Introduction
Toxoplasma gondii (T. gondii) is an
opportunistic protozoan pathogen, which is capable of invading and
replicating in all nucleated cells. T. gondii infection can
cause neurological problems, including encephalitis, in
immunocomprised individuals (1).
Of note, toxoplasmic infection in the prenatal period is a common
cause of brain malformation, and can critically affect the central
nervous system and retinal development, culminating in long-term
cognitive deficits (2).
Histopathological lesions in the frontal lobe have been found to be
more prevalent than in other areas of the brain of BALB/c mice
infected with T. gondii (3). T. gondii is defined by the
presence of an apical complex, including secretory organelles
(4,5). The rhoptries are one type of apical
secretory organelle of T. gondii, and they have been shown
to be closely associated with the parasites' pathogenesis, host
cell invasion and host cell interaction (6). At present, >30 rhoptry proteins
have been identified, with several being involved in the invasive
process, and important for growth and survival in the host cell
(4). ROP16 has serine-threonine
kinase activity, a molecular weight of 96 kD and comprises 707
amino acids. The uniqueness of ROP16 to the apicomplexa is crucial
in the host-pathogen interaction (7). ROP16 is a key virulence determinant
in terms of being a member of the ROP2 family and being able to
invade the host cell nucleus rapidly following parasite infection.
This protein invades host cell and accumulates in the host cell
nucleus due to the nucleus localized sequence (8). ROP16, a regulator of host cell
transcription, subverts the host functions by direct tyrosine
phosphorylation of the signal transducer and activator of
transcription (STAT) pathways. Evidence obtained has confirmed that
ROP16 from type I or III strains of T. gondii can affect the
activation of the STAT3/6 signaling pathways, with consequent
downstream effects on the key host cytokine, interleukin-12
(9). In addition, ROP16 induces
the phosphorylation and nuclear translocation of STAT5 to generate
protective immunity (10). These
findings show that ROP16 can affect and subvert gene functions in
the nucleus. The present study investigated the gene expression
profile in the SH-SY5Y human neuroblastoma cell line transfected
with ROP16 in order to improve understanding of the molecular
function of ROP16 in the host neurocyte.
Materials and methods
Cell culture
The SH-SY5Y cells (American Type Culture Collection,
Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's
medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan UT, USA)
supplemented with 10% heat-inactivated fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cells
were incubated at 37°C in a humidified air atmosphere containing 5%
CO2 and were passaged every 2–4 days by trypsinization.
Transfection of cells with ROP16
The T. gondii ROP16 gene (preserved in house)
was amplified via polymerase chain reaction (PCR) analysis using
the following primers: ROP16, forward
5′-GAGAATTCCATGAAAGTGACCACGAAAGG-3′, containing EcoRI, and
reverse
5′-GCGATATCCTTGTCATCGTCGTCCTTGTAGTCCATCCGATGTGAAGAAAGTTC-3′,
containing the Flag-tag gene sequence EcoRV, and ligated
using the NEB kit (New England Biolabs, Inc., Ipswich, MA, USA)
with the target vector, pPEXPR-IBA105IP (preserved in house). The
cycling conditions for PCR were as follows: i) Initial denaturation
at 94°C for 5min, ii) denaturation at 94°C for 30 sec, iii)
annealing at 65°C for 30 sec, iv) elongation at 72°C for 150 sec
(repeat steps ii-iv for 30 cycles), vi) final extension at 72°C for
10 min. The vector DNA and Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) were mixed together following incubation
at room temperature for 5 min. The mixture was added to 6-well
culture plates (density, ~80%) following incubation at room
temperature for 20 min. The mixture was then incubated in 5% CO2 at
37°C. Puromycin (1 µg/ml) was used to select stably transfected
cells following 24–48 h of culture. The SH-SY5Y-ROP16 cell line was
cultured in the same conditions as the SH-SY5Y cell line. Finally,
the expression level of ROP16 in the SH-SY5Y was determined using
western blot analysis.
RNA isolation and microarray
analysis
The total RNA of each cell line was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The RNA was further
purified using a NucleoSpin® RNA clean-up kit
(Macherey-Nagel, Düren, Germany) and was quantified using an
ultraviolet spectrophotometer and formaldehyde denaturing agarose
gel electrophoresis. The RNA samples were sent to the Bioassay
Laboratory of CapitalBio Corporation (Beijing, China) for
microarray hybridization and analysis. Briefly, total RNA from each
sample was reverse transcribed by denaturation at 94°C for 5 min,
94°C for 30 sec, annealing at 55°C for 30 sec and elongation at
68°C for 150 sec, with a final extension at 68°C for 5 min. The
cDNA synthesized was then labeled with CY3-dCTP (cat. no. PA 53021;
GE Healthcare Life Sciences) using klenow enzyme (New England
Biolabs, Inc.) according to the manufacturer's protocol. The
hybridized slides were scanned on an Agilent G2565CA microarray
scanner (Agilent Technologies, Inc., Santa Clara, CA, USA). The
resulting text files were then obtained from the scanned image
using Agilent Feature Extraction image analysis software and
imported to Agilent GeneSpring GX software (Agilent Technologies,
Inc.). The microarray data was normalized using the percentile
shift method.
Analysis of differentially expressed
genes (DEGs)
In the present study, a 2-fold change (log2
fold-change ≥1 or ≤-1) and a flag tag of ‘Detected’ were used as
the thresholds to identify the DEGs. A gene was considered to be
differentially expressed by transfection of ROP16 if its expression
increased or decreased by ≥2-fold, compared with the control
sample. Gene Ontology(GO) analysis was performed using Protein
Analysis Through Evolutionary Relationships (PANTHER) for
biological processes (11,12). The ToppGene Suite was used in order
to select candidate genes of T. gondii from the microarray
data (13). DEGs from the
microarray data were defined as the ‘test gene set’ and genes
associated with T. gondii were defined as the ‘training gene
set’ according to the records of Online Mendelian Inheritance in
Man (OMIM; http://omim.org/) and GeneCards
(http://www.genecards.org/) databases.
The Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING; http://www.string-db.org/) online
database was used to evaluate information on protein-protein
interactions (PPIs) of the DEGs (14). Interactions with a high required
confidence with a combined score ≥0.7 were selected. According to
these interactions, PPI networks were constructed using Cytoscape
software (3.2.1; http://cytoscape.org/) (15). Further module analysis of the
networks was performed using Molecular Complex Detection (MCODE),
which is an application in Cytoscape (16). The parameters used to identify the
enriched functional modules were as follows: Degree cutoff, 2; Node
score cutoff, 0.2; K-Core, 2; Maximum depth, 100.
