Introduction
Mature articular cartilage is unable to heal
spontaneously and, consequently, lesions eventually progress to
osteoarthritis. This lack of capacity for self-repair has prompted
intensive research into methods of articular cartilage
regeneration, including cell-based cartilage tissue engineering
(1,2). The use of stem cells (SCs) may help
to overcome the drawbacks of autologous chondrocytes, which include
the limited number of chondrocytes available for cell culture,
preservation of the cells' chondrogenic potential, and
re-differentiation of cells during tissue formation following
implantation. Human mesenchymal stem cells and human induced
pluripotent stem cells (hiPSCs) may be useful for cartilage
regeneration (3–5). It is possible to use defined
transcription factors to transorm terminally-differentiated cells,
including fibroblasts, into hiPSCs, which share characteristics
with human embryonic SCs (hESCs) (6). However, this strategy is not without
risk, given that hESCs and hiPSCs are potentially tumorigenic and
must therefore be monitored carefully if they are to be applied
safely (7). Patient-derived hiPSCs
differentiate into derivatives of three germ layers, ecto-, meso-
and endoderm, and may be ideal autologous cells for chondrocyte
generation because they are not subject to immune rejection and are
easily expanded prior to chondrocyte generation (8).
Numerous techniques are available for chondrogenic
differentiation of SCs, although the most common and efficient
method of obtaining chondrocyte-like cells from hiPSCs is the
formation of embryoid bodies (EB). Depending on the specific
approach, chondrogenic differentiation may require the use of
selected growth factors, scaffolds, or other biomaterials, as well
as specific culture dishes (2- or 3-dimensional culture). Although
it is possible to use a variety of mediums for chondrogenic
differentiation, the optimal medium remains unclear (9–12).
Similarly, during the chondrogenic differentiation process, a wide
range of markers are available to assess cell differentiation, but
the relative utility of these markers is not well-understood, in
particular in the context of hiPSC differentiation, which is a
novel approach in regenerative medicine (13,14).
Given this context, the primary aims of the present
study were as follows: To determine the gene expression profile of
chondrogenic-like cells derived from hiPSCs cultured in mediums
conditioned with HC-402-05a cells or supplemented with transforming
growth factor β3 (TGF-β3), and to assess the relative utility of
the most commonly used chondrogenic markers as indicators of cell
differentiation.
The cells were differentiated in chondrogenic
mediums supplemented with either TGF-β3, the member of the TGF-β
superfamily with the most chondrogenic potential (15) or conditioned with growth factors
from the human primary chondrocyte cell line HC-402-05a. The gene
expression profile of the chondrogenic-like cells derived from the
hiPSCs cultured in the TGF-β-supplemented medium (TGF-β medium)
were them compared with the cells cultured in the
HC-402-05a-conditioned medium (condtioned medium). Notably, the
type of medium had a notable impact on gene expression profiles. A
total of 20 markers of chondrogenic differentiation were also
evaluated, and paired box 9 (PAX9), sex determining region
Y-box 5 (SOX5), sex determining region Y-box 6
(SOX6), sex determining region Y-box 9 (SOX9) and
cartilage oligomeric matrix protein (COMP) were demonstrated
to be good markers of hiPSC differentiation, whereas insulin-like
growth factor 1 (IGF-1), Tenascin-C (TNC), and
β-catenin were less valuable indicators of cell
differentiation. Furthermore, the origin (mesoderm) of fibroblasts
and chondrocytes should be taken into consideration, due to the
fact that several genes are common for stem cell-derived
chondrocytes and human fibroblasts (e.g., SMAD3 and
BMP-2), decreasing their utility in the evaluation of
chondrogenic process in vitro.
The findings of the present study demonstrated that
cells differentiated in the conditioned medium present features
that are characteristic of mature chondrocytes, whereas the
features of cells cultured in the presence of TGF-β3 are
characteristic of hypertrophic chondrocytes, thus underscoring the
potential of the HC-402-05a-conditioned medium for in vitro
chondrogenesis. The present study contributes to an improved
understanding of the changes in gene expression that occur during
the in vitro chondrogenic process and short-term culture of
stem-derived chondrocytes, in addition to helping to clarify the
relative value of a wide range of chondrogenic differentiation
markers.
The present study is a two-part study. Part A,
presented here, describes the markers that are characteristic for
pluripotency state and early-stage chondrogenesis (Table I). The second part of the study
(16) focused on markers that are
characteristic of late stage chondrogenesis, hypertrophy and
ossification.
 | Table I.Assessment of selected markers for
early hiPSC chondrogenic differentiation in vitro. |
Table I.
Assessment of selected markers for
early hiPSC chondrogenic differentiation in vitro.
| Marker | Function of marker
(stage of presentation) | Influence on
chondrogenesis: positive (+) or negative (−) | Utility of the
marker to assess chondrogenic progression (+, ++, +++) |
|---|
| NANOG | Maintenance of
pluripotency (SCs) | − | +++ |
| OCT-4 | Maintenance of
pluripotency (SCs) | − | +++ |
| SOX2 | Maintenance of
pluripotency (SCs) | − | +++ |
| E-CADHERIN | Maintenance of
pluripotency (SCs) | − | +++ |
| BRACHYURY | Cells from
mesodermal stage | −/+ | +++ |
| CXCR4 | Cells from
mesodermal and endodermal stage | − | +++ |
| TENASCIN-C | ECM of articular
cartilage/condensation stage | + | + |
| PAX9 | Induction of
chondrogenesis (chondroprogenitors) | + | +++ |
| NCAM | ECM/osteoblasts
(condensation stage) | −/+ | ++ |
| NKX3.2 |
Chondroprogenitors | + | ++ |
| The SOX trio: SOX5,
6 and 9 | Chondrogenesis | + | +++ |
| IGF-1 |
Pluripotency/chondrocytes/hypertrophic
chondrocytes/osteoblasts | + | + |
| CD44 | Cell-surface
glycoprotein | + | ++ |
| COMP | Cartilage ECM | + | +++ |
| AGGRECAN | Cartilage ECM | + | ++ |
| β-CATENIN |
Pluripotency/mesoderm/chondrocytes/hypertrophic
chondrocytes/osteoblasts | + | + |
| EGF | SCs/cell
proliferation/chondrogenesis | +/− | + |
| FGFR3 | SCs/cell
proliferation/chondrogenesis | +/− | + |
Materials and methods
Culturing human induced pluripotent
stem cells
The hiPSCs obtained during the reprogramming process
as previously described (17) were
seeded on 10 cm Petri dishes in Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA) that had previously been coated with inactivated
murine embryonic fibroblasts as a feeder layer (1×106).
Following 24 h preparation of the feeder layer, hiPSCs were seeded
at 2×106 in hiPSC growth medium: Dulbecco's modified
Eagle's medium (DMEM) F12 with L-glutamine (Merck Millipore,
Darmstadt, Germany), 20% knockout serum replacement (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 1% non-essential amino acids
(Merck Millipore), 0.1 mM β-mercaptoethanol (Merck Millipore), 1%
penicillin/streptomycin (P/S; Merck Millipore). Prior to use, the
medium was supplemented with fibroblast growth factor 2 (FGF-2; 10
ng/ml; Merck Millipore). The complete hiPSC growth medium was
supplemented with ciprofloxacin (0.5 µg/ml; Sigma Aldrich; Merck
Millipore) to avoid Mycoplasma spp. contamination for the first 7
days of culture. The culture medium was changed daily. Experiments
using hiPSCs do not need approval from a local ethics
committee.
