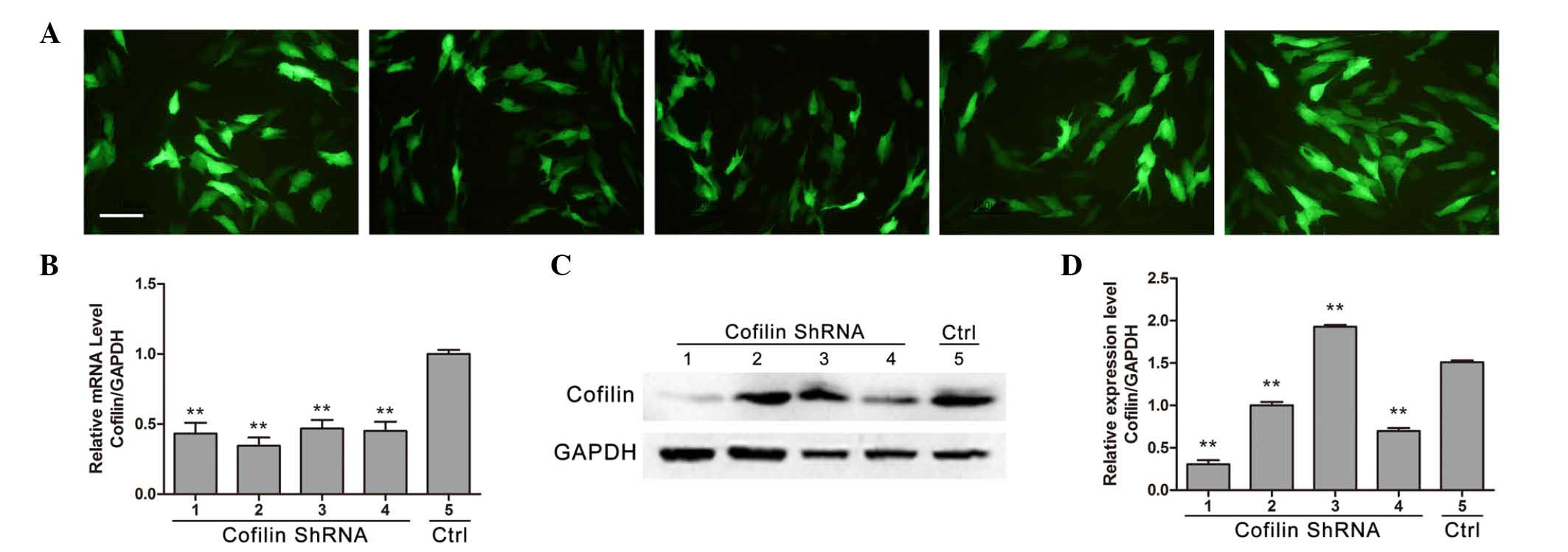
Stretch-induced increases in
osteoblast-associated genes are suppressed by cofilin gene
knockdown
Immunofluorescence was preformed to detect cofilin
protein expression levels in osteoblast-like MG-63 cells
transfected with different shRNA fragments (Fig. 3A). Compared with control shRNA, the
four different cofilin shRNAs significantly reduced the expression
of cofilin at the mRNA level (P<0.01; Fig. 3B), whereas no significant
difference between each shRNA was observed (P>0.05; Fig. 3B). Western blot results
demonstrated that shRNAs no. 1 and 4 significantly reduced the
protein expression level of cofilin (P<0.01), indicating that
shRNAs no. 1 and 4 effectively inhibited the expression of cofilin
(Fig. 3C and D). Therefore, shRNA
no. 1 was used to knockdown cofilin expression in osteoblast-like
cells. As demonstrated in Fig. 4,
cofilin inhibition by shRNA significantly decreased the ALP mRNA
expression levels compared with the levels under mechanical stretch
(P<0.01). Furthermore, the mRNA expression levels of OCN, Runx2
and COL-1 were also significantly decreased by cofilin knockdown
compared with the levels under mechanical stretch.