RT-qPCR analysis
RT-qPCR analysis was used to confirm the microarray
results in the present study. The cDNA was synthesized in a 20 µl
reaction system. Genomic DNA was extracted using 2.0 µl 5xgDNA
Eraser Buffer, 1.0 µl gDNA Eraser, 1 µg total RNA and complemented
RNase-free dH2O up to 10 µl. The above mixture was incubated at
42°C for 2 min. Subsequently, 4.0 µl 5X PrimeScript®
Buffer 2 (for Real Time), 1.0 µl PrimeScript® RT Enzyme
mix I, 1.0 µl RT Primer mix and complemented RNase-free H2O up to
20 µl were added. The mixture was incubated at 37°C for 15 min and
85°C for 5 sec to generate cDNA. The prepared cDNA was stored at
−20°C. SYBR Green Real-time PCR Master mix (Takara Bio, Inc., Otsu,
Japan) was used to perform the reaction, a 10 µl volume contained
2.5 µl SYBR® Premix Ex TaqTM II (2X), 0.2 µl PCR forward
primer (10 µM), 0.2 µl PCR reverse primer (10 µM), 0.1 µl ROX
reference dye (50X), 0.4 µl cDNA and 1.6 µl dH2O. RT-qPCR analysis
was performed on a StepOnePlus Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The housekeeping gene,
glycerldehyde-3-phosphate dehydrogenase (GAPDH) was selected as an
internal reference, and 10 additional genes were selected at random
from the DEGs, the primers of which are shown in Table I. Triplicate RT-qPCR amplifications
were performed for each gene. The amplification parameters were set
as follows: 95°C for 30 sec, 95°C for 5 sec, 60°C for 31 sec over
40 cycles. Finally, the quantification values were calculated and
analyzed using the 2-∆∆Cq method (17).
 | Table I.Primers used in Reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primers used in Reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene symbol | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| ABCA1 |
ACCCACCCTATGAACAACATGA |
GAGTCGGGTAACGGAAACAGG |
| STAT4 |
GCTTAACAGCCTCGATTTCAAGA |
GAGCATGGTGTTCATTAACAGGT |
| BMP2 |
ACTACCAGAAACGAGTGGGAA |
GCATCTGTTCTCGGAAAACCT |
| S1PR5 |
GCGCACCTGTCCTGTACTC |
SGTTGGTGAGCGTGTAGATGATG |
| TPM2 |
AGTTTGCCGAGAGGTCTGTG |
TGGTCCAAGGTCTGGTGAATC |
| ISL1 |
GCGGAGTGTAATCAGTATTTGGA |
GCATTTGATCCCGTACAACCT |
| SDK2 |
TCAAGTGGCTCCACAACAACA |
GCACGATGCAACGGTAAAAGC |
| SEMA3D |
TCAGAGCACTACTGGCTCAAT |
ATCGGAGGTACTGCCTTCTTG |
| SP2 |
CTCAGCCCCGGCAAGAATAG |
TTGATCGGGTCCCTTTGTTGA |
| ACSBG1 |
AGTACATCGCTTATGACTGCTGC |
TGGGTGTCAATGATGGCGTC |
| GAPDH |
ACAACTTTGGTATCGTGGAAGG |
GCCATCACGCCACAGTTTC |
Results
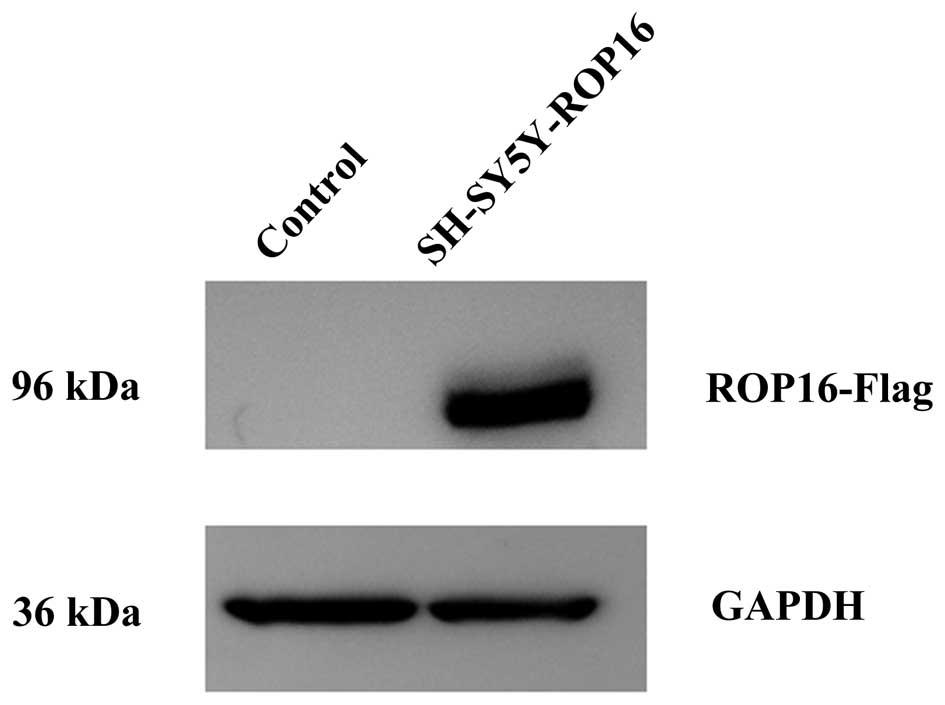
Transfection of cells with ROP16
The present study used GAPDH as an internal
reference for western blot analysis in order to compare the
transcriptional levels of ROP16 in the SH-SY5Y-ROP16 and SH-SY5Y
cell lines. The result demonstrated that ROP16 was successfully
transfected into the SH-SY5Y cells (Fig. 1).