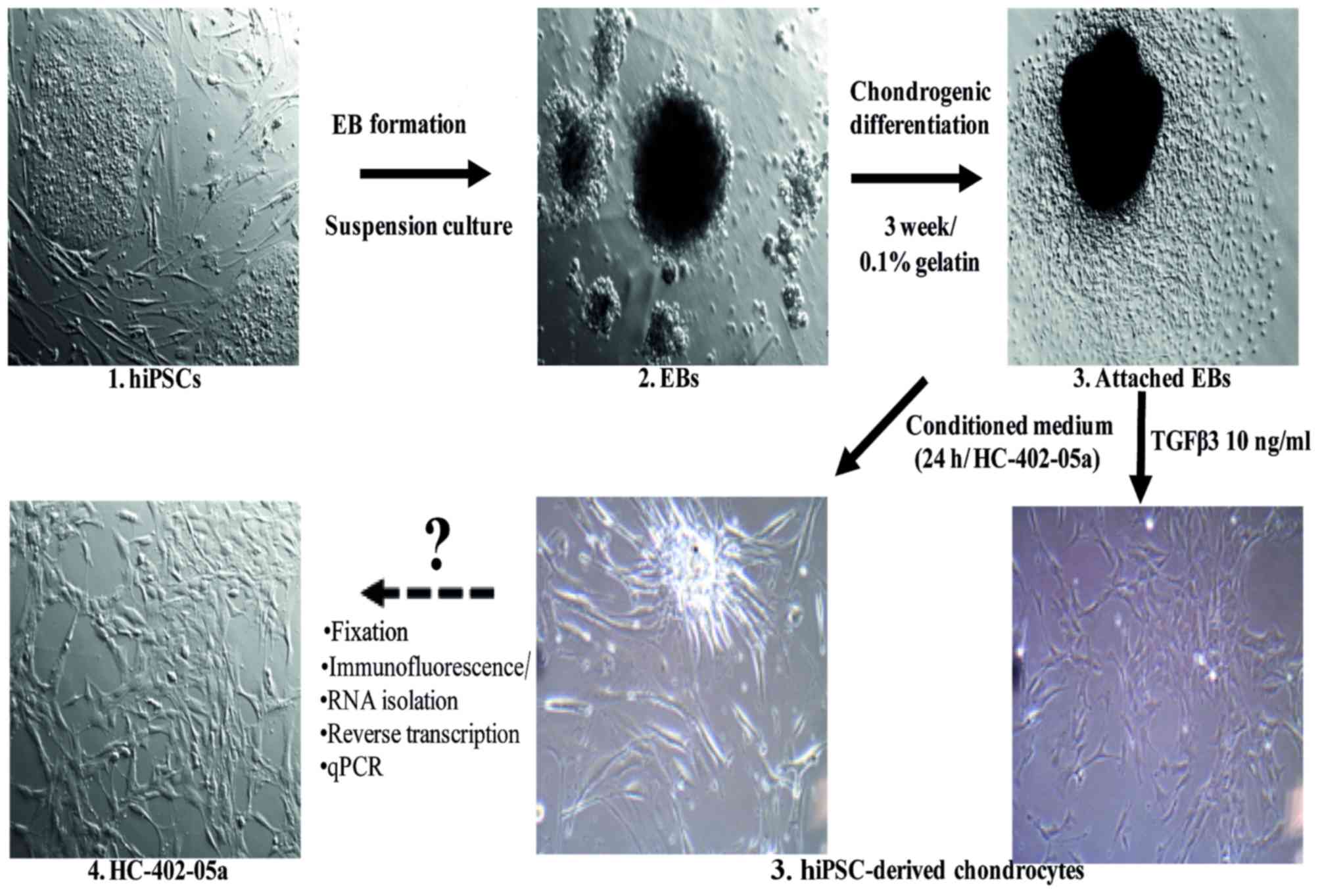
Embryoid body formation
At 80% confluency, hiPSC colonies were passaged and
dissociated into clumps with 0.1% collagenase IV solution (Thermo
Fisher Scientific, Inc.). The cells were centrifuged (300 × g, 5
min, room temperature) in order to remove the collagenase and
transferred into non-adherent 96-well plates (1,000 cells per well;
Brand GmbH, Wertheim, Germany) in EB growth medium, which is a
hiPSC growth medium without FGF-2. Embryoid bodies (EBs) formed
within 24 h and were observed as free-floating aggregates. The
culture medium was changed every 48 h. On day 7 the EBs were used
for chondrogenic differentiation.
Chondrogenesis in vitro
A standard chondrogenic medium was used: DMEM F12
with L-glutamine (Merck Millipore), 10% fetal bovine serum (FBS;
Biowest, Nuaillé, France), 50 µM L-proline (Sigma Aldrich; Merck
Millipore), 50 µM ascorbic acid (Sigma Aldrich; Merck Millipore), 1
mM sodium pyruvate (Biowest), 1% ITS + Premix (Corning Life
Sciences, Big Flats, NY, USA), 1% P/S (Merck Millipore) and
10−7 M dexamethasone (Sigma Aldrich; Merck
Millipore).
Medium conditioning
Standard chondrogenic medium was used: DMEM F12 with
L-Glutamine (Merck Millipore), 10% FBS (Biowest), 50 µM L-proline
(Sigma Aldrich; Merck Millipore), 50 µM ascorbic acid (Sigma
Aldrich; Merck Millipore), 1 mM sodium pyruvate (Biowest), 1% ITS +
Premix (Corning Life Sciences), 1% P/S (Merck Millipore) and
10−7 M dexamethasone (Sigma Aldrich; Merck Millipore),
which was conditioned on the HC-402-05a cell line (up to 3
passages). Medium was collected following 24 h conditioning and
administered to the differentiated EBs.
Chondrogenesis using embryoid
bodies
The mature EBs were transferred onto 6-well plates
(10 EBs per well) previously coated with 0.1% gelatin (Merck
Millipore) and allowed to adhere for the next 24 h, following which
the medium was replaced with a chondrogenic medium. This was either
supplemented with TGF-β3 (10 ng/ml; ImmunoTools GmbH, Friesoythe,
Germany), as a growth factor with the most chondrogenic potential,
or conditioned on the HC-402-05a cell line as above. The positive
influence of standard chondrogenic medium with the addition of
exogenous TGF-β3 (10 ng/ml) on pluripotent SCs was previously
tested and confirmed (18). The
chondrogenic medium was changed every 48 h. The culture period
lasted 21 days. In order to confirm that chondrocyte-like cells had
been obtained, immunofluorescence analysis was performed on passage
0 (p0). Next, to evaluate the expression profile of chondrogenic
markers (p3), reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) analysis was performed (Fig. 1). In all analyses, the stable adult
human articular chondrocyte cell line (HC-402-05a) served as a
positive control, as the European Collection of Authenticated Cell
Cultures recommended it for the evaluation of the differentiation
process in in vitro model systems.
Culture of differentiated cells
The derived stem cells were cultured in 0.1% gelatin
(Merck Millipore) in DMEM F12 with L-glutamine (Merck Millipore),
10% FBS (Biowest), and 1% P/S (Merck Millipore) up to 3
passages.
Immunofluorescence analysis
The cells (p0; 0.5×105) were transferred
into a gelatin-coated (1:50) 48-well plate for 48 h. The cells were
washed with PBS (Sigma Aldrich; Merck Millipore) and fixed for 20
min in 100% methanol (intercellular antigens; CHEMPUR, Piekary
Śląskie, Poland) or 4% formaldehyde (extracellular antigens;
CHEMPUR; 400 µl methanol/formaldehyde per well). Then, the cells
were rinsed with PBS containing 1% FBS (Sigma Aldrich; Merck
Millipore) and incubated for 30 min in PBS containing 1% FBS and
0.2% Triton X-100 (Sigma Aldrich; Merck Millipore) at room
temperature. The cells were subsequently washed with PBS containing
1% FBS. The cells were incubated overnight at 4°C with the
following primary antibodies: COMP (1:100; cat. no. ab128893), type
II collagen (COL2A1; 1:100; cat. no. ab34712), type IX collagen
(COL9A1; 1:100; cat. no. ab134568), agreccan (AGC1; 1:85; cat. no.
ab3778), SOX6 (1:50; cat. no. ab30455), SOX9 (1:50; cat. no.
ab59252); all from Abcam, Cambridge, UK), Nanog (1:50; cat. no.