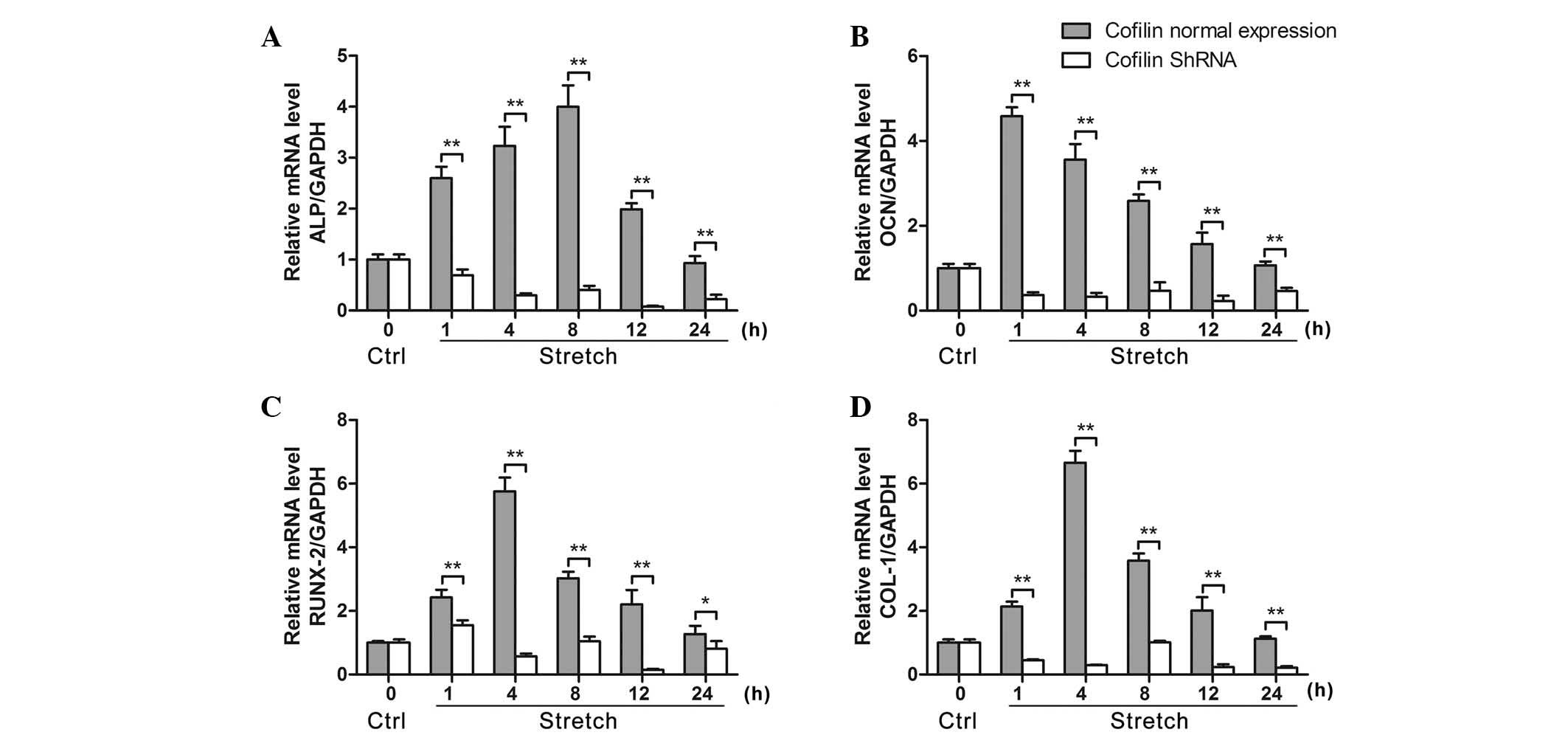 | Figure 4Inhibition of cofilin suppresses
stretch-induced decreased expression of osteogenesis-associated
genes ALP, OCN, Runx2 and COL-1 in osteoblast-like MG-63 cells. (A)
shRNA knockdown of cofilin suppressed the cyclic mechanical
stretch-induced mRNA expression of (A) ALP, (B) OCN, (C) Runx2 and
(D) COL-1 under cyclic stretch for 1, 4, 8, 12 and 24 h, measured
by reverse transcription-quantitative polymerase chain reaction
(n=6). Data are presented as the mean ± standard deviation.
