Introduction
Colon carcinoma (CC) is the second most common type
of malignant tumor and ranks as the third highest cause of
cancer-associated mortality worldwide (1–3).
Although early CC detection and treatment can lead to a good
prognosis, the survival rate is low when metastasis occurs
(4–6). Due to the numerous contributing
factors in the development of CC, the pathogenesis remains unclear,
therefore, the investigation of novel therapeutic strategies is a
key focus in CC research.
Investigations of CC have focused on the
identification of dysregulated genes, protein markers, non-encoding
RNA, including miroRNA-145 (7) and
additional prognostic molecular markers. The aim of these
investigation has been to formulate novel strategies for the
treatment of CC on the basis of identifying abnormal genes, key
molecular targets and CC-associated signaling pathways (8).
It is widely known that the metastasis of a
malignant tumor from the primary source to other tissues and organs
is a serious complication in cancer, and is key in the treatment of
malignant tumors (9,10). Cancer cells acquire motility
through the remodeling of the actin cytoskeleton, which is a key
process involved in the invasion and metastasis of cancer cells
(11).
Gelsolin (GSN) is a widely expressed actin
regulator, which is important in regulating cell motility by
severing actin (12,13). In addition, GSN is able to regulate
cell morphology, proliferation and apoptosis (14). A previous study demonstrated that
the expression levels of GSN are reduced in breast, urinary
bladder, colon, kidney, ovary, prostate, gastric and urinary system
cancer (15).
However, whether there is a direct association
between the expression of GSN and tumor development remains to be
fully elucidated (16–18). Previous studies have reported that
the overexpression of GSN promotes the motility of tumor cells and
enhances their invasiveness by regulating various signaling
pathways, including the phosphoinositide 3-kinase (PI3K) and
Ras-PI3K-Rac pathways (19,20).
However, it has been demonstrated that GSN can inhibit
epithelial-mesenchymal cell transformation in breast cancer
(15), and act as a suppressor of
metastasis in B16 melanoma cells (21).
In the present study, the role of GSN in the
proliferation and invasion of human CC cells was investigated in
order to determine whether the overexpression of GSN attenuates the
invasiveness of these cells, and whether a reduction in the
expression of GSN is associated with the invasiveness of human CC
cells or the prognosis of CC patients. This may determine whether
stabilizing the expression of GSN inhibits the invasiveness of CC
cells.
The signal transducer and activator of transcription
(STAT) protein family regulates the expression of several genes,
which are involved in cell survival, proliferation and apoptosis
(22–25). STAT3 is associated with tumor
occurrence via promotion of the proliferation and invasion of
several types of cancer cell, including human CC cells (26–29).
The inhibition of STAT3 was observed to inhibit the proliferation
of human CC cells, indicating that STAT3 may be a potential target
in the treatment of CC (30–34).
To further elucidate the role of GSN in CC cells,
the present study investigated the effect of the expression of GSN
on the STAT signaling pathway, to determine how GSN coordinates
with STAT3 to regulate metastasis in CC.
Materials and methods
Tissue specimens and cell culture
A total of 30 paired primary colon tumors and
corresponding normal colon tissue specimens were obtained from
patients with CC (gender, 13 men and 17 women; mean age, 64.43
years; age range, 23–93 years) who were admitted to Zhongshan
Hospital of Fudan University (Shanghai, China) between 2009 and
2011, and from whom informed written consent was obtained.
The selection of CC cases was based on a clear
pathological diagnosis and follow-up data in patients who had not
previously received local or systemic treatment. Tumor stages were
defined, according to the 2002 American Joint Committee on
Cancer/International Union against Cancer tumor-node-metastasis
classification system (35). The
present study was approved by the Institutional Research Ethics
Committee of Zhongshan Hospital of Fudan University. The SW480 and
HT29 CC cell lines were cultured in RRMI medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The SW620 and
HCT116 CC, and the normal CCD-18Co colon cell lines were maintained
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS. All cell lines were
purchased from the Shanghai Cell Institute Country Cell Bank.
(Shanghai, China). During tumor resection surgery, fresh tissue
samples were harvested from the recruited patients; tumor tissues
were obtained from the center of the tumor and adjacent normal
tissues from 5 cm away from the tumor margin. The tissue samples
were snap-frozen and preserved at −80°C.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized
using a PrimeScript RT Reagent kit (Promega Corporation, Madison,
WI, USA). The expression level of GSN was analyzed relative to the
level of the β-actin gene transcript using an Applied Biosystems
7300 PCR system (Thermo Fisher Scientific, Inc.). First-strand cDNA
(2 μl) was amplified in a 20 μl PCR reaction mixture,
containing 10 μl 2X SYBR green PCR master mix (Qiagen,
Hilden, Germany), 0.4 μl 50X ROX Reference Dye (Thermo
Fisher Scientific, Inc.), 0.4 μl of each specific primer set
and ddH2O added to a total volume of 20 μl. The
primer sequences were as follows: β-actin, forward
5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse 5′-GGGCACGAAGGCTCATCATT-3′;
and GSN, forward 5′-GGTGTGGCATCAGGATTCAAG-3′ and reverse
5′-TTTCATACCGATTGCTGTTGGA-3′. The primer sequences were purchased
from (Invitrogen; Thermo Fisher Scientific, Inc.). The
amplification was performed under the following conditions: 10 min
at 95°C for one cycle, 40 cycles of 95°C for 15 sec and 60°C for 60
sec. The RT-qPCR data were quantified using the comparative Cq
method (36).