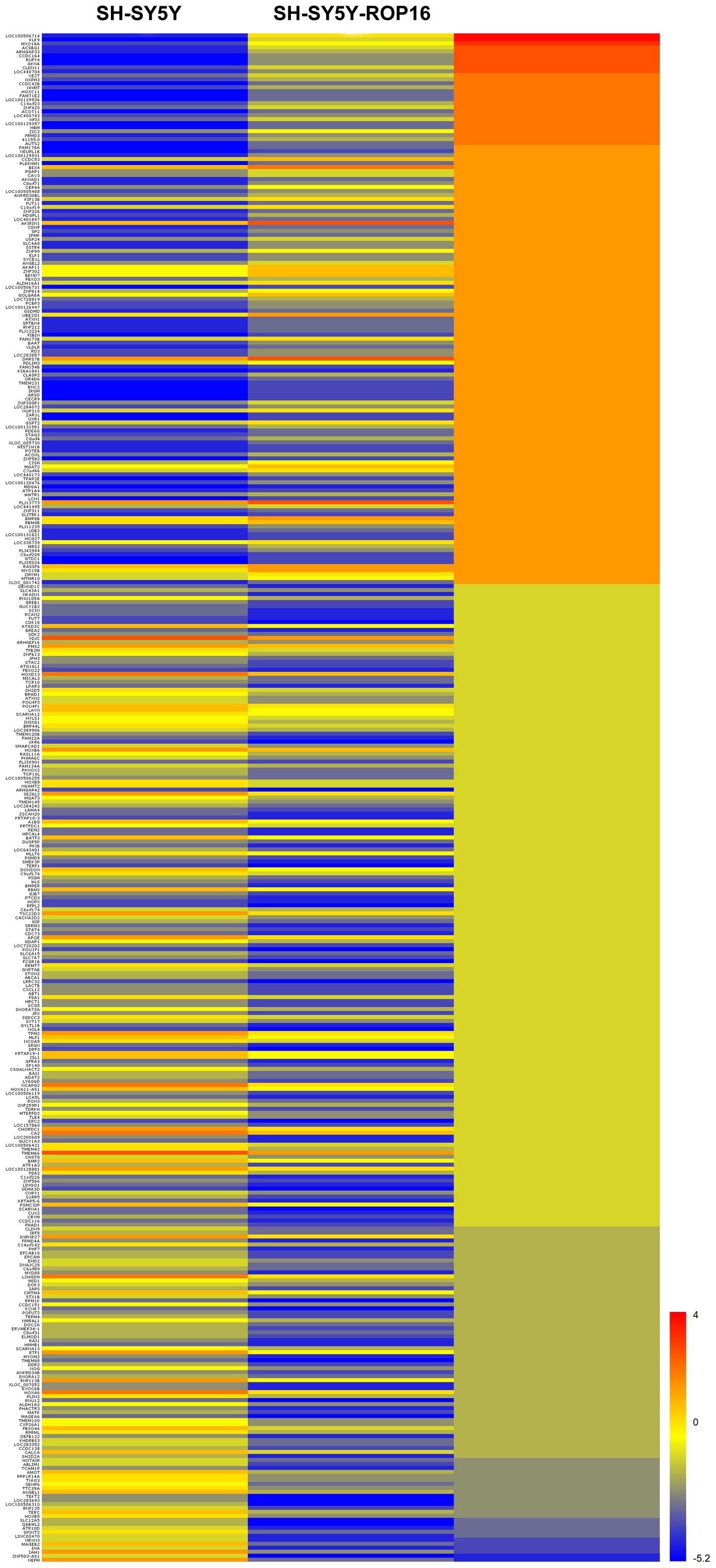
Response of DEGs to transfection with
ROP16
The microarray data revealed that the host cell
genes exhibited significant variety in expression level due to the
transfection. A total of 383 genes were significantly altered as
DEGs, which included 138 upregulated genes and 245 downregulated
genes (Fig. 2).
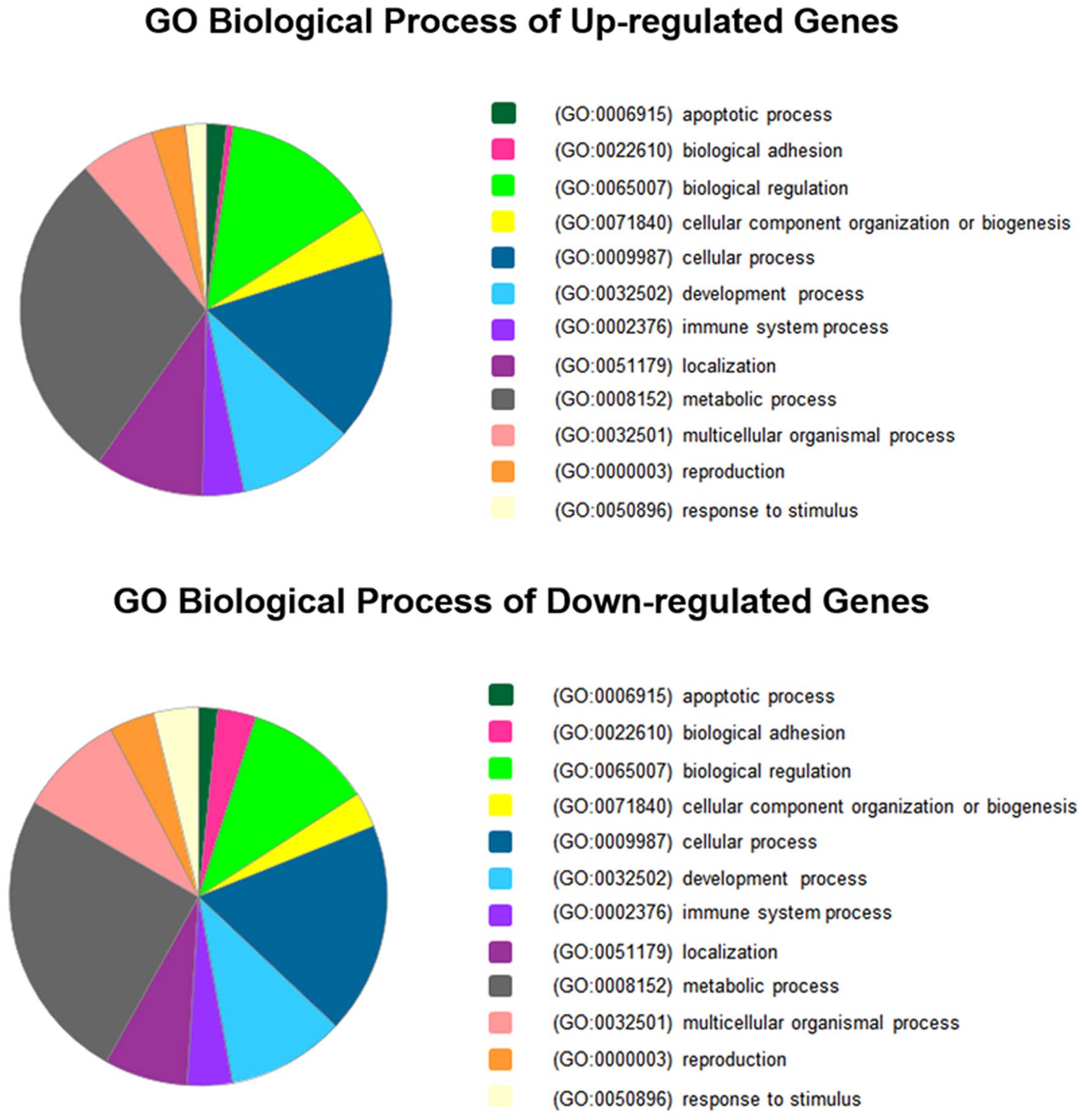
Functional categories of DEGs
The DEGs were analyzed using PANTHER in accordance
with the biological process of PANTHER GO slim (a subset of GO
terms providing a broad overview of GO) to elucidate the
correlation between genes and physiology in host cells. As revealed
by the results, transfection of cells with ROP16 affected several
biological processes, including apoptotic process, biological
regulation, development process, metabolic process and response to
stimulus (Fig. 3). The details of
the biological processes of these genes are listed in Table II. Genes associated with neural
system development, apoptosis and transcriptional regulation are
listed in Table III according to
the functional categories.
 | Table II.Biological processes categories of
differentially expressed genes. |
Table II.