*P<0.05, **P<0.01, comparisons
indicated by brackets. ALP, alkaline phosphatase; OCN, osteocalcin;
Runx2, runt-related transcription factor 2; COL-1, collagen 1;
shRNA, short hairpin RNA; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; Ctrl, control. |
Discussion
Cyclic strain has previously been demonstrated to
regulate various aspects of cellular activity, including cell
proliferation, migration and cytoskeletal arrangement in a number
of cell types, including fibroblasts, endothelial cells and smooth
muscle cells (4,19,20).
It has also been demonstrated that cyclic stretch can regulate
osteoblast activities, including increasing cell proliferation, DNA
synthesis and prostaglandin E2 production and secretion
(6). However, the mechanisms by
which bone cells sense the external mechanical stimuli and
transduce them into intracellular biochemical signals remain poorly
understood (21). Understanding
mechanotransduction in bone cells has significant clinical
implications for diseases involving dysfunctional bone remodeling
(including osteoporosis and osteopetrosis), and is also beneficial
for understanding the mechanisms that underlie treatments
associated with mechanical stimulation (such as distraction
osteogenesis).
Several previous studies have demonstrated that the
deformation of cytoskeletal components, including F-actin and
microtubules, is an essential early perception event for bone cells
to sense external mechanical stimulations (8,22,23).
Cofilin is a regulator of actin filament non-equilibrium assembly
and disassembly (24,25). The promotional effects of cofilin
on actin assembly or disassembly depend upon the concentration of
cofilin relative to actin and other actin-binding proteins
(26). The results of these
previous studies reveal that cofilin may be closely associated with
intracellular signal transduction. However, the function of cofilin
in osteoblastic activities and osteoblastic mechanotransduction has
rarely been investigated. The findings of the present study
demonstrate that cyclic strain significantly promotes cofilin
activation, and inhibition of cofilin significantly reduced the
expression of osteogenesis-associated genes in response to
mechanical loading. Thus, the results of the current study clearly
demonstrated that cofilin was involved in mechanical load-induced
osteogenesis and, to the best of our knowledge, provides the first
evidence demonstrating that cofilin is important for osteoblastic
mechanotransduction.
Mechanical loading regulates the gene expression of
cells in various organs, including bone, cartilage and ligaments
(27). Several previous studies
have also demonstrated that mechanical stretch stimulates the
expression of a variety of osteogenesis-associated genes, including
OCN, Runx2, ALP, and COL-I and III (28,29).
In accordance with these previous findings, the present results
also demonstrated that cyclic mechanical stretch with 12%
elongation significantly promoted the mRNA expression of
osteogenesis-associated genes, including ALP, OCN, COL-1 and Runx2.
Furthermore, significant increases in ALP and OCN mRNA expression
were detected even at 1 h post-mechanical stimulation, indicating
that ALP and OCN acted as major early sensitive genes for detecting
and responding to external mechanical signals. The COL-1 and Runx2
mRNA expression levels peaked at 4 h following the application of
cyclic stretch, which suggested that the changes in COL-1 and Runx2
gene expression levels may not be early intracellular biochemical
events in the response of cyclic stretch. These findings are
consistent with previous investigations (4,28,30).