Construction of the pGEM-T-GSN
vector
GSN target fragments (Wegene) were recovered and
purified using 1% low melting agarose gel electrophoresis
(Sigma-Aldrich, St. Louis, MO, USA) and a DNA purification kit
(K0512; Thermo Fisher Scientific, Inc.). The purified gene
fragments and pGEM-T vector (Promega Corporation, Madison, WI, USA)
were combined according to the manufacturer's instructions, and
transformed into JM101 competent cells (prepared by the calcium
chloride method). Single colonies were randomly selected and added
to lysogeny broth (InvivoGen, San Diego, CA, USA) liquid medium
(containing ampicillin; Sigma-Aldrich) at 37°C and agitated for 12
h. Following plasmid extraction, the products were identified by
restriction enzyme digestion using HindIII and KpanI
(New England BioLabs, Inc., Ipswich, MA, USA). The positive
plasmids of the double digested results were sent to Shanghai
Invitrogen Biotechnology Co., Ltd. (Shanghai, China) for forward
and reverse sequencing. The correct plasmid was identified from the
sequencing results and termed pGEM-T-GSN.
DNA transfection
Cells were seeded uniformly into 6-well plates at a
density of 3×105 cells/well. When the cells were 80%
confluent, the recombined pGEM-T-GSN plasmids (1 μl) were
transfected into the four CC cell lines and the CCD-18Co normal
cells using 3–5 μl Lipofectamine® 2000 per
200,000-1,000,000 cells (Thermo Fisher Scientific, Inc.).
Invasion and metastasis assays
Tumor cell invasion and metastasis were assessed
using a Transwell insert (7 μm; Corning, Inc., Corning, NY,
USA). The SW480 and HT29 cells were grown to 85% confluence and
then transfected with pGEM-T-GSN or the empty vector. Following
transfection for 24 h, the cells (5×104) were harvested,
washed with phosphate-buffered saline, resuspended in 200 μl
serum-free medium and seeded into the upper chamber of the
Transwell insert. A total of 600 μl DMEM, containing 10% FBS
as a chemoattractant, was added to the lower chamber. For the
invasion assay, the inserts were precoated with 30 μl
Corning Matrigel Matrix (Corning Inc.) and 6×104 cells
were added to the upper chamber. Following incubation for 24 h at
37°C in a humidified atmosphere of 5% CO2, non-migrating
(non-invading) cells were removed from the upper surface of the
filter with a cotton-tipped swab. The cells on the lower surface of
the filter were fixed in 4% formaldehyde (Sigma-Aldrich) and
stained with crystal violet staining solution (Sigma-Aldrich).
Following staining, five randomly-selected fields were counted at a
magnification of ×100 using Eclipse E200-LED microscope (Nikon
Corporation, Tokyo, Japan). All obtained data were from a minimum
of three independent experiments performed in duplicate.
Western blot analysis
The cells and tissues were homogenized in
radioimmunoprecipitation assay buffer, containing 20 mM Tris-HCl
(pH 7.5), 150 mM NaCl, 1 mM Na2 EDTA, 1 mM EGTA, 1%
NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM
β-glycerophosphate, 1 mM Na3VO4 and 1
μg/ml leupeptin (Cell Signaling Technology, Inc., Danvers,
MA USA). The protein concentration was determined using a
Bicinchoninic Acid Protein Assay kit (Pierce Biotechnology, Inc.,
Rockford, IL, USA). Protein (20 μg) was subjected to a 4–12%
gradient Bis-Tris Gel (Invitrogen; Thermo Fisher Scientific, Inc.)
and electrotransferred onto a Hybond-enhanced chemiluminescence
nitrocellulose membrane (GE Healthcare Life Sciences, Chalfont,
UK). Following the transfer, the membrane was washed with 25 ml
Tris-buffered saline (TBS; Cell Signaling Technology, Inc.) for 5
min at room temperature, incubated in 25 ml blocking buffer (Cell
Signaling Technology, Inc.) for 1 hr at room temperature, washed
three times for 5 min each with 15 ml TBS Tween 20 (TBST; Cell
Signaling Technology, Inc.). The membrane was then incubated with
rabbit anti-GSN monoclonal antibody (1:25,000, cat. no. ab7583;
Abcam, Cambridge, MA, USA) overnight at 4°C, washed three times for
5 min with 15 ml TBST, and incubated with the goat anti-rabbit
horseradish peroxidase-conjugated secondary antibodies (1:500) for
1 h at room temperature. Finally, it was washed three times for 5
min with 15 ml TBST. The reaction was detected using an enhanced
chemiluminescence system (GE Healthcare Life Sciences). The
membranes were then reprobed using a mouse monoclonal anti-GAPDH
antibody (cat. no. ab9484; Abcam) as an internal control.
Immunohistochemistry
Immunohistochemical analysis was performed on
paraffin-embedded sections using an Envision kit (Dako, Glostrup,
Denmark), according to the manufacturer's protocol. The sections
were autoclaved for 10 min at 121°C for antigen retrieval. Anti-GSN
monoclonal antibodies (Upstate Biotechnology Inc.) were applied to
the sections at 1:100. The presence of staining was evaluated by a
single pathologist, according to the overall level of the
immunostaining.