MABD24) and octamer-binding transcription factor 3/4 (OCT3/4; 1:50;
cat. no. MABD76); from BD Biosciences). The primary antibodies were
diluted in PBS containing 1% FBS and 0.2% Triton X-100. Following
conjugation with the primary antibodies, the cells were rinsed
three times with PBS containing 1% FBS. The following Alexa Fluor
488 conjugated secondary antibodies were diluted with 1% FBS in PBS
and were incubated in the dark for 1 h at 37°C: Mouse monoclonal
anti-immunoglobulin G (cat. no. 715-545-150), mouse monoclonal
anti-immunoglobulin M (cat. no. 715-545-140) and rabbit polyclonal
antibody (cat. no. 711-546-152; 1:500; Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA). Following washing three
times with 1% FBS in PBS, the cells were stained for 5 min with
diamidino-2-phenylindole dye (Sigma Aldrich; Merck Millipore)
solution in water (1:10,000) followed by washing with PBS and
fluorescent microscopic analysis. The intensity of the signals was
evaluated using the bioinformatics programme ImageJ, version 1.49j
(developed by National Institutes of Health, Bethesda, MD,
USA).
RT-qPCR
Total RNA was extracted from cells (p3;
2×106 cells) with TRIzol (Sigma Aldrich; Merck
Millipore). Total RNA (1 µg per 20 µl reaction volume) free of
genomic DNA contamination was reverse-transcribed using the
iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) according to the manufacturer's protocol (25°C for 5 min,
42°C for 30 min, 85°C for 5 min). cDNA was prepared three times for
each repetition. qPCR reactions were performed using the
LightCycler 480 Probes Master mix and appropriate probes labeled
with fluorescein for each primer (Roche Diagnostics, Basel,
Switzerland). The reaction conditions for all amplicons were as
follows: Initially 95°C for 10 min, followed by 45 cycles at 94°C
for 10 sec, 60°C for 15 sec and 72°C for 1 sec. All reactions were
performed in the presence of 3.2 mM MgCl2. cDNA samples
(2.5 µl for a total volume of 10 µl) were analyzed for genes of
interest and for the reference gene glyceraldehyde 3-phosphate
dehydrogenase, which were selected based on the latest literature
data concerning chondrogenic differentiation of hiPSCs (19). The level of expression of each
target gene was calculated as −2ΔΔCq (20). The reaction was performed in
triplicate for the genes of interest. Primer information is
available upon request.
Statistical analysis
All experiments were performed a minimum of three
times. The results are reported as the mean ± standard deviation.
Comparisons between the study groups and controls were performed
using one-way analysis of variance. Where the analysis of variance