The mechanism by which osteoblasts convert mechanical strain into
biochemical signals remains unclear. A previous proteomic analysis
on low shear stress-induced vascular remodeling demonstrated that
Rho GDP dissociation inhibitor α responds to shear stress, and
modulates vascular smooth muscle cell migration and apoptosis
(9). Furthermore, Lin et al
(31) demonstrated that shear
stress activates the Rho-ROCK-LIMK-cofilin pathway and, thus,
regulates the activity of endothelial cells. Therefore, it was
hypothesized that the Rho-ROCK-LIMK-cofilin pathway may be involved
in intracellular signal transduction of osteoblasts in response to
external mechanical signals. The findings of the current study
clearly demonstrated that cyclic strain for 1 and 4 h significantly
upregulated the protein expression levels of cofilin in
osteoblast-like MG-63 cells subjected to cyclic stretch compared
with the control group, whereas the cofilin expression was
downregulated after 8 h cyclic stretch compared with the 4 h group
but remained higher than the control group. Additionally, cofilin
phosphorylation was downregulated after 1, 4, 8 and 12 h, and
upregulated at 24 h of mechanical stretch compared with the control
group. Pan et al (27)
demonstrated that cyclic strain increases the phosphorylation of
cofilin over 24 h. The findings of the current study demonstrated
that cyclic strain for 24 h increased the level of phospho-cofilin,
whereas at the early time points of cyclic strain, the level of
phospho-cofilin was decreased. This may be a result of mechanical
strain activating cofilin expression at the early stages, but with
the increasing cofilin levels, Rho and ROCK upregulated LIMK to
increase the phosphorylation of cofilin in response to mechanical
strain, which caused the reduced expression of cofilin over time
(15,31,32).
Using shRNA-targeted transfection technology, decreased cofilin
expression resulted in a significant reduction in the mRNA
expression levels of osteogenesis-associated genes (ALP, OCN, COL-1
and Runx2). The increase in osteogenesis-associated gene expression
upon the application of mechanical strain in the present study
further confirms the close association between cofilin and
osteogenic functions induced by mechanical strain. In addition, the
simultaneous increases in cofilin phosphorylation activation
indicate that the Rho-ROCK-LIMK-cofilin signaling pathway is
important for mechanotransduction and the regulation of
osteogenesis functions in response to cyclic strain. However, it
will be important to further explore the specific functions and
effects of cofilin on osteoblastic cytoskeletal deformation and
reconstruction in response to mechanical signals in future
studies.
Actin is recognized to be essential for chromatin
remodeling, formation of heterogeneous nuclear ribonucleoprotein
complexes and gene expressions (33,34).
However, the actin amino acid sequence lacks a nuclear
translocation signal, and as a 42-kDa protein, is unlikely to enter
the nucleus by diffusion. Thus, it relies on transporter proteins
(such as cofilin) to mediate this entry, which may be promoted by a
variety of adverse cellular conditions, including heat shock
(35,36) and ATP depletion (37). Thus, the current study considered
that the ability to enable actin nuclear functions is one of the
crucial cellular functions of cofilin in response to external
stimuli, such as cyclic strain. Using immunofluorescent staining,
the present study observed that cofilin was translocated from the
cytoplasm to the nucleus after 1 h mechanical loading, indicating
that cofilin may bind actin and enter the nucleus Thus, mechanical
strain, as an external stimuli, induced a protective effect via
cofilin cytoskeletal-binding protein. However, more detailed
analyses of these protein events are necessary to improve our
understanding of cofilin functions.
In summary, the findings of the present study
demonstrated that cyclic strain promotes the mRNA expression of
osteogenesis-associated genes and also regulates cofilin protein
expression levels. Furthermore, stretch-induced increases of
osteogenesis-associated genes, including ALP, OCN, Runx2 and COL1,
are reduced by cofilin gene knockdown. These results demonstrate
that cofilin is involved in the regulation of mechanical
load-induced osteogenesis, and to the best of our knowledge,
provide the first evidence that cofilin is involved in osteoblastic
mechanotransduction.
Acknowledgments
The present study was supported by grants from the
National Science Foundation of China (grant nos. 31070836 and
81100750).