Statistical analysis
Statistical analysis was performed using SPSS
software, version 16.0 (SPSS, Inc., Chicago, IL, USA). The
differences between variables were assessed by the χ2
test or Fisher's exact test. The survival rates of patients with CC
were analyzed using Kaplan-Meier analysis, and a log rank test was
used to compare the survival curves. Data derived from the cell
line experiments are presented as the mean ± standard deviation and
assessed using a two-tailed Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of GSN in the human CC
specimens
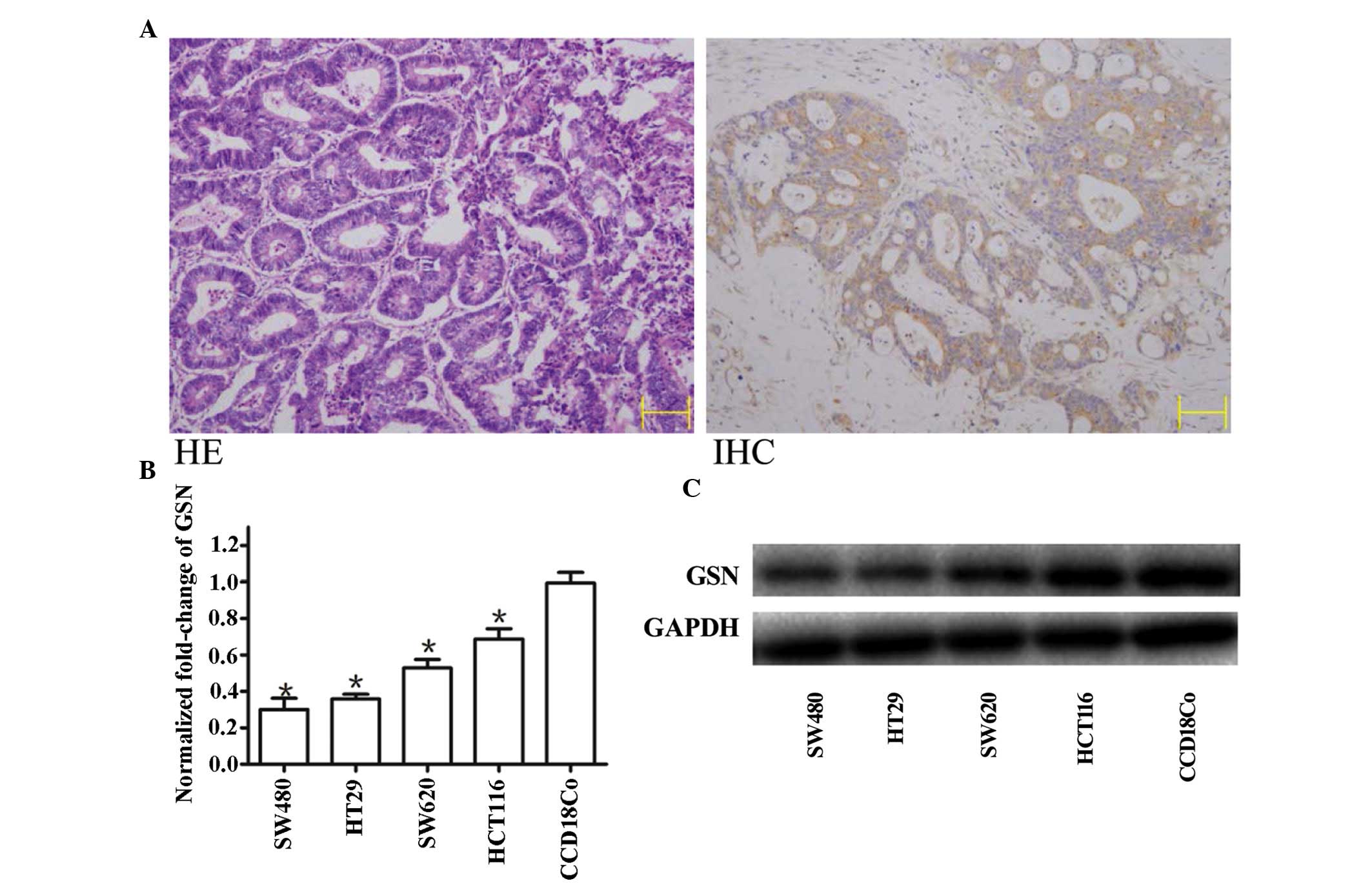
The expression of GSN was detected in human CC
specimens and CC cell lines (Fig.
1). A total of 30 paired primary colon tumor and corresponding
normal colon tissue samples were screened using immunohistochemical
staining with anti-GSN antibodies (Fig. 1A; right panel) and RT-qPCR. Partial
sections from the same tissues were then stained with HE to confirm
the presence of tumorous tissue (Fig.
1A; left panel). Tissues containing >90% tumor cells were
defined as tumors for further quantitative assessment of the
expression of GSN using RT-qPCR.
Low expression levels of GSN are detected
in CC cell lines
The expression levels of GSN were measured using
RT-qPCR in the SW480, HT29, SW620 and HCT116 cell lines, and 30
paired CC and adjacent non-neoplastic colon tissues. The results
demonstrated that the expression levels of GSN in the four CC cell
lines were significantly lower, compared with those in the normal
CCD-18Co cell line, with the lowest level in the SW480 cell line
(Fig. 1B).