Biological processes categories of
differentially expressed genes.
| Biological
process | Count | Percentage gene
hits, vs. total | Process hits
(%) |
|---|
| Upregulated
differentially expressed genes |
|
|
|
|
Cellular component
organization or biogenesis (GO:0071840) | 7 | 0.06 | 0.04 |
|
Cellular process
(GO:0009987) | 28 | 0.24 | 0.17 |
|
Localization (GO:0051179) | 16 | 0.14 | 0.10 |
|
Apoptotic process
(GO:0006915) | 3 | 0.02 | 0.02 |
|
Reproduction (GO:0000003) | 5 | 0.04 | 0.03 |
|
Biological regulation
(GO:0065007) | 23 | 0.20 | 0.14 |
|
Response to stimulus
(GO:0050896) | 3 | 0.03 | 0.02 |
|
Developmental process
(GO:0032502) | 17 | 0.15 | 0.10 |
|
Multicellular organismal
process (GO:0032501) | 11 | 0.10 | 0.07 |
|
Biological adhesion
(GO:0022610) | 1 | 0.01 | 0.01 |
|
Metabolic process
(GO:0008152) | 49 | 0.42 | 0.29 |
| Immune
system process (GO:0002376) | 6 | 0.06 | 0.04 |
| Downregulated
differentially expressed genes |
|
|
|
|
Cellular component
organization or biogenesis (GO:0071840) | 11 | 0.05 | 0.03 |
|
Cellular process
(GO:0009987) | 66 | 0.30 | 0.18 |
|
Localization (GO:0051179) | 26 | 0.12 | 0.07 |
|
Apoptotic process
(GO:0006915) | 6 | 0.03 | 0.02 |
|
Reproduction (GO:0000003) | 14 | 0.06 | 0.04 |
|
Biological regulation
(GO:0065007) | 40 | 0.18 | 0.11 |
|
Response to stimulus
(GO:0050896) | 14 | 0.06 | 0.04 |
|
Developmental process
(GO:0032502) | 37 | 0.17 | 0.10 |
|
Multicellular organismal
process (GO:0032501) | 33 | 0.15 | 0.09 |
|
Biological adhesion
(GO:0022610) | 12 | 0.06 | 0.03 |
|
Metabolic process
(GO:0008152) | 92 | 0.42 | 0.25 |
| Immune
system process (GO:0002376) | 14 | 0.06 | 0.04 |
 | Table III.Genes associated with nervous system
development, apoptosis and transcriptional regulation. |
Table III.
Genes associated with nervous system
development, apoptosis and transcriptional regulation.
|
| Gene symbol |
|---|
|
|
|
|---|
| Functional category
of interest | Upregulated
genes | Downregulated
genes |
|---|
| Nervous system
development | HOXC11, NEURL1B,
BNC2, ZIC2, RBM4B, SLITRK1, PCBP3 | HOXB6, SDK2, HOXB5,
IGDCC3, SH2D5, HOXD13, SEMA3D, SCG5, HOXB8, TPM2, RASL11A, ATXN2,