References
|
1
|
Harter LV, Hruska KA and Duncan RL: Human
osteoblast-like cells respond to mechanical strain with increased
bone matrix protein production independent of hormonal regulation.
Endocrinology. 136:528–535. 1995.PubMed/NCBI
|
|
2
|
Frost HM: Bone's mechanostat: A 2003
update. Anat Rec A Discov Mol Cell Evol Biol. 275:1081–1101. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan H, Biechler S, Junor L, Yost MJ, Dean
D, Li J, Potts JD and Goodwin RL: Fluid flow forces and rhoA
regulate fibrous development of the atrioventricular valves. Dev
Biol. 374:345–356. 2013. View Article : Google Scholar
|
|
4
|
Yano Y, Geibel J and Sumpio BE: Cyclic
strain induces reorga-nization of integrin alpha 5 beta 1 and alpha
2 beta 1 in human umbilical vein endothelial cells. J Cell Biochem.
64:505–513. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Archambault J, Tsuzaki M, Herzog W and
Banes AJ: Stretch and interleukin-1beta induce matrix
metalloproteinases in rabbit tendon cells in vitro. J Orthop Res.
20:36–39. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mayr M, Hu Y, Hainaut H and Xu Q:
Mechanical stress-induced DNA damage and rac-p38MAPK signal
pathways mediate p53-dependent apoptosis in vascular smooth muscle
cells. FASEB J. 16:1423–1425. 2002.PubMed/NCBI
|
|
7
|
Wang JH and Thampatty BP: An introductory
review of cell mechanobiology. Biomech Model Mechanobiol. 5:1–16.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan J, Wang T, Wang L, Chen W and Song M:
Cyclic strain-induced cytoskeletal rearrangement of human
periodontal ligament cells via the Rho signaling pathway. PLoS One.
9:e915802014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qi YX, Qu MJ, Long DK, Liu B, Yao QP,
Chien S and Jiang ZL: Rho-GDP dissociation inhibitor alpha
downregulated by low shear stress promotes vascular smooth muscle
cell migration and apoptosis: A proteomic analysis. Cardiovasc Res.
80:114–122. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sit ST and Manser E: Rho GTPases and their
role in organizing the actin cytoskeleton. J Cell Sci. 124:679–683.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bamburg JR and Bernstein BW: ADF/cofilin.
Curr Biol. 18:R273–R275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bamburg JR and Wiggan OP: ADF/cofilin and
actin dynamics in disease. Trends Cell Biol. 12:598–605. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bamburg JR: Proteins of the ADF/cofilin
family: Essential regulators of actin dynamics. Annu Rev Cell Dev
Biol. 15:185–230. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maekawa M, Ishizaki T, Boku S, Watanabe N,
Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K and Narumiya S:
Signaling from Rho to the actin cytoskeleton through protein
kinases ROCK and LIM-kinase. Science. 285:895–898. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Edwards DC, Sanders LC, Bokoch GM and Gill
GN: Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase
signalling to actin cytoskeletal dynamics. Nat Cell Biol.
1:253–259. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bernstein BW and Bamburg JR: ADF/cofilin:
A functional node in cell biology. Trends Cell Biol. 20:187–195.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dumas V, Ducharne B, Perrier A, Fournier
C, Guignandon A, Thomas M, Peyroche S, Guyomar D, Vico L and
Rattner A: Extracellular matrix produced by osteoblasts cultured
under low-magnitude, high-frequency stimulation is favourable to
osteogenic differentiation of mesenchymal stem cells. Calcif Tissue
Int. 87:351–364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Torcasio A, van Lenthe GH and Van
Oosterwyck H: The importance of loading frequency, rate and
vibration for enhancing bone adaptation and implant
osseointegration. Eur Cell Mater. 16:56–68. 2008.PubMed/NCBI
|
|
20
|
Zeichen J, van Griensven M and Bosch U:
The proliferative response of isolated human tendon fibroblasts to
cyclic biaxial mechanical strain. Am J Sports Med. 28:888–892.