Expression of GSN is downregulated in the
majority of CC specimens
The expression levels of GSN was high in the
non-cancerous tissues. The mean expression levels of GSN in the CC
tissue were significantly reduced, compared with the non-cancerous
tissue (P<0.01; Fig. 2A).
Western blot analysis was performed to measure the protein
expression levels of GSN in the four CC cell lines, and was
compared with the results of the RT-qPCR. It was observed that the
mRNA expression of GSN was reduced, with the greatest reduction in
the SW480 cells (Fig. 1B). This
was reflected by the western blotting, which indicated the greatest
reduction in protein expression levels of GSN in the SW480 cells
(Fig. 1C).
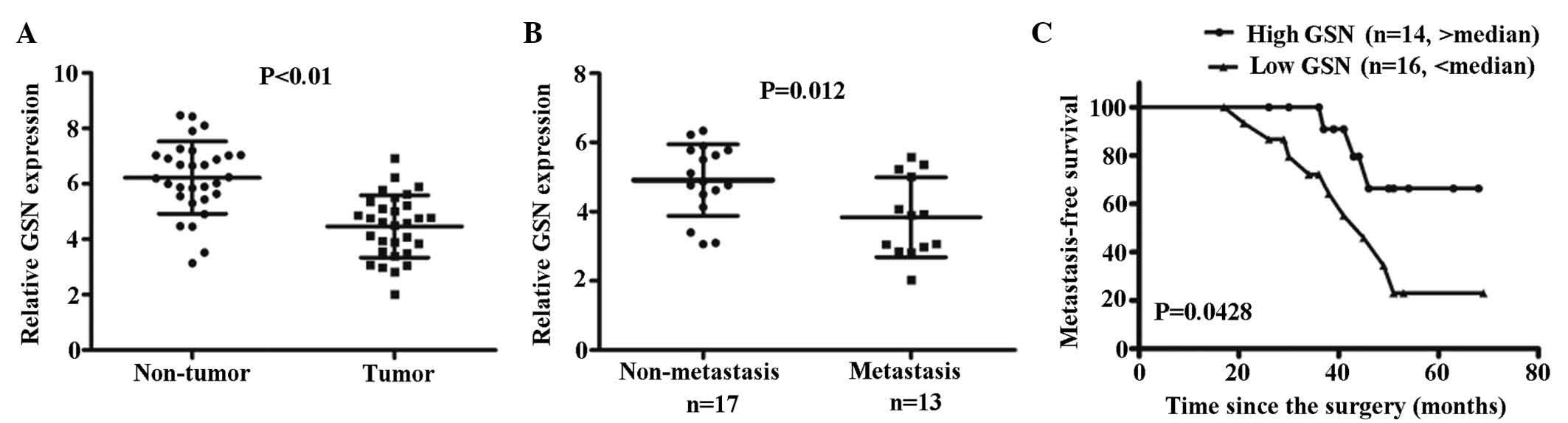
Low expression levels of GSN are
associated with a poor metastasis-free survival (MFS) rate
To investigate the correlation between the
clinicopathological parameters and the expression of GSN in
patients with CC, expression levels of GSN in the 30 CC tissue
specimens were measured using RT-qPCR. Low or high levels of GSN in
the tumor were defined when the normalized expression of GSN
resided in the <50% or >50% of the tumor, respectively.
Accordingly, a low level of GSN was detected in 16/30 CC specimens
(53.3%), whereas a high level of GSN was detected in the remaining
14/30 CC specimens (46.6%; Fig.
2C). Correlation analysis indicated that low expression levels
of GSN were significantly associated with tumor metastasis
(P=0.012; Fig. 2B). Kaplan-Meier
analysis indicated that a low expression levels of GSN were
associated with reduced MFS in the patients with CC (P=0.0428;
Fig. 2C).