LINGO1, BAI1, SYT17, RBMX, CDK18, ABLIM1, PHACTR3, EHD2, ISL1,
SLC6A15, REM2, NRXN3, HOXA6, DOC2A, LAMA4 |
| Apoptosis | CISH, RASSF6 | MAGEA6, SDK2,
IGDCC3, MAGEB2, STAT4, DPF3 |
| Transcriptional
regulation | HOXC11, USP24,
ZIC2, TFAP2E, PDLIM3, WWTR1, ZMYM1, ZNF582, ANGEL2, ZNF814, POTEB,
ZNF99, LDB2, NFIX, GSPT2, MRS2, RBM4B, ELK1, SLITRK1, HDGFL1, SP2,
PCBP3 | ZNF566, GNPTAB,
BATF3, HOXB6, POU4F3, HOXB5, CUX2, ZSCAN20, SMARCAD1, HOXD13,
ZNF613, KHDRBS3, TLE4, HOXB8, POU4F1, RAI1, TSC22D3, PKNOX2, TFB2M,
PMS2, LINGO1, RBMX, IRF8, IARS, ISL1, POU1F1, GDAP1, STAT4, PIGM,
MLLT6, ANGEL1, HOXA6, SP140, DPF3 |
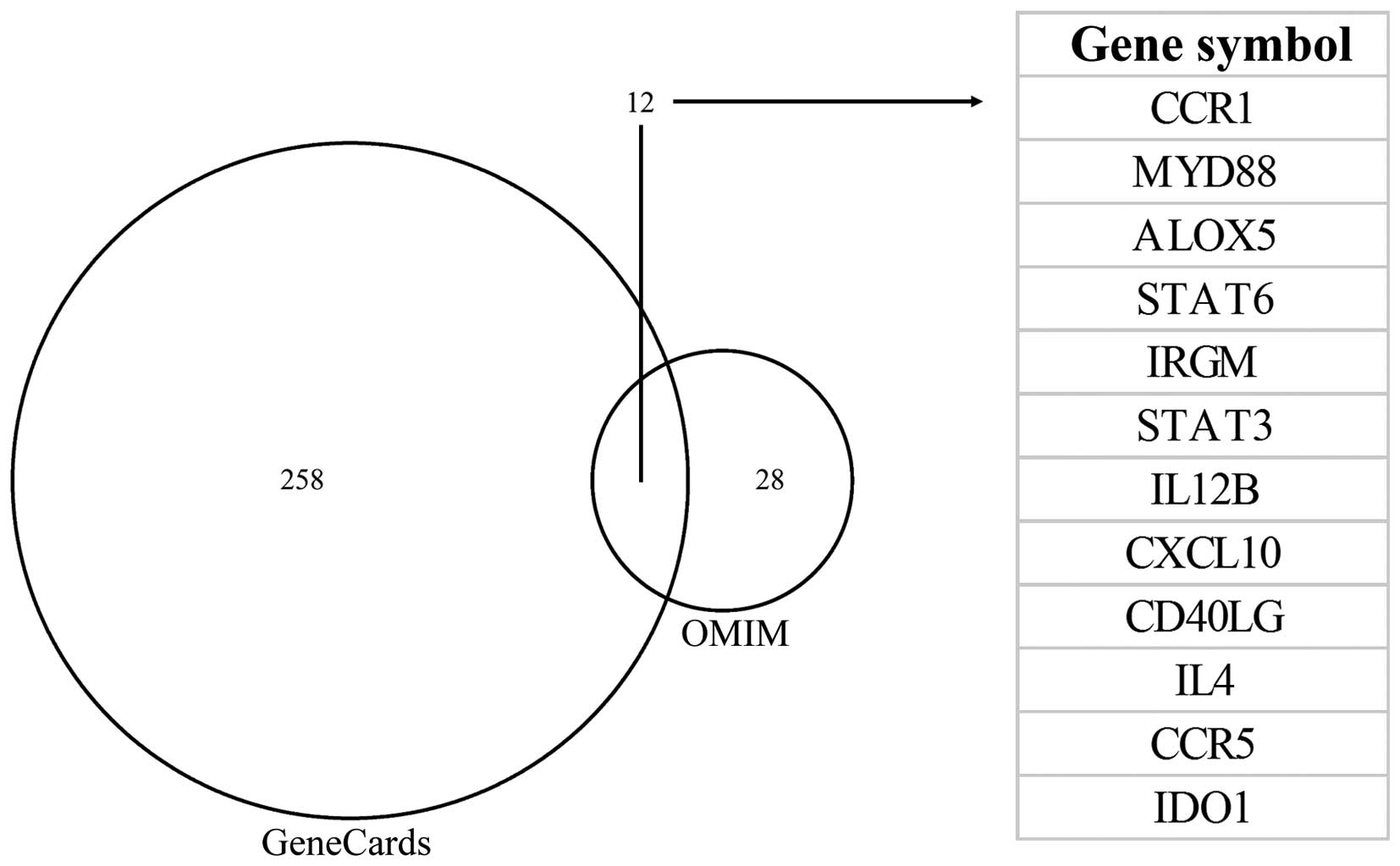
Candidate genes derived from DEGs
According to the instructions of ToppGene and
ToppNet, the ‘training gene set’ contained 12 genes associated with
Toxoplasma, which were collected from the OMIM and GeneCards
database searches (Fig. 4).
Prioritization of candidate genes was obtained using the ToppGene
method and ToppNet method. Candidate genes selected from the top 30
of the ToppGene and ToppNet ranks are shown in Table IV. In addition, 15 genes (CXCL12,
APOE, STAT4, ABCA1, CISH, BMP2, ATG16L1, IGF8, PMS2, ATXN2, ELK1,
PSMD9, AMOT, IARS and ISL1) were present in both sets of
results.
 | Table IV.Top 30 candidate genes of ToppGene
and ToppNet prioritization results. |
Table IV.
Top 30 candidate genes of ToppGene
and ToppNet prioritization results.
| ToppGene | ToppNet |
|---|
|
|
|---|
| Rank | Gene symbol | Average score | Overall
P-value | Rank | Gene symbol | Interactant
count | Score |
|---|
| 1 | CXCL12 | 0.95112 | 4.83E-09 | 1 | STAT4 | 14 | 2.34E-03 |
| 2 | APOE | 0.72009 | 5.09E-07 | 2 | TERF1 | 299 | 7.37E-04 |
| 3 | STAT4 | 0.78586 | 1.08E-05 | 3 | ATXN1 | 255 | 4.48E-04 |
| 4 | ABCA1 | 0.73670 | 1.91E-05 | 4 | CXCL12 | 10 | 3.94E-04 |
| 5 | CISH | 0.70228 | 2.13E-04 | 5 | CISH | 27 | 3.82E-04 |
| 6 | SSTR4 | 0.72088 | 4.10E-04 | 6 | SRGN | 11 | 2.47E-04 |
| 7 | BMP2 | 0.67482 | 4.48E-04 | 7 | RBMX | 89 | 1.61E-04 |
| 8 | ATG16L1 | 0.64074 | 4.59E-04 | 8 | APOE | 54 | 1.56E-04 |
| 9 | CALCA | 0.57179 | 4.87E-04 | 9 | SMARCAD1 | 119 | 1.43E-04 |
| 10 | IRF8 | 0.54026 | 7.56E-04 | 10 | HOXC11 | 13 | 1.27E-04 |