results were significant, post hoc analysis was performed via
Tukey's multiple comparison test with a single pooled variance.
Statistical tests were performed with GraphPad Prism (version 5.0a;
GraphPad Software, Inc., San Diego, CA, USA). *P<0.05 was
considered to indicate a statistically significant difference.
Results
Immunofluorescence analysis confirmed
that chondrocyte-like cells were obtained
To confirm the presence of markers characteristic of
chondrocytes, the cells (p0) following chondrogenic differentiation
in the TGF-β3 and conditioned media were analyzed by
immunofluorescent staining. These cells indicated the occurence
COMP, COL2A1, COL9A1, AGC1, SOX6 and SOX9 at levels similar to
those established in the HC-402-05a chondrocyte cell line (Figs. 2 and 3). Furthermore, the chondrocyte-like
cells did not demonstrate the presence of pluripotency markers:
Nanog and OCT3/4/OCT4 (Figs. 2 and
3). These results confirm that the
obtained chondrocyte-like cells were fully differentiated from
hiPSCs. Furthermore, they express the chondrocyte specific
markers.
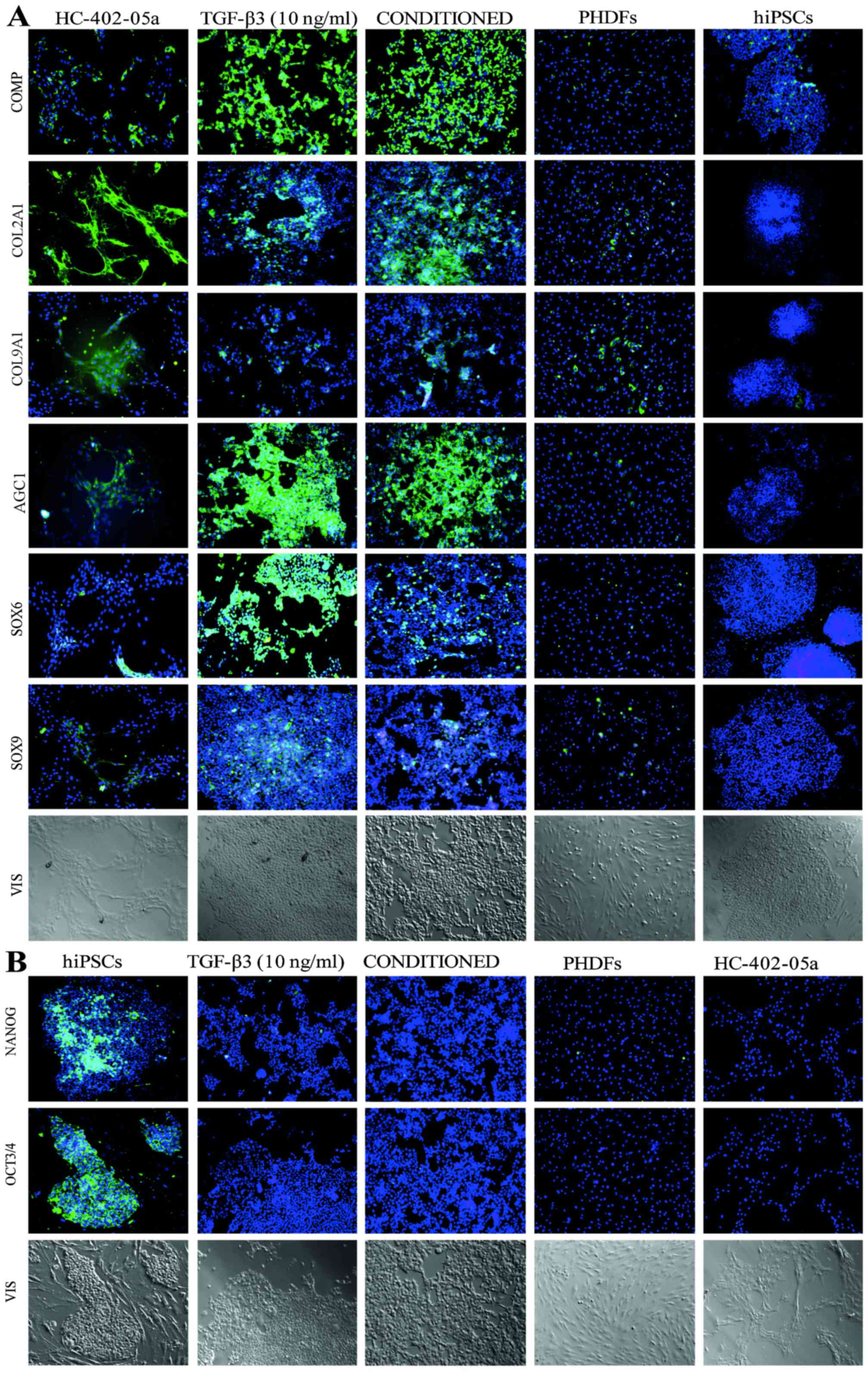 | Figure 2.Immunofluoresence analysis of the
chondrocyte-like cells instantly following the differentiation
process, identified the presence of (A) markers characteristic of
mature chondrocytes and (B) the simultaneous lack of pluripotency
markers. hiPSCs, human induced pluripotent stem cells; TGF-β3,
transforming growth factor β3; PHDFs, primary human dermal
fibroblasts; COMP, cartilage oligomeric matrix protein; COL2A1,
type II collagen; COL9A1, type IX collagen; AGC1, aggrecan; SOX,
sex determining region Y-box; OCT3/4, octamer-binding transcription
factor 3/4. |
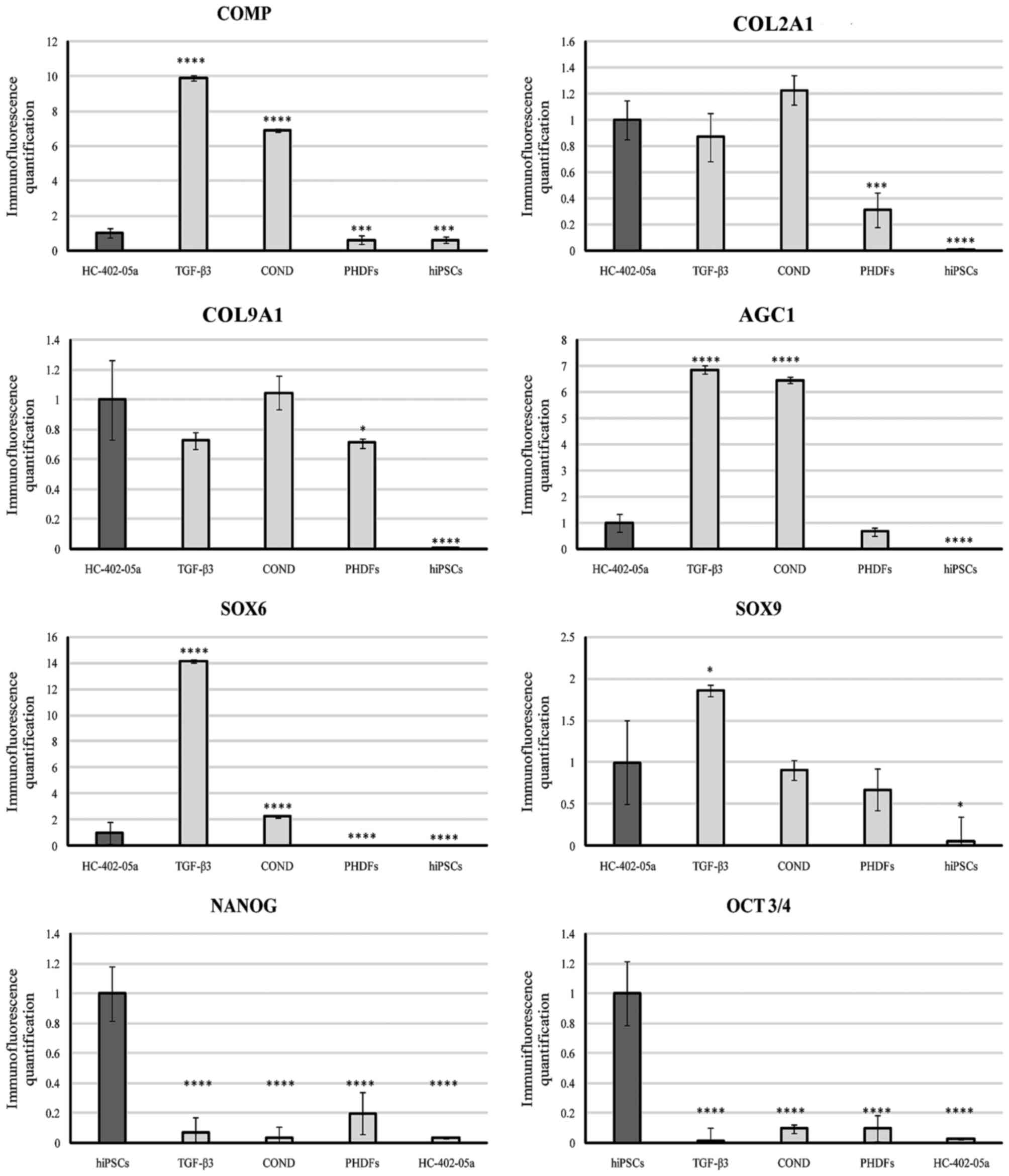 | Figure 3.Quantified immunofluorescence
analysis of chondrogenic markers and pluripotency markers.
****P<0.0001 vs. HC0402-05a. COMP, cartilage oligomeric matrix
protein; COL2A1, type II collagen; COL9A1, type IX collagen; AGC1,
aggrecan; SOX, sex determining region Y-box; OCT3/4,
octamer-binding transcription factor 3/4; TGF-β3, transforming
growth factor β3; COND, conditioned medium; PHDFs, primary human
dermal fibroblasts; hiPSCs, human induced pluripotent stem
cells. |
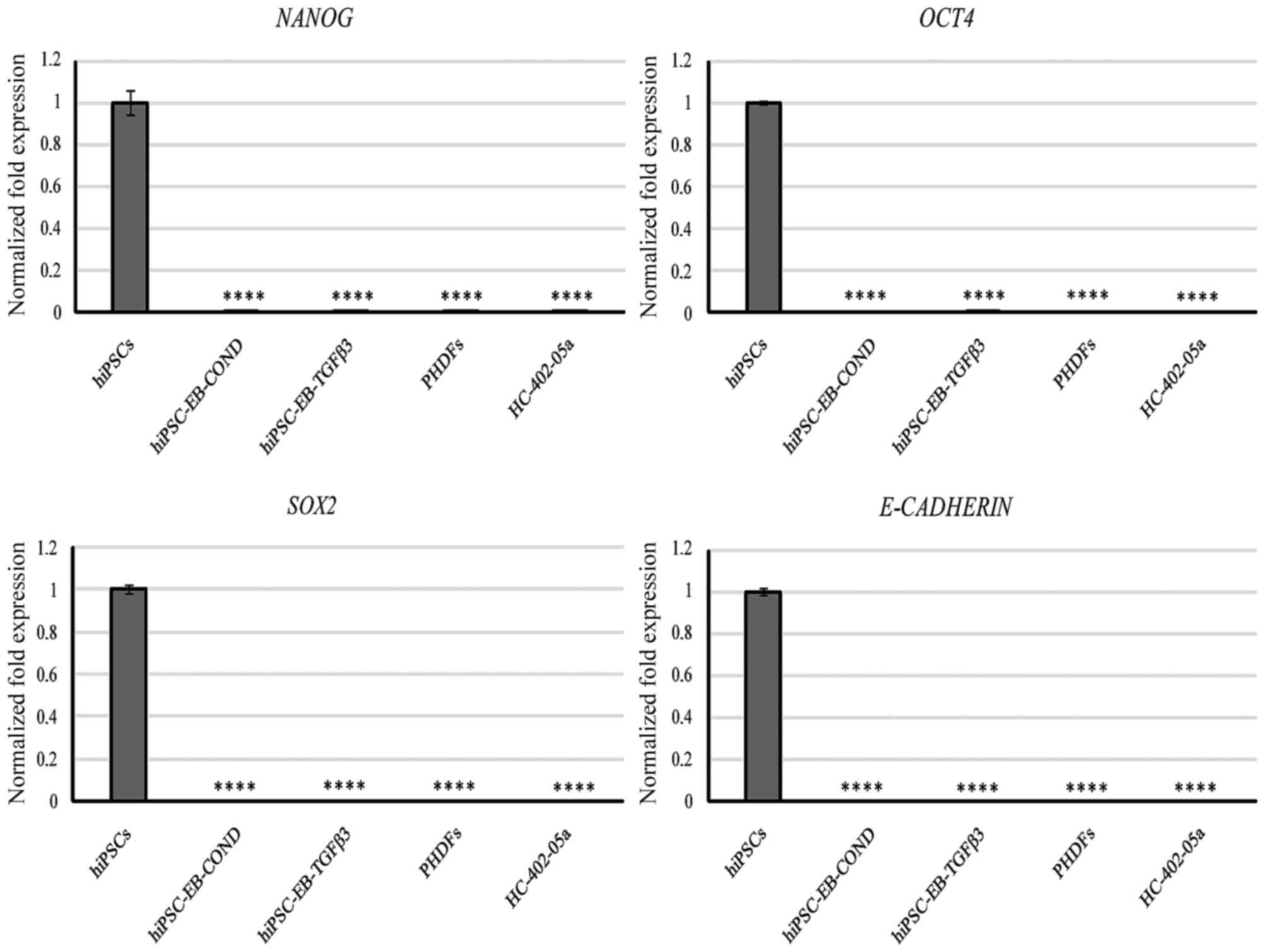
Pluripotency and mesodermal markers
were not observed in the gene expression profiles of stem
cell-derived chondrocytes
All cells were collected and analyzed following the
third passage. Neither the cells differentiated in the TGF-β3
medium nor those differentiated in the conditioned medium expressed
any of the following protein-coding genes assigned to pluripotency
state: Nanog, OCT4 and SOX2 (Fig.