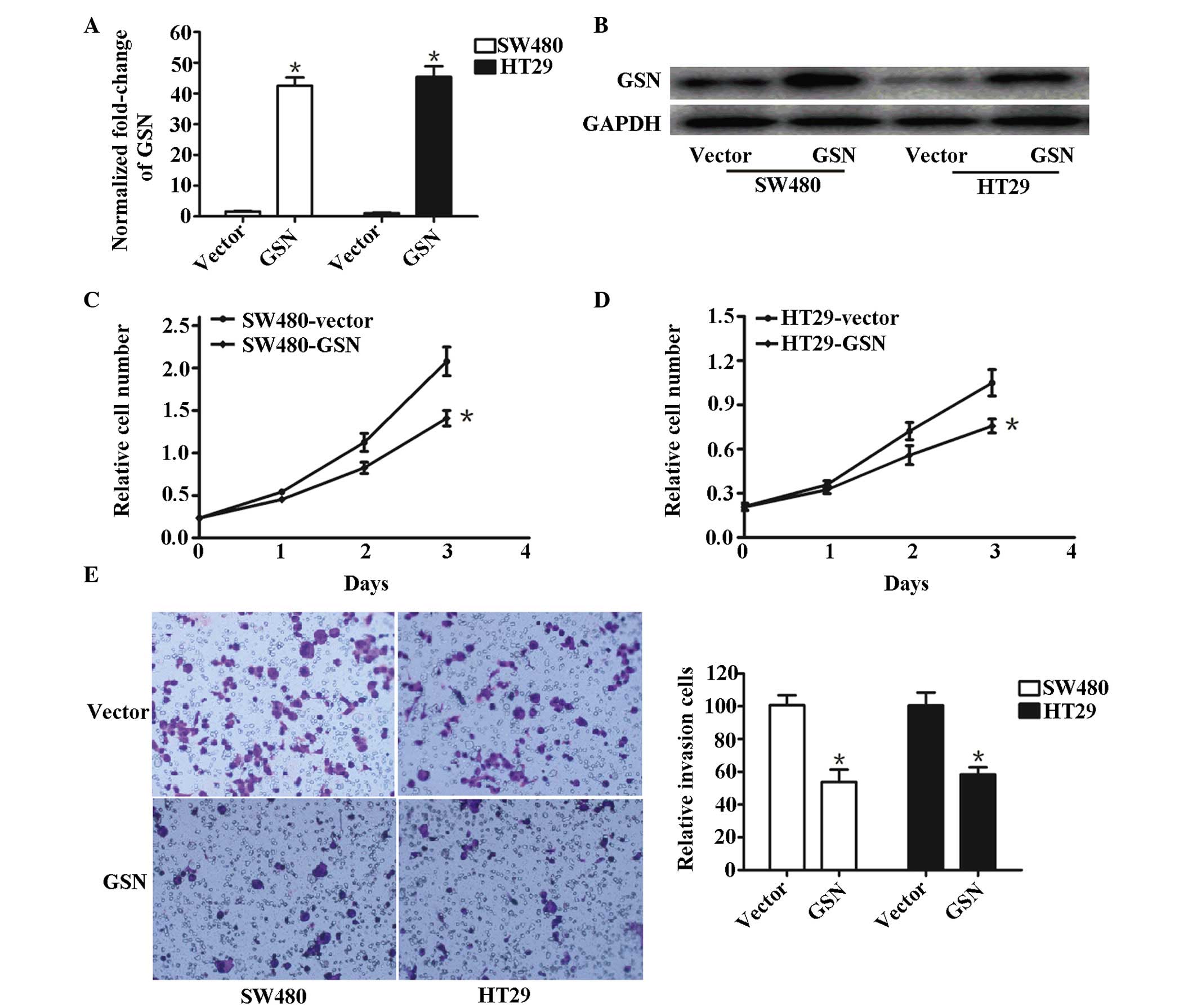
High expression levels of GSN suppress
the invasion of SW480 and HT29 cells in vitro
Overexpression of GSN in GSN transfected SW480 and
HT29 cells, and vector-transfected SW480 and HT29 cells, measured
using RT-qPCR (Fig. 3A) and
western blotting (Fig. 3B). The
expression of GSN in GSN-transfected SW480 and HT29 cells was
higher compared with that in the control group (P<0.05). It was
observed that the proliferation of SW480 and HT29 cells remained
almost the same initially (Fig. 3C and
D). Following transfection for 24 h with p-GEM-T-GSN or vector
controls, vector-SW480 and vector-HT29 cells proliferated at a
higher rate, compared with the GSN-transfected SW480 and HT29
cells. At 3 days post-transfection, the rate of proliferation was
significantly different between the vector-transfected SW480 and
HT-29 cells and their corresponding GSN-transfected cells (P=0.0312
and P=0.0217, respectively; Fig. 3C
and D), suggesting that the overexpression of GSN inhibited the
proliferation of SW480 and HT29 cells.
Furthermore, the relative number of invasive cells
in the vector-transfected cells was significantly lower, compared
with the GSN-transfected SW480 cells (P=0.023) and HT29 cells
(P=0.011; Fig. 3E), suggesting
that increased expression levels of GSN suppressed the invasion of
the GSN-transfected SW480 and HT29 cells.
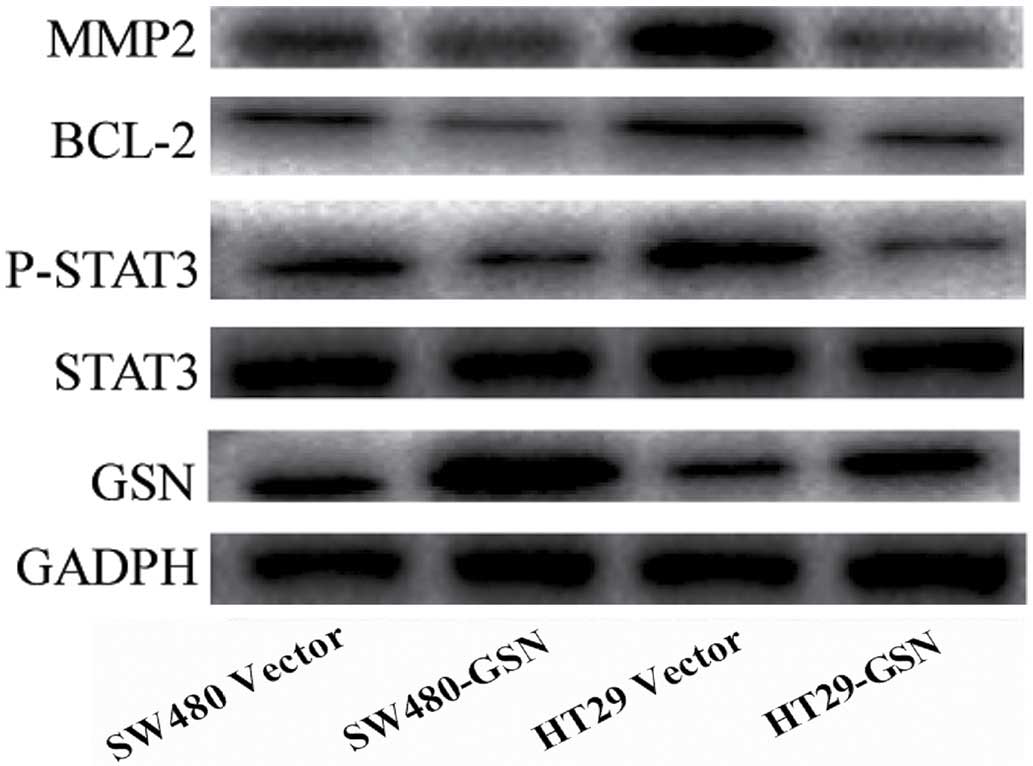
Overexpression of GSN reduces the
expression levels of matrix metalloproteinase 2 (MMP2), BCL-2 and
phosphorylated (p)-STAT3 in SW480 and HT29 cells
As presented in Fig.