| 11 | PMS2 | 0.58553 | 7.61E-04 | 11 | ABCA1 | 48 | 1.05E-04 |
| 12 | LAMA4 | 0.53700 | 9.43E-04 | 12 | AMOT | 69 | 9.70E-05 |
| 13 | BATF3 | 0.65302 | 0.001017 | 13 | HIST1H1B | 42 | 9.64E-05 |
| 14 | ATXN2 | 0.54901 | 0.001293 | 14 | TPM2 | 35 | 9.59E-05 |
| 15 | ELK1 | 0.45828 | 0.001894 | 15 | ELK1 | 41 | 9.49E-05 |
| 16 | WLS | 0.61716 | 0.002097 | 16 | IARS | 69 | 9.14E-05 |
| 17 | MGAT2 | 0.38112 | 0.002123 | 17 | PTCD3 | 32 | 8.18E-05 |
| 18 | MYO18A | 0.47104 | 0.003082 | 18 | CDC73 | 61 | 8.06E-05 |
| 19 | PSMD9 | 0.43535 | 0.003175 | 19 | PSMD9 | 38 | 7.48E-05 |
| 20 | WWTR1 | 0.53933 | 0.003483 | 20 | SH2D2A | 12 | 6.98E-05 |
| 21 | AMOT | 0.49193 | 0.003639 | 21 | ATG16L1 | 35 | 6.76E-05 |
| 22 | LPAR3 | 0.58023 | 0.003655 | 22 | BMP2 | 19 | 6.06E-05 |
| 23 | IARS | 0.47188 | 0.003673 | 23 | IRF8 | 22 | 6.05E-05 |
| 24 | GFRA3 | 0.69785 | 0.003804 | 24 | RTF1 | 26 | 5.94E-05 |
| 25 | DDR2 | 0.54028 | 0.003883 | 25 | KHDRBS3 | 29 | 5.75E-05 |
| 26 | ISL1 | 0.64497 | 0.003949 | 26 | ATXN2 | 38 | 5.75E-05 |
| 27 | S1PR5 | 0.60005 | 0.004607 | 27 | ISL1 | 20 | 5.30E-05 |
| 28 | EPCAM | 0.51005 | 0.004612 | 28 | MID1 | 30 | 5.27E-05 |
| 29 | VLDLR | 0.53509 | 0.004849 | 29 | PMS2 | 44 | 5.20E-05 |
| 30 | FBXO22 | 0.45987 | 0.005457 | 30 | PDE6G | 8 | 5.04E-05 |
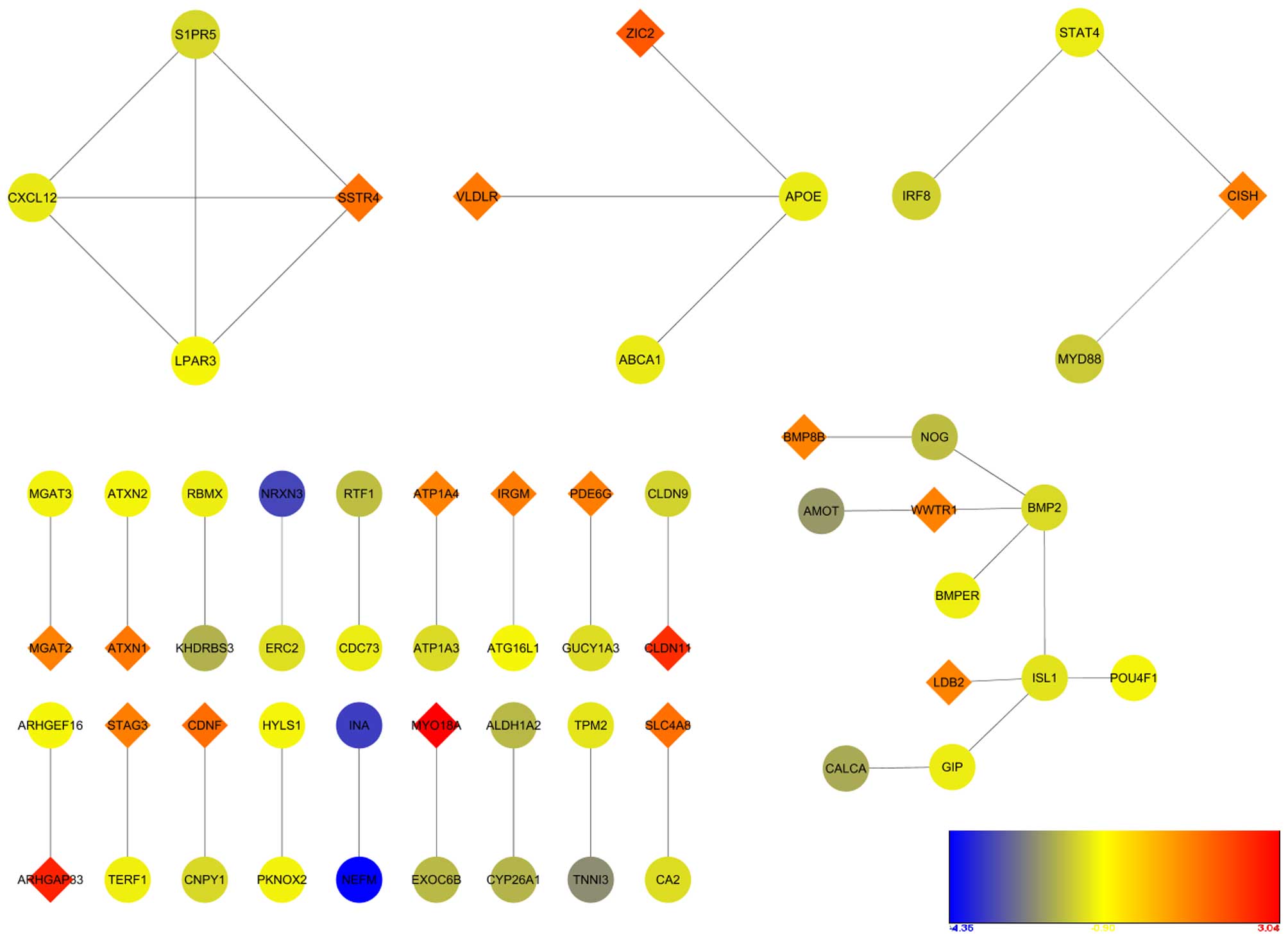
PPI network construction
A total of 40 PPI associations were obtained using
STRING, and networks were constructed using Cytoscape (Fig. 5). Typically, an individual network
comprised a small gene count. S1PR5, CXCL12, LPAR3 and SSTR4 showed
a high degree of connectivity, which was the only cluster examined
by MCODE. The network was selected as a significant module by
MCODE, with an MCODE score of four.
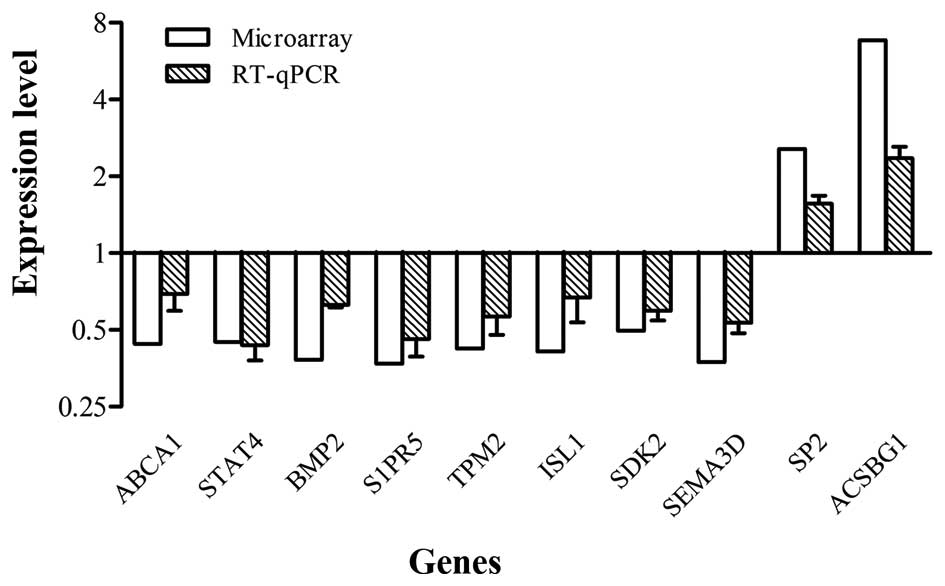
Validation of microarray results
To confirm the microarray results, RT-qPCR analysis
was performed on 10 selected genes (ABCA1, STAT4, BMP2, S1PR5,
TPM2, ISL1, SDK2, SEMA3D, SP2 and ACSBG1). These genes were all in
accordance with the microarray data, although individual values
were not identical due to differences in sensitivity between the
two methods (Fig. 6).