4). Furthermore, E-cadherin, a glycoprotein that is involved in
embryogenesis by mediating cell-cell contact in hESCs, was not
expressed either (Fig. 4). In
contrast, the positive control hiPSCs expressed these markers at a
high level (Fig. 4). This finding
indicates that these cells lost their pluripotent state. These
markers are specific and may be useful to evaluate the loss of
pluripotency state.
None of the investigated cells expressed the
Bra gene (coding brachyury; data not shown), which is
present in cells from the primitive streak or nascent mesoderm.
This may indicate that the differentiated cells did not stop
differentiating in the early stage of chondrogenesis. It is
possible to assume that they had lost their pluripotent nature and
were no longer mesodermal precursors. The forced chondrogenesis
in vitro may have given rise to chondrocyte-like cells
lacking mesodermal features. Furthermore, brachyury is a
particularly specific marker because none of the controls
[HC-402-05a, primary human dermal fibroblasts (PHDFs), and hiPSCs]
expressed the Bra gene.
Likewise, expression of C-X-C motif chemokine
receptor 4 (CXCR4) was not observed, which is active in the
primitive streak, the endoderm and in later stages of
embryogenesis, including intermediated and lateral plate mesoderm.
CXCR4 expression was present in HC-402-05a and hiPSCs
(Fig. 5). This finding confirmed
that the obtained cells did not present features characteristic of
the mesoderm.
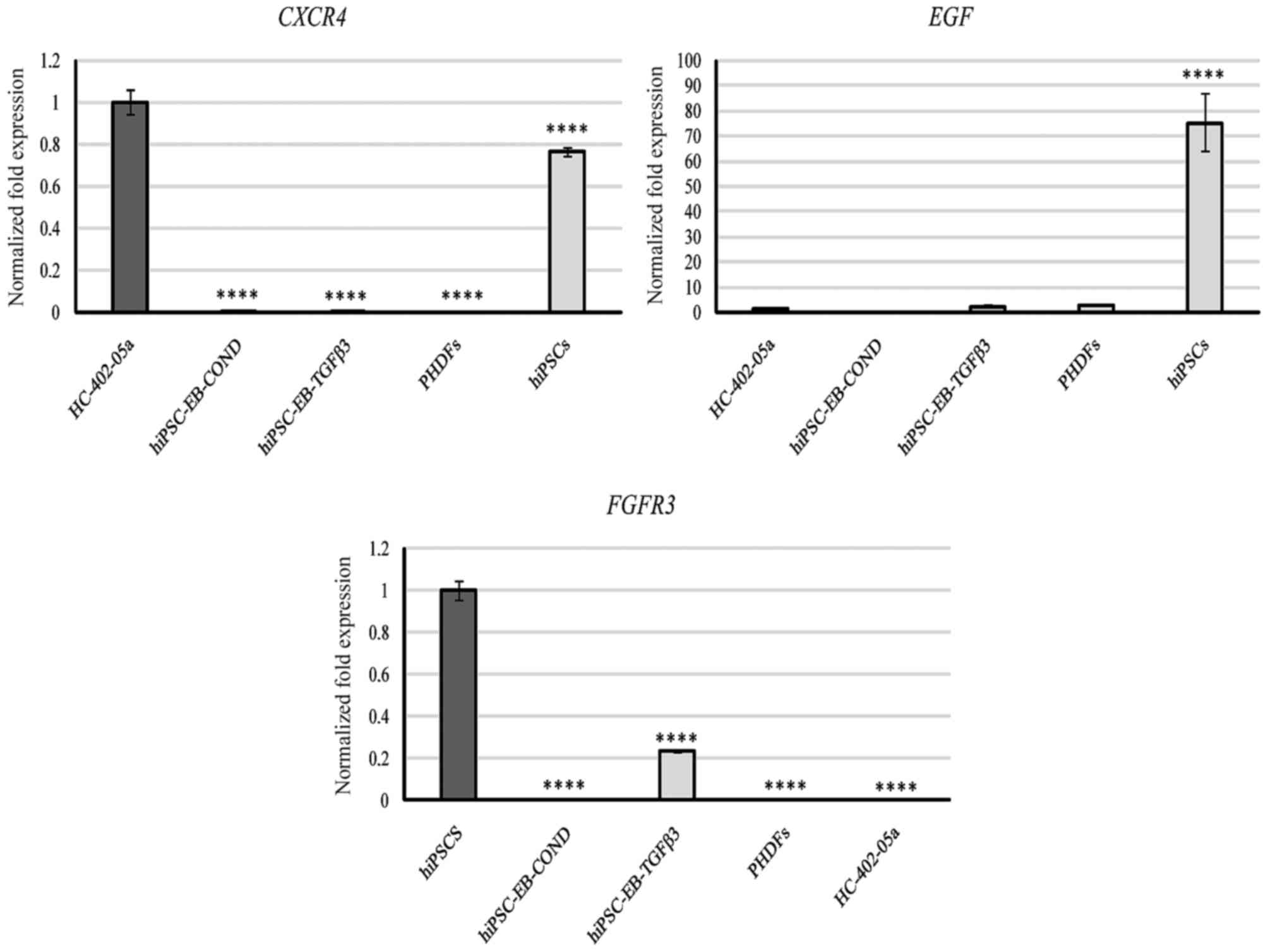 | Figure 5.Based on quantitative polymerase
chain reaction analysis, the hiPSC-derived chondrocytes
differentiated in chondrogenic medium with TGF-β3 (10 ng/ml) or in
medium conditioned with HC-402-05a cells did not remain in the
first stage of chondrogenesis because of the lack of expression of
CXCR4. Furthermore, EGF and FGFR3 mRNA levels
were low. The HC-402-05a cell line served as a positive control.
PHDFs and hiPSCs were used as negative controls. ****P<0.0001
vs. HC-402-05a cells. hiPSCs, human induced pluripotent stem cells;
TGF-β3, transforming growth factor β3; CXCR4, C-X-C motif chemokine
receptor 4; EGF, epidermal growth factor; FGFR3, fibroblast growth
factor receptor 3; PHDFs, primary human dermal fibroblasts; hiPSCs,
human induced pluripotent stem cells; EB, embryoid bodies; COND,
conditioned medium. |
Assessment of markers engaged in
induction of chondrogenesis
The presence of several markers necessary to induce
chondrogenesis was also assessed: PAX9, neural cell adhesion
molecule (NCAM) and NK-related homeodomain protein (NKX3.2). PAX9
was observed only in HC-402-05a cells and in cells differentiated
in the conditioned medium (Fig.
6). NCAM was expressed by all the studied cells, but at varying
levels, with the most prominent expression observed in PHDFs and
cells differentiated in the TGF-β3 medium (Fig. 6). NKX3.2 was also present at a more
stable level in all the study cells, in contrast to the more
variable NCAM. NKX3.2 expression was highest in HC-402-05a cells
(Fig. 6).