4, the relative protein expression levels of STAT3 were not
significantly reduced in the GSN-transfected SW480 and HT29 cells,
compared with the vector-transfected SW480 and HT29 cells. However,
the expression levels of MMP2, BCL-2 and p-STAT3 in the
GSN-transfected SW480 and HT29 cells were significantly reduced,
compared with the vector-transfected SW480 and HT29 cells
(P=0.0231, P=0.0326 and P=0.0176, respectively; Fig. 4). This indicated that the increased
expression of GSN reduced the expression levels of MMP2, BCL-2 and
p-STAT3 in the SW480 and HT29 CC cells.
Discussion
The abnormal regulation of cell migration is the
primary cause of several diseases, including the invasion and
migration of tumor cells. The migration of tumor cells across
tissue barriers requires the degradation of specific components of
the extracellular matrix, which triggers alterations in the
interaction between the actin cytoskeleton and extracellular matrix
proteins (37,38). This process is affected by multiple
factors, is dependant on adhesion molecule receptors and is
regulated by specific actin binding factors (39). The involvement of specific actin
binding factors in the migration of tumor cells has received
increased attention.
GSN is a protein that is widely expressed
intracellularly, including in the cytoplasm and mitochondria, and
extracellularly, including the plasma (40). GSN can inhibit apoptosis by
stabilizing mitochondria (41).
Previous studies have demonstrated that GSN is involved in the
regulation of epithelial-mesenchymal cell transformation (12), and that the expression of GSN
expression can inhibit the migration potential of several types of
human cancer cells (42).
In the present study, RT-qPCR was used to analyze
the mRNA expression levels of GSN in CC cells, and the results
revealed that there were significant reductions in the expression
levels of GSN, compared with the levels of expression in the normal
colon tissue (Fig. 1). In
addition, the correlation between the expression levels of GSN and
clinicopathological parameters was investigated, which indicated
that the expression levels of GSN were reduced in 16/30 patients
diagnosed with metastatic CC, and were higher in the remaining 14
patients without metastatic CC. The mean expression level of GSN in
the CC tissue was significantly reduced, compared with that in the
non-cancerous tissue (P<0.01; Fig.
2A). Additionally, western blot analysis was performed in the
present study to measure the expression levels of GSN in SW480,
HT29, SW620 and HCT116 cell lines, which was observed to be
downregulated in all four CC cell lines (Fig. 1C). These results suggested that GSN
may be associated with the invasiveness of tumor cells, however,
further investigations with a larger sample size are required to
confirm this conclusion.
The results of the correlation analysis indicated
that lower expression levels of GSN were associated with metastasis
in the patients with CC (P=0.012; Fig.
2B). Kaplan-Meier analysis indicated that low expression levels
of GSN were associated with the reduced rates of survival in
patients with metastatic CC (P=0.0428; Fig. 2C). These results suggested that
reduced expression of GSN may assist in the assessment of prognosis
in patients with CC, and may represent a novel prognostic marker in
CC.
A previous study reported the persistent activation
of STAT proteins in several human cancer cell lines, including
leukemia, multiple myeloma, breast cancer and prostate cancer
(43). Cross-Knorr et al
(44) reported that the low
expression levels of P-STAT3, MMP-2 and BCL-2 were associated with
the invasiveness of tumor cells. Notably, the present study
indicated that the overexpression of GSN in SW480 and HT29 cell
lines downregulated the expression levels of P-STAT3, MMP-2 and
BCL-2. In addition, it was observed that the invasiveness of the
SW480 and HT29 cells was reduced when GSN was overexpressed
(Fig. 3). Therefore, the present
study hypothesized that a low expression level of GSN results in
persistent activation of P-STAT3, MMP-2 and BCL-2 via a specific
molecular mechanisms, which further induces CC metastasis. By
contrast, the overexpression of GSN may reduce this activation,
thereby inhibiting CC metastasis.
Low or depleted expression levels of GSN appear to
reflect the degree of CC malignancy, however, further
investigations are required to validate the predictive value of GSN
on the prognosis of CC.