Discussion
Identifying novel candidate genes of certain
diseases provides clues for the investigation of novel biomarkers
or target genes for diagnosis or therapy, and improves
understanding of pathogenesis. ToppGene Suite is an advanced
bioinformatics tool for examining and prioritizing candidate genes
through comprehensive assessment of factors, including GO
annotation, phenotype, signaling pathway and protein interaction,
from a specific gene list (13,18).
In the present study, 383 genes were found to have significantly
altered expression through microarray-based detection comparing the
expressional profiles of SH-SY5Y-ROP16 with those of SH-SY5Y. A
total of 138 genes were upregulated and 245 genes were
downregulated due to transfection with ROP16. The present study
attempted to screen the most likely candidate gene associated with
Toxoplasma in the DEGs following transfection with ROP16.
The results of ToppGene and ToppNet revealed which DEGs caused by
transfection with ROP16 were the most likely candidate genes
associated with Toxoplasma and may be further investigated
as target genes. The intersecting genes from ToppGene and ToppNet
included CXCL12, APOE, STAT4, ABCA1, CISH, BMP2, ATG16L1, IGF8,
PMS2, ATXN2, ELK1, PSMD9, AMOT, IARS and ISL1. Intersection
indicated that the gene warranted high prioritization. The data in
the present study showed that CXCL12 had the highest prioritization
with a downregulated expression level, which may indicate that
CXCL12 was a critical factor associated with the state of host cell
development in response to the ROP16 transfection. CXCL12 is a key
innate immune mediator, which is involved in embryonic neuronal
survival and migration. CXCL12 is generally expressed in the brain
of the central nervous system. A previous study noted that CXCL12
maintains the development of motor neurons and axons (19), and also contributes to the
development of glioma (20).
CXCL12 was involved in a significant network module with S1PR5,
SSTR4 and LPAR3 according to the PPI network analysis. This
suggested that these four genes may be considered as potential
targets for the interaction between Toxoplasma and host.
All DEGs analyzed by PANTHER in the present study
were involved in several biological processes, including cellular
component organization or biogenesis, cellular process,
localization, apoptotic process, reproduction, biological
regulation, response to stimulus, developmental process,
multicellular organismal process, biological adhesion, metabolic
process and immune system process. The human nervous system is
vital in maintaining homeostasis, comprising the central nervous
system and peripheral nervous system. The brain is a preferred
target during infection, particularly for chronic T. gondii
infections. T. gondii may interfere with host neuronal
function and modulate signaling pathways in the host brain
(21). The expressional profiles
revealed 23 genes (six upregulated and 17 downregulated) associated
with nervous system development, a subclass of developmental
process. The expression of BAI1 was downregulated by the
transfection with ROP16 among these genes. BAI1 is known as an
angiogenesis inhibitor, which can be directly induced by wild-type
p53 and has been reported to be specifically expressed in the human
brain. BAI1 is typically expressed in the cerebral cortex of the
human adult brain, and regulates synaptic plasticity and signal
transduction in the neurological system (22,23).
The ZIC2 gene is upregulated as it is a vital gene in human brain,
which is involved in neural development (24). RBMX is a protein-coding gene for
the production of an RNA-binding protein involved in the
tissue-specific regulation of gene transcription and alternative
splicing of several pre-mRNAs. A previous study showed that a low
expression level of RBMX resulted in poor development of the
zebrafish head (25). The
downregulated expression of RBMX was also shown in the present
study, which may indicate that the central nervous system was
disturbed in the SH-SY5Y cells due to transfection with ROP16.
Taken together, the decreased expression levels of BAI1 and RBMX,
and increased expression of ZIC2 following transfection with ROP16
disrupted nervous system development.
Apoptosis or ‘programmed’ cell death is an essential
process in cells. Apoptosis is involved in healthy cells and
diseased cells. It occurs during the normal development of cells
and maintains the population of cells in tissues. Apoptosis acts as
a defense mechanism, which removes toxins and contributes to
homeostasis when immune reactions are activated. Previous studies
have revealed that T. gondii infection results in cell
apoptosis through disturbing signaling pathways of host cells
(26,27). In the present study, nine genes
associated with apoptosis were detected using microarray analysis,
including three upregulated genes and six downregulated genes. It
has been found that the upregulated gene, RASSF6, can interact with
MDM2 and stabilize p53, promoting apoptosis and G1/S arrest in
HCT116 cell lines (28). The
present study showed that RASSF6 was upregulated, which indicate
that similar pathways may be involved in the apoptotic process of
SH-SY5Y cells. It is well known that MAGE-A proteins can switch off
the association between p53 and its responsive genes by inhibiting
p53-dependent transcription. For example, the MAGE-A6 protein can
inhibit expression of the p21 promoter. Low expression levels of
MAGE-A results in the upregulation of genes, including p21, MDM2
and PUMA (29). As the present
study found MAGE-A6 was downregulated by transfection of ROP16, it
was hypothesized that, in SH-SY5Y cells, the downregulated
expression of MAGE-A6 contributed to the expression of p21,
therefore, p53-dependent apoptosis may have been partly
stimulated.
The typical characteristic of ROP16 is that it can
rapidly migrate into the nucleus of host cells following the
invasion of T. gondii. It is essential to investigate the
effect of ROP16 on gene transcription as the nucleus has the
ability to control vital activities of the cell through the
regulation of gene transcription and translation. Several DEGs were
found to be involved in transcriptional regulation. The homeobox
(HOX) genes encode conserved transcription regulators and govern
processes, including morphogenesis and cell differentiation in
vertebrate embryonic development and development of the central
nervous system, where their expression is restricted and complex.
It has been reported that, in human adipose tissue, the HOX gene
network regulates the transcription of adipogenesis (30). In the present study, it was found
that the majority of genes were upregulated, with the exception of
HOXC11, which was downregulated. As HOX genes were differentially
expressed due to the transfection of ROP16 in the present study, it
was hypothesized that HOX genes may function critically in SH-SY5Y.
However, the mechanism underlying the transcriptional regulation of
HOX genes and their individual functions requires further
investigation. The variation in gene expression in host cells under
different physiological condition has been a key focus of
investigations. The use of microarray technology assists in
detecting alterations at a genome-wide level, which provides
substantial benefits for experiments. In the process of altering
gene expression in host cells, it is clear that pathogen proteins
located in the nucleus are important. Thus, the examination of
ROP16, a parasitic protein injected into the nucleus, contributed
to improving comprehension of the pathogenic mechanisms of
Toxoplasma.