2000.PubMed/NCBI
|
|
21
|
Watson PA: Function follows form:
Generation of intracellular signals by cell deformation. FASEB J.
5:2013–2019. 1991.PubMed/NCBI
|
|
22
|
Mayr M, Hu Y, Hainaut H and Xu Q:
Mechanical stress-induced DNA damage and rac-p38MAPK signal
pathways mediate p53-dependent apoptosis in vascular smooth muscle
cells. FASEB J. 16:1423–1425. 2002.PubMed/NCBI
|
|
23
|
Pan J, Wang T, Wang L, Chen W and Song M:
Cyclic strain-induced cytoskeletal rearrangement of human
periodontal ligament cells via the Rho signaling pathway. PLoS One.
9:e915802014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Van Troys M, Huyck L, Leyman S, Dhaese S,
Vandekerkhove J and Ampe C: Ins and outs of ADF/cofilin activity
and regulation. Eur J Cell Biol. 87:649–667. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nishida E, Iida K, Yonezawa N, Koyasu S,
Yahara I and Sakai H: Cofilin is a component of intranuclear and
cytoplasmic actin rods induced in cultured cells. Proc Natl Acad
Sci USA. 84:5262–5266. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin MC, Galletta BJ, Sept D and Cooper JA:
Overlapping and distinct functions for cofilin, coronin and Aip1 in
actin dynamics in vivo. J Cell Sci. 123:1329–1342. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan JS, Han Y, Chen DP, Xu L, Qi YX and
Yan ZQ: Cyclic strain promotes migration of human periodontal
ligament cell via extracellular signal-regulated kinase (ERK)
signaling pathway. Zhonghua Kou Qiang Yi Xue Za Zhi. 45:80–84.
2010.In Chinese. PubMed/NCBI
|
|
28
|
Liu X, Zhang X and Luo ZP: Strain-related
collagen gene expression in human osteoblast-like cells. Cell
Tissue Res. 322:331–334. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Larsson T, Aspden RM and Heinegård D:
Effects of mechanical load on cartilage matrix biosynthesis in
vitro. Matrix. 11:388–394. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jing D, Cai J, Wu Y, Shen G, Li F, Xu Q,
Xie K, Tang C, Liu J, Guo W, et al: Pulsed electromagnetic fields
partially preserve bone mass, microarchitecture and strength by
promoting bone formation in hindlimb-suspended rats. J Bone Miner
Res. 29:2250–2261. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin T, Zeng L, Liu Y, DeFea K, Schwartz
MA, Chien S and Shyy JY: Rho-ROCK-LIMK-cofilin pathway regulates
shear stress activation of sterol regulatory element binding
proteins. Circ Res. 92:1296–1304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Campbell JJ, Blain EJ, Chowdhury TT and
Knight MM: Loading alters actin dynamics and up-regulates cofilin
gene expression in chondrocytes. Biochem Biophys Res Commun.
361:329–334. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pederson T: As functional nuclear actin
comes into view, is it globular, filamentous, or both? J Cell Biol.
180:1061–1064. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng B, Han M, Bernier M and Wen JK:
Nuclear actin and actin-binding proteins in the regulation of
transcription and gene expression. FEBS J. 276:2669–2685. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iida K, Matsumoto S and Yahara I: The
KKRKK sequence is involved in heat shock-induced nuclear
translocation of the 18-kDa actin-binding protein, cofilin. Cell
Struct Funct. 17:39–46. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Burgos-Rivera B, Ruzicka DR, Deal RB,
McKinney EC, King-Reid L and Meagher RB: ACTIN DEPOLYMERIZING
FACTOR9 controls development and gene expression in Arabidopsis.
Plant Mol Biol. 68:619–632. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pendleton A, Pope B, Weeds A and Koffer A:
Latrunculin B or ATP depletion induces cofilin-dependent
translocation of actin into nuclei of mast cells. J Biol Chem.
278:14394–14400. 2003. View Article : Google Scholar : PubMed/NCBI
|