In conclusion, to the best of our knowledge, the
present study is the first to report the association between the
mRNA expression of GSN and the survival rate of patients with
metastatic CC. In addition, the present study indicated the
potential diagnostic value of low expression levels of GSN as a
prognostic factor in postoperative patients with CC. The findings
indicate the potential of using the mRNA expression level of GSN as
a biomarker to assess the degree of tumor malignancy. However,
whether GSN may be used as a treatment target or a marker of
clinical tumor treatment requires further investigation.
References
|
1
|
Landherr L and Nagykálnai T: Chemotherapy
of elderly patients with colorectal cancer. Magy Onkol. 53:97–105.
2009.In Hungarian. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meyerhardt JA and Mayer RJ: Systemic
therapy for colorectal cancer. N Engl J Med. 352:476–487. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Murray T, Ward E, Samuels A,
Tiwari RC, Ghafoor A, Feuer EJ and Thun MJ: Cancer statistics,
2005. CA Cancer J Clin. 55:10–30. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar
|
|
5
|
Ohman U: Prognosis in patients with
obstructing colorectal carcinoma. Am J Surg. 143:742–747. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moreaux J and Catala M: Carcinoma of the
colon: Long-term survival and prognosis after surgical treatment in
a series of 798 patients. World J Surg. 11:804–809. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar
|
|
8
|
Addeo R, Montella L, Baldi A, Cennamo G,
Guarrasi R, Faiola V, Caraglia M and Del Prete S: Atypical
cutaneous lymphoid hyperplasia induced by chemotherapy in a patient
with advanced colon carcinoma. Clin Colorectal Cancer. 6:728–730.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dimitroff CJ, Sharma A and Bernacki RJ:
Cancer metastasis: A search for therapeutic inhibition. Cancer
Invest. 16:279–290. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fidler IJ: Critical determinants of cancer
metastasis: Rationale for therapy. Cancer Chemother Pharmacol.
43(Suppl): S3–S10. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamaguchi H and Condeelis J: Regulation of
the actin cytoskeleton in cancer cell migration and invasion.
Biochim Biophys Acta. 1773:642–652. 2007. View Article : Google Scholar
|
|
12
|
Yin HL and Stossel TP: Control of
cytoplasmic actin gel-sol transformation by gelsolin, a
calcium-dependent regulatory protein. Nature. 281:583–586. 1979.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun HQ, Yamamoto M, Mejillano M and Yin
HL: Gelsolin, a multifunctional actin regulatory protein. J Biol
Chem. 274:33179–33182. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kwiatkowski DJ: Functions of gelsolin:
Motility, signaling, apoptosis, cancer. Curr Opin Cell Biol.
11:103–108. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanaka H, Shirkoohi R, Nakagawa K, Qiao H,
Fujita H, Okada F, Hamada J, Kuzumaki S, Takimoto M and Kuzumaki N:
siRNA gelsolin knockdown induces epithelial-mesenchymal transition
with a cadherin switch in human mammary epithelial cells. Int J
Cancer. 118:1680–1691. 2006. View Article : Google Scholar
|
|
16
|
Thor AD, Edgerton SM, Liu S, Moore DH 2nd
and Kwiatkowski DJ: Gelsolin as a negative prognostic factor and
effector of motility in erbB-2-positive epidermal growth factor
receptor-positive breast cancers. Clin Cancer Res. 7:2415–2424.
2001.PubMed/NCBI
|
|
17
|
Rao J, Seligson D, Visapaa H, Horvath S,
Eeva M, Michel K, Pantuck A, Belldegrun A and Palotie A: Tissue
microarray analysis of cytoskeletal actin-associated biomarkers
gelsolin and E-cadherin in urothelial carcinoma. Cancer.
95:1247–1257. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dosaka-Akita H, Hommura F, Fujita H,
Kinoshita I, Nishi M, Morikawa T, Katoh H, Kawakami Y and Kuzumaki
N: Frequent loss of gelsolin expression in non-small cell lung
cancers of heavy smokers. Cancer Res. 58:322–327. 1998.PubMed/NCBI
|
|
19
|
Chen P, Murphy-Ullrich JE and Wells A: A
role for gelsolin in actuating epidermal growth factor
receptor-mediated cell motility. J Cell Biol. 134:689–698. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lader AS, Lee JJ, Cicchetti G and
Kwiatkowski DJ: Mechanisms of gelsolin-dependent and -independent
EGF-stimulated cell motility in a human lung epithelial cell line.
Exp Cell Res. 307:153–163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fujita H, Okada F, Hamada J, Hosokawa M,
Moriuchi T, Koya RC and Kuzumaki N: Gelsolin functions as a
metastasis suppressor in B16-BL6 mouse melanoma cells and
requirement of the carboxyl-terminus for its effect. Int J Cancer.
93:773–780. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao SP and Bromberg JF: Touched and moved
by STAT3. Sci STKE. 2006:pe302006.PubMed/NCBI
|
|
23
|
Costantino L and Barlocco D: STAT 3 as a
target for cancer drug discovery. Curr Med Chem. 15:834–843. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aggarwal BB, Kunnumakkara AB, Harikumar
KB, Gupta SR, Tharakan ST, Koca C, Dey S and Sung B: Signal
transducer and activator of transcription-3, inflammation, and
cancer: How intimate is the relationship? Ann N Y Acad Sci.