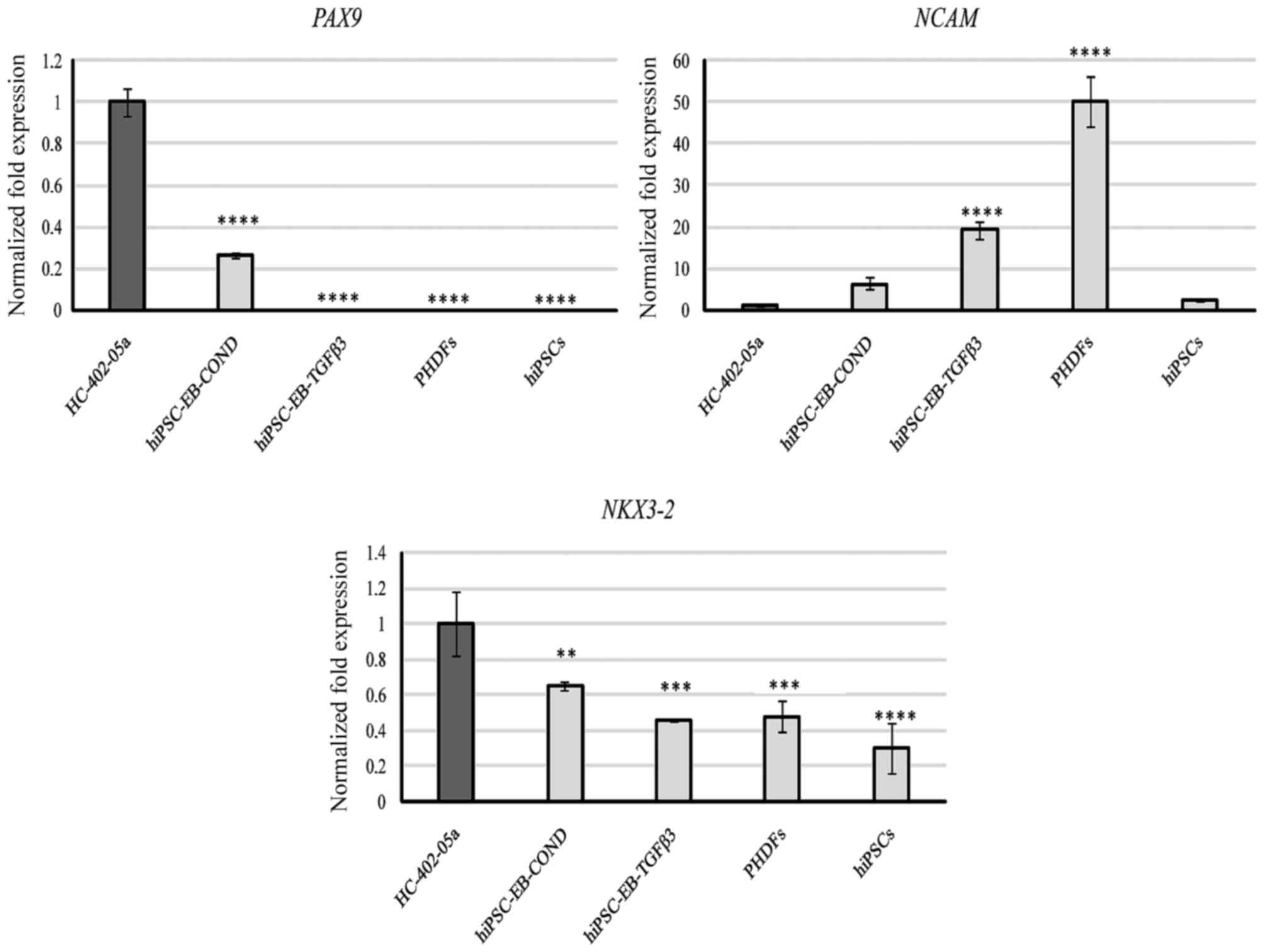 | Figure 6.Based on quantitative polymerase
chain reaction analysis, the hiPSC-derived chondrocytes
differentiated in chondrogenic medium with TGF-β3 (10 ng/ml) or in
medium conditioned with HC-402-05a cells revealed the expression of
genes responsible for induction of chondrogenesis. The HC-402-05a
cell line served as a positive control. PHDFs and hiPSCs were used
as negative controls. **P<0.01, ***P<0.001, ****P<0.0001
vs. HC-402-05a cells. hiPSCs, human induced pluripotent stem cells;
TGF-β3, transforming growth factor β3; PHDFs, primary human dermal
fibroblasts; PAX9, paired box 9; NCAM, neural cell adhesion
molecule; NKX3.2, NK-related homeodomain protein; EB, embryoid
bodies; COND, conditioned medium. |
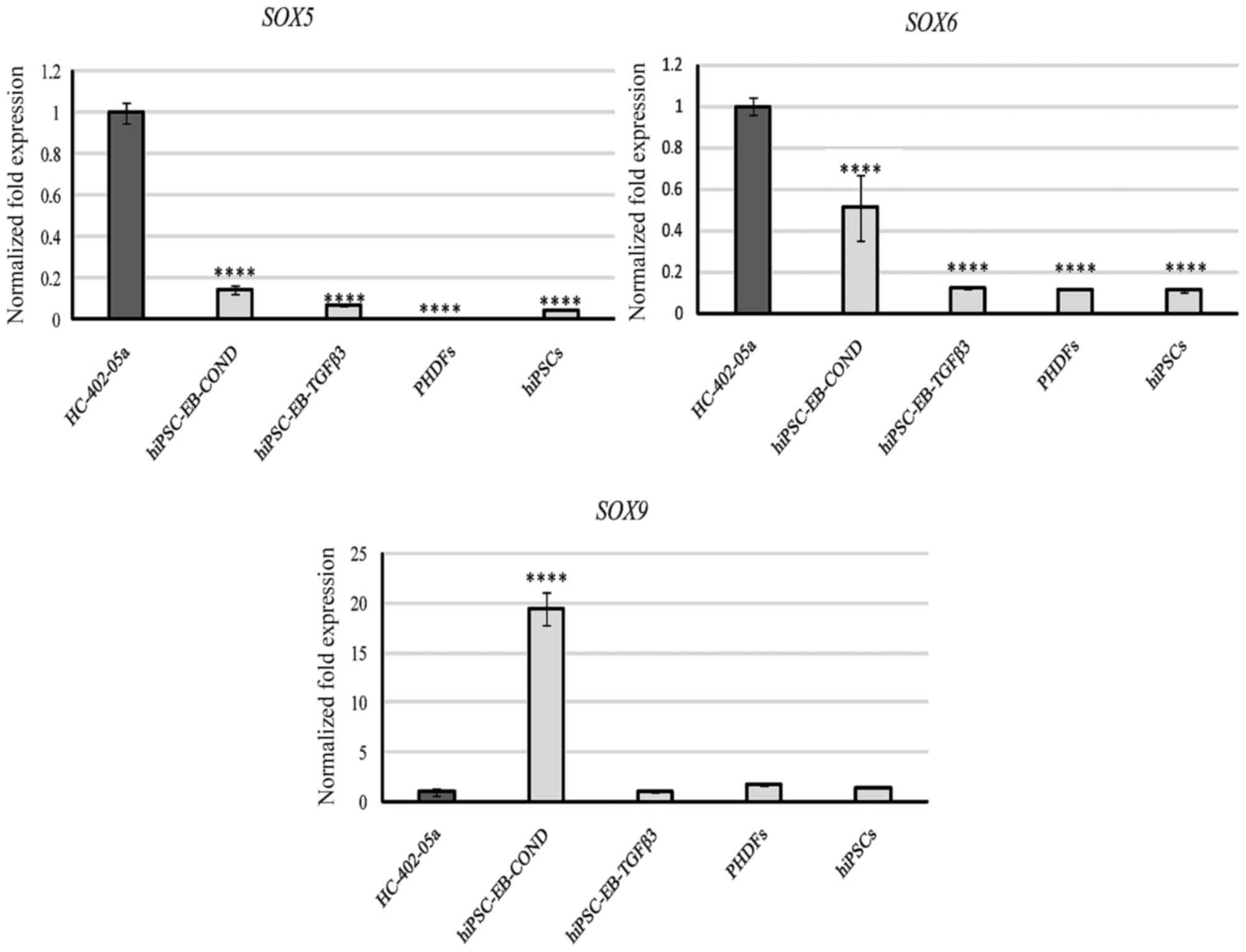
Assessment of SOX gene expression
Next, the expression of a trio of transcription
factors (SOX5, also called L-SOX5 or SOX5L;
SOX6 and SOX9) belonging to the SRY family (encoded
by the sex-determining region on the Y chromosome) was assessed.
SOX5 was expressed at low levels by all cells except for
HC-402-05a (Fig. 7). Similar
results were observed for SOX6, although cells
differentiated in the conditioned medium expressed this marker at
significantly higher levels than cells cultured in TGF-β3 (Fig. 7). The results obtained in the cells
cultured in the conditioned medium were promising because
expression of SOX9, one of the most important markers of
chondrogenesis, was expressed highly in these cells compared with
all other groups (Fig. 7).