In conclusion, the present study suggested that
ROP16 transfection caused host cell alterations in the expression
of multiple genes. These genes, particularly those involved in the
processes of nervous system development, apoptosis and
transcriptional regulation, may be vital in the response of host
cells against T. gondii infection, and contribute to the
pathogenesis of Toxoplasma. Novel candidate genes obtained
through microarray analysis may provide insight for further
investigations of its pathogenesis and therapeutic methods.
References
|
1
|
John B, Ricart B, Wojno ED Tait, Harris
TH, Randall LM, Christian DA, Gregg B, De Almeida DM, Weninger W,
Hammer DA and Hunter CA: Analysis of behavior and trafficking of
dendritic cells within the brain during toxoplasmic encephalitis.
PLoS Pathog. 7:e10022462011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Montoya JG and Remington JS: Management of
Toxoplasma gondii infection during pregnancy. Clin Infect Dis.
47:554–566. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tanaka S, Nishimura M, Ihara F, Yamagishi
J, Suzuki Y and Nishikawa Y: Transcriptome analysis of mouse brain
infected with Toxoplasma gondii. Infect Immun. 81:3609–3619. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boothroyd JC: Have it your way: How
polymorphic, injected kinases and pseudokinases enable Toxoplasma
to subvert host defenses. PLoS Pathog. 9:e10032962013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dubremetz JF: Rhoptries are major players
in Toxoplasma gondii invasion and host cell interaction. Cellular
Microbiol. 9:841–848. 2007. View Article : Google Scholar
|
|
6
|
Mueller C, Klages N, Jacot D, Santos JM,
Cabrera A, Gilberger TW, Dubremetz JF and Soldati-Favre D: The
Toxoplasma protein ARO mediates the apical positioning of rhoptry
organelles, a prerequisite for host cell invasion. Cell Host
Microbe. 13:289–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor S, Barragan A, Su C, Fux B,
Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, et
al: A secreted serine-threonine kinase determines virulence in the
eukaryotic pathogen Toxoplasma gondii. Science. 314:1776–1780.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boothroyd JC and Dubremetz JF: Kiss and
spit: The dual roles of Toxoplasma rhoptries. Nat Rev Microbiol.
6:79–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saeij JP, Coller S, Boyle JP, Jerome ME,
White MW and Boothroyd JC: Toxoplasma co-opts host gene expression
by injection of a polymorphic kinase homologue. Nature.
445:324–327. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jensen KD, Hu K, Whitmarsh RJ, Hassan MA,
Julien L, Lu D, Chen L, Hunter CA and Saeij JP: Toxoplasma gondii
rhoptry 16 kinase promotes host resistance to oral infection and
intestinal inflammation only in the context of the dense granule
protein GRA15. Infect Immun. 81:2156–2167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mi H, Muruganujan A and Thomas PD: PANTHER
in 2013: Modeling the evolution of gene function, and other gene
attributes, in the context of phylogenetic trees. Nucleic Acids
Res. 41:(Database Issue). D377–D386. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mi H, Muruganujan A, Casagrande JT and
Thomas PD: Large-scale gene function analysis with the PANTHER
classification system. Nature Protoc. 8:1551–1566. 2013. View Article : Google Scholar
|
|
13
|
Chen J, Bardes EE, Aronow BJ and Jegga AG:
ToppGene suite for gene list enrichment analysis and candidate gene
prioritization. Nucleic Acids Res (Web Server Issue). 37:W305–W311.
2009. View Article : Google Scholar
|
|
14
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen J, Xu H, Aronow BJ and Jegga AG:
Improved human disease candidate gene prioritization using mouse
phenotype. BMC Bioinformatics. 8:3922007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dugas JC, Mandemakers W, Rogers M, Ibrahim
A, Daneman R and Barres BA: A novel purification method for CNS
projection neurons leads to the identification of brain vascular
cells as a source of trophic support for corticospinal motor
neurons. J Neurosci. 28:8294–8305. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oh JW, Olman M and Benveniste EN:
CXCL12-mediated induction of plasminogen activator inhibitor-1
expression in human CXCR4 positive astroglioma cells. Biol Pharm
Bull. 32:573–577. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carruthers VB and Suzuki Y: Effects of
Toxoplasma gondii infection on the brain. Schizophr Bull.
33:745–751. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mori K, Kanemura Y, Fujikawa H, Nakano A,
Ikemoto H, Ozaki I, Matsumoto T, Tamura K, Yokota M and Arita N:
Brain-specific angiogenesis inhibitor 1 (BAI1) is expressed in
human cerebral neuronal cells. Neurosci Res. 43:69–74. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu D, Li C, Swanson AM, Villalba RM, Guo
J, Zhang Z, Matheny S, Murakami T, Stephenson JR, Daniel S, et al:
BAI1 regulates spatial learning and synaptic plasticity in the
hippocampus. J Clin Invest. 125:14975082015. View Article : Google Scholar
|
|
24
|
Nagai T, Aruga J, Minowa O, Sugimoto T,
Ohno Y, Noda T and Mikoshiba K: Zic2 regulates the kinetics of
neurulation. Proc Natl Acad Sci USA. 97:1618–1623. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsend-Ayush E, O'Sullivan LA, Grützner FS,
Onnebo SM, Lewis RS, Delbridge ML, Graves JA Marshall and Ward AC:
RBMX gene is essential for brain development in zebrafish. Dev Dyn.
234:682–688. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang T, Zhou J, Gan X, Wang H, Ding X,
Chen L, Wang Y, DU J, Shen J and Yu L: Toxoplasma gondii induce
apoptosis of neural stem cells via endoplasmic reticulum stress
pathway. Parasitology. 141:988–995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nyoman AD and Lüder CG: Apoptosis-like
cell death pathways in the unicellular parasite Toxoplasma gondii
following treatment with apoptosis inducers and chemotherapeutic
agents: A proof-of-concept study. Apoptosis. 18:664–680. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iwasa H, Kudo T, Maimaiti S, Ikeda M,
Maruyama J, Nakagawa K and Hata Y: The RASSF6 tumor suppressor
protein regulates apoptosis and the cell cycle via MDM2 protein and
p53 protein. J Biol Chem. 288:30320–30329. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marcar L, MacLaine NJ, Hupp TR and Meek
DW: Mage-A cancer/testis antigens inhibit p53 function by blocking
its interaction with chromatin. Cancer Res. 70:10362–10370. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cantile M, Procino A, D'armiento M,
Cindolo L and Cillo C: HOX gene network is involved in the
transcriptional regulation of in vivo human adipogenesis. J Cell
Physiol. 194:225–236. 2003. View Article : Google Scholar : PubMed/NCBI
|