1171:59–76. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aggarwal BB, Sethi G, Ahn KS, Sandur SK,
Pandey MK, Kunnumakkara AB, Sung B and Ichikawa H: Targeting
signal-transducer-and-activator-of-transcription-3 for prevention
and therapy of cancer: Modern target but ancient solution. Ann N Y
Acad Sci. 1091:151–169. 2006. View Article : Google Scholar
|
|
26
|
Berry DC, Levi L and Noy N:
Holo-retinol-binding protein and its receptor STRA6 drive oncogenic
transformation. Cancer Res. 74:6341–6351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ung N, Putoczki TL, Stylli SS, Ng I,
Mariadason JM, Chan TA, Zhu HJ and Luwor RB: Anti-EGFR therapeutic
efficacy correlates directly with inhibition of STAT3 activity.
Cancer Biol Ther. 15:623–632. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen A, Chen Y, Hong F, Lin J, Wei L, Hong
Z, Sferra TJ and Peng J: Pien Tze Huang suppresses IL-6-inducible
STAT3 activation in human colon carcinoma cells through induction
of SOCS3. Oncol Rep. 28:2125–2130. 2012.PubMed/NCBI
|
|
29
|
Geinguenaud F, Souissi I, Fagard R, Motte
L and Lalatonne Y: Electrostatic assembly of a DNA
superparamagnetic nano-tool for simultaneous intracellular delivery
and in situ monitoring. Nanomedicine (Lond). 8:1106–1115. 2012.
|
|
30
|
Lerner I, Hermano E, Zcharia E, Rodkin D,
Bulvik R, Doviner V, Rubinstein AM, Ishai-Michaeli R, Atzmon R,
Sherman Y, et al: Heparanase powers a chronic inflammatory circuit
that promotes colitis-associated tumorigenesis in mice. J Clin
Invest. 121:1709–1721. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zugowski C, Lieder F, Müller A, Gasch J,
Corvinus FM, Moriggl R and Friedrich K: STAT3 controls matrix
metalloproteinase-1 expression in colon carcinoma cells by both
direct and AP-1-mediated interaction with the MMP-1 promoter. Biol
Chem. 392:449–459. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsareva SA, Wagner S, Müller A, Corvinus F
and Friedrich K: Cell-cell contacts induce STAT3 activity in colon
carcinoma cells through an autocrine stimulation loop. J Cancer Res
Clin Oncol. 137:857–863. 2011. View Article : Google Scholar
|
|
33
|
Fenton JI and Birmingham JM: Adipokine
regulation of colon cancer: Adiponectin attenuates
interleukin-6-induced colon carcinoma cell proliferation via
STAT-3. Mol Carcinog. 49:700–709. 2010.PubMed/NCBI
|
|
34
|
Tadlaoui Hbibi A, Laguillier C, Souissi I,
Lesage D, Le Coquil S, Cao A, Metelev V, Baran-Marszak F and Fagard
R: Efficient killing of SW480 colon carcinoma cells by a signal
transducer and activator of transcription (STAT) 3 hairpin decoy
oligodeoxynucleotide–interference with
interferon-gamma-STAT1-mediated killing. FEBS J. 276:2505–2515.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
O'Connell JB, Maggard MA and Ko CY: Colon
cancer survival rates with the new American Joint Committee on
Cancer sixth edition staging. J Natl Cancer Inst. 96:1420–1425.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
37
|
Marín O and Rubenstein JL: Cell migration
in the forebrain:. Annu Rev Neurosci. 26:441–483. 2003. View Article : Google Scholar
|
|
38
|
Martin TA, Ye L, Sanders AJ, Lane J and
Jiang WG: Cancer Invasion and Metastasis: Molecular and Cellular
Perspective. Madame Curie Bioscience Database [Internet] Landes
Bioscience. 2000, Austin: Available from: http://www.ncbi.nlm.nih.gov/books/NBK164700/.
|
|
39
|
Linder S: The matrix corroded: Podosomes
and invadopodia in extracellular matrix degradation. Trends Cell
Biol. 17:107–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wen D, Corina K, Chow EP, Miller S, Janmey
PA and Pepinsky RB: The plasma and cytoplasmic forms of human
gelsolin differ in disulfide structure. Biochemistry. 35:9700–9709.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Koya RC, Fujita H, Shimizu S, Ohtsu M,
Takimoto M, Tsujimoto Y and Kuzumaki N: Gelsolin inhibits apoptosis
by blocking mitochondrial membrane potential loss and cytochrome c
release. J Biol Chem. 275:15343–15349. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tanaka M, Müllauer L, Ogiso Y, Fujita H,
Moriya S, Furuuchi K, Harabayashi T, Shinohara N, Koyanagi T and
Kuzumaki N: Gelsolin: A candidate for suppressor of human bladder
cancer. Cancer Res. 55:3228–3232. 1995.PubMed/NCBI
|
|
43
|
Buettner R, Mora LB and Jove R: Activated
STAT signaling in human tumors provides novel molecular targets for
therapeutic intervention. Clin Cancer Res. 8:945–954.
2002.PubMed/NCBI
|
|
44
|
Cross-Knorr S, Lu S, Perez K, Guevara S,
Brilliant K, Pisano C, Quesenberry PJ, Resnick MB and Chatterjee D:
RKIP phosphorylation and STAT3 activation is inhibited by
oxaliplatin and camptothecin and are associated with poor prognosis
in stage II colon cancer patients. BMC Cancer. 13:4632013.
View Article : Google Scholar : PubMed/NCBI
|