Assessment of markers that are
activated throughout the entirety of chondrogenesis
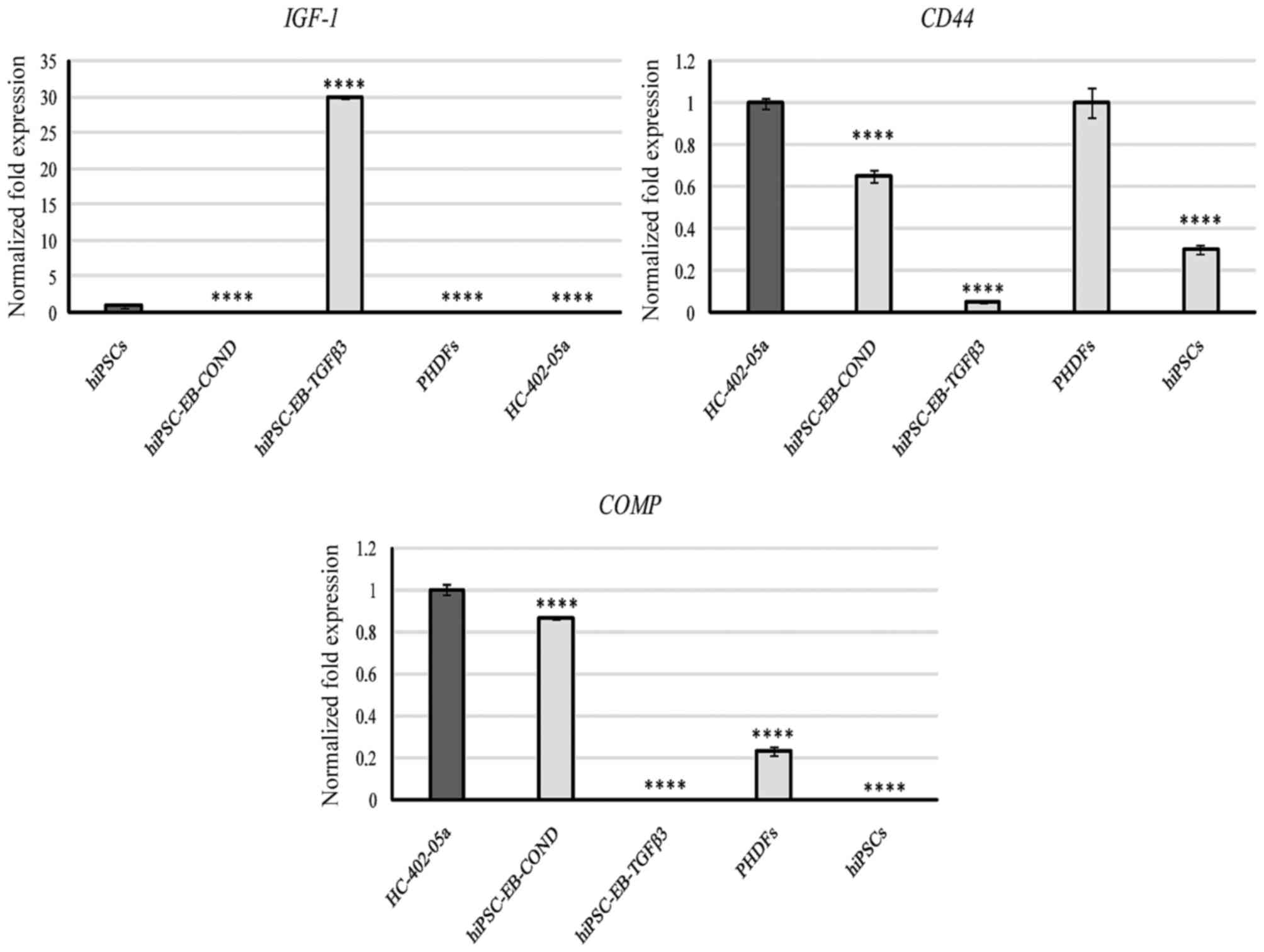
Next, the expression of markers involved in the
entire chondrogenic process were assessed, including IGF-1,
CD44, β-catenin and the components of the cartilage
extracellular matrix (ECM; TNC and COMP). Cells
differentiated in the conditioned medium expressed genes required
for production of CD44, COMP, and β-catenin, while the cells
cultured with TGF-β3 expressed β-catenin and, in particular, IGF-1
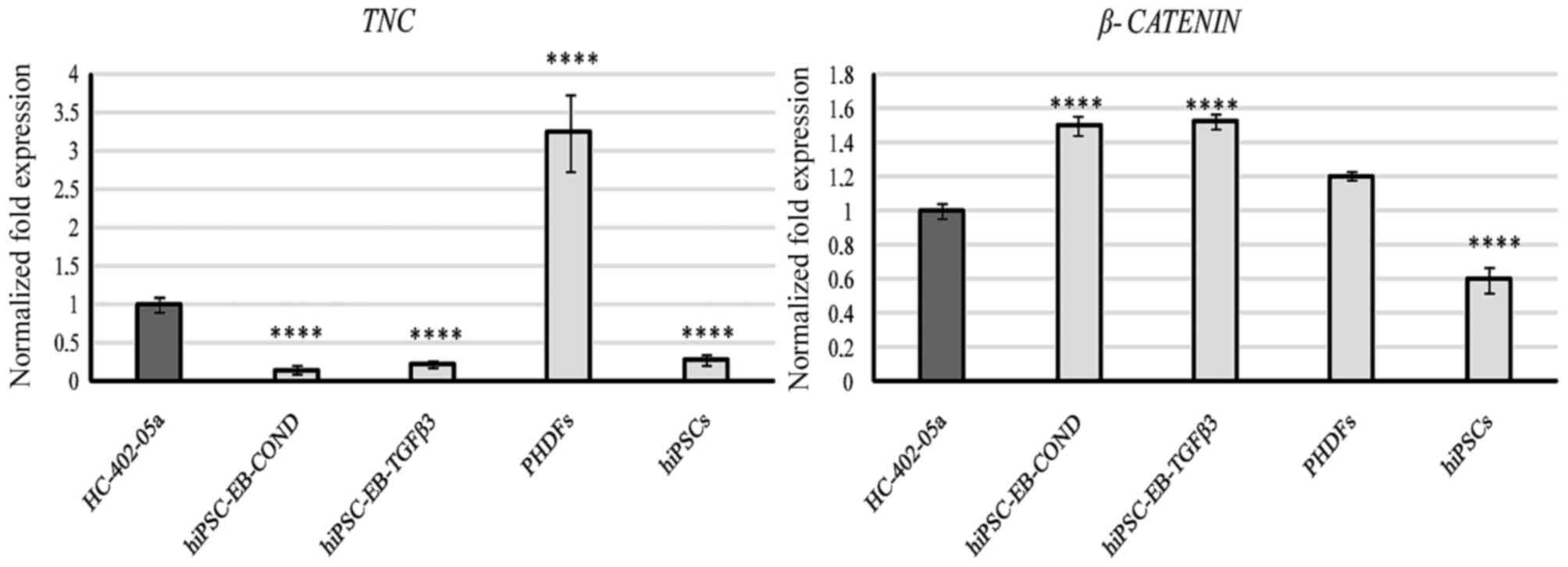
(Figs. 8 and 9). TNC was expressed by all cells,
but expression was significantly higher only in PHDFs (Fig. 9).
Assessment of markers responsible for
cell proliferation rate and the inhibition or enhancement of
chondrogenesis
Finally, two markers used to evaluate the
proliferation rate of cultured cells were examined: fibroblast
growth factor receptor 3 (FGFR3) and epidermal growth factor
(EGF). FGFR3 expression was detected in cells
differentiated in TGF-β3 medium and in hiPSCs (Fig. 5). The hiPSCs demonstrated
significantly higher levels of EGF expression than all other
groups (Fig. 5), suggesting a
strong proliferative potential. EGF was also observed in the
cells differentiated with TGF-β3 and in PHDFs, although EGF
expression was significantly decreased in those cells compared with
hiPSCs (Fig. 5).
Discussion
Current methods of differentiating hiPSCs into
chondrocyte-like cells are not efficient and require further
improvement. The present study evaluated and compared two different
mediums used to differentiate hiPSCs into chondrocyte-like cells,
and revealed that medium conditioned with human cartilage
chondrocytes was a highly effective chondrogenic stimulator.
Furthermore, the chondrogenic properties were demonstrated to
change, even during short-term culture (passage 0 vs. 3).
Immunofluorescence analysis confirmed that chondrocyte-like cells
were obtaine, and. qPCR analysis assessed the relative utility of
the most commonly used chondrogenic markers as indicators of cell
differentiation. The main aim of the present study was to evaluate
the relative value of a wide range of chondrogenic markers to
assess the progress of chondrogenic differentiation. Among the 20
different chondrogenic markers evaluated, it was possible to
identify those that were the most useful as indicators of
differentiation (Table I). This
finding will help to improve and accelerate research involving
hiPSC and chondrogenic differentiation.
The results of the present study confirmed that all
the differentiated cells lost their pluripotent nature (Fig. 4). Furthermore, the results
indicated that these cells did not preserve properties
characteristic of early-phase differentiation involving the
mesodermal stage (Fig. 5). Below,
the markers of early chondrogenesis are discussed.
PAX1 and PAX9 belong to the PAX family
and are involved in the formation of the axial skeleton. They are
characterized by the presence of a highly-conserved DNA binding
domain, the paired box. PAX1 and PAX9 are the main
mediators of Sonic hedgehog, which belongs to the Indian hedgehog
(IHH) family, and are required to induce chondrogenesis. Once
chondrogenesis has been initiated, expression of the PAX
genes is downregulated (21,22).
PAX9 is required to induce the chondrogenic process, and
PAX9 expression was demonstrated to be associated with
IHH expression. The presence of PAX9 mRNA was visible
in cells differentiated in the conditioned medium, and these cells
also exhibited a high level of IHH expression (16). In cells differentiated in the
TGF-β3 medium, expression of IHH was lower (16) and PAX9 expression was lower
compared with cells cultured in the conditioned medium. This
observation indicated that these cells originated from late stages
of chondrogenesis, during which PAX9 expression is
downregulated (Fig. 6). These two
signaling pathways are associated and were previously confirmed to
be dependent on each other (21,22).
PAX genes offer a promising strategy to evaluate the
progression of in vitro chondrogenesis, because PAX9
is not expressed by hiPSCs nor by PHDFs.
Condensation, the first stage of chondrogenesis,
depends on expression of the cell-cell adhesion proteins N-cadherin
and NCAM. Expression of these molecules is rapidly reduced when
cells shift into the differentiation phase, resulting in the
release of cells from strong interactions with each other (23). In healthy cartilage, there are no
cell-cell contacts, however there are functional cell-matrix
contacts that are primarily integrin-mediated (24). In addition, as NCAM is
expressed in osteoprogenitor cells and osteoblasts but not in
chondrocytes, chondroprogenitor cells, or chondroblasts, NCAM is
involved in the induction of secondary chondrogenesis (25). NCAM mediates not only cell-to-cell
binding, but also the interaction between cells and components of
the ECM, including heparin sulfate proteoglycans and collagens. It
is relevant to the regenerative process, where fibroblasts serve an
important function (26).
Francavilla et al (27) demonstrated that NCAM has the
ability to repress several FGF-induced processes, including signal
transduction and cell proliferation. The negative effect of NCAM
depends on its capacity to compete with FGF to bind to the FGF
receptor. However, the data from the present contradict the results
from the study by Francavilla et al (27): Cells differentiated in the presence
of TGF-β3 demonstrated high levels of FGFR3 and NCAM
in the present study. Due to the presence of other hypertrophic and
osteogenic markers, the high level of NCAM observed in cells
cultured in TGF-β3 is likely to be associated with secondary
chondrogenesis rather than the condensation stage. The cells
differentiated in the conditioned medium additionally presented
with a relatively high level of NCAM expression (Fig. 6). Nevertheless, due to the fact
that other hypertrophic and osteogenic markers were observed at
very low levels, it is possible to assume that these cells scarcely
shifted to advanced chondrogenesis or the hypertrophic stage. NCAM
is characteristic of fibroblasts, thus it is not surprising that
these cells presented with high levels of these markers, thereby
reducing the usefulness of NCAM as a marker of chondrogenesis.
The pro-chondrogenic NK-related homeodomain protein
NKX3.2 is required to activate the master chondrogenic
transcriptional regulator SOX9. The presence of this protein
results in the expression of chondrocyte phenotypic genes including
AGC1, COL2A1, and components of cartilaginous ECM. A
feedback loop exists between NKX3.2 and runt-related transcription
factor 2 (RUNX2), leading to repression of RUNX2. In
osteoprogenitor cells, the ability of NKX3.2 to repress RUNX2 is
abrogated, suggesting the existence of a switching mechanism from
chondrogenesis to osteoblast formation (28,29).
The initial induction of NKX3.2 in chondrocyte precursor cells
during early-stage chondrocyte formation and its downregulation in
the terminal-stage of chondrogenesis is controlled by the IHH
pathway, a key regulator of chondrocyte hypertrophy (30).
NKX3.2 was expressed by all the investigated
cells. The presence of NKX3.2 would appear to suggest that
differentiated cells underwent chondrogenesis. Unfortunately,
because NKX3.2 was also observed at low levels in hiPSCs and
PHDF, its use in the evaluation of chondrogenesis may be limited
(Fig. 6).
The SOX trio of transcription factors (SOX5, SOX6,
and SOX9) belong to the SRY family (encoded by the sex-determining
region on the Y chromosome). They are expressed in proliferating
and prehypertrophic chondrocytes. However, in hypertrophic
chondrocytes, expression of SOX genes is turned off. In
contrast to SOX5 and SOX6, SOX9 is required for
chondrogenesis. Nevertheless, the lack of SOX5 and
SOX6 results in defective skeletogenesis. SOX9 is
also upregulated via FGFR3 signaling, and there is a positive
regulatory loop between these (31). SOX9 is required to activate
other cartilage genes including COL2A1 and AGC1.
SOX5 and SOX6 increase the binding efficiency of SOX9
to other cartilage-specific enhancers. In the absence of
SOX5 and SOX6, the expression of COL2A1,
AGC1 and other chondrocyte markers is either very low or
undetectable. Without the presence of SOX5 and SOX6, SOX9 has a
limited capacity to bind to the other cartilage enhancers (32). Yamamizu et al (33) examined the involvement of SOX9 in
the repression of SOX2, and reported that SOX9 has a significant
influence on the cyclin dependent kinase inhibitor 1A (CDKN1A)-SOX2
pathway. SOX9 activity induces the formation of p21, which
subsequently binds to the SRR2 enhancer of SOX2, inhibiting its
expression and facilitating differentiation. Furthermore, SOX9 has
the ability to compete with T-cell factor/lymphoid enhancer factor
(Tcf-Lef) to bind to β-catenin, resulting in its degradation. This
suggests that the chondrogenic process is regulated by the
interaction between SOX9 and the WNT/β-catenin signaling pathway.
SOX9 also inhibits the activity of the cyclin D1 promoter, which
has a high affinity for the β-catenin/Tcf-Lef complex. The
WNT/β-catenin signaling pathway inhibits the differentiation of
chondrocyte precursors and initiates the progression of mature
chondrocytes towards hypertrophy (34). SOX9 prevents osteogenic bone
morphogenetic protein-2 (BMP-2) and RUNX2-induced osteogenic
differentiation and endochondral ossification, respectively. BMP-2
has a high capacity to induce chondrogenic differentiation but also
undesirable hypertrophic differentiation. The forced overexpression
of SOX9 in BMP-2-mediated chondrogenic differentiation seems to be
a promising strategy for cartilage tissue engineering (35).
Based on the results of the present study, it
appears that cells cultured in the conditioned medium may originate
from early or advanced chondrogenesis, in which the expression of
SOX9 is most prominent (Fig.
7). In addition, these cells did not express hypertrophic
markers, which include RUNX2 (16). In contrast, the cells
differentiated in the TGF-β3 medium expressed low levels of
SOX9 and high levels of RUNX2, an observation that
suggests these cells underwent hypertrophy. The low levels of
SOX5 and SOX6 mRNA explain the lack of expression of
the COL2A1 and AGC1 genes in all differentiated
cells. This result is supported by the fact that the mRNA level of
COL2A1 and AGC1 in cultured chondrocytes abruptly
decreased following the first passage while, by contrast, the
expression of genes that code for type I and X collagen increased
or remained unchanged, respectively (36). This may also explain the lack of
expression of COL2A1 and AGC1 genes in the cell
samples obtained following the third passage. Growth factors
including TGF-β and bone morphogenetic protein-7, in addition to
growth and differentiation factor 5, promote AGC1 synthesis while
simultaneously preventing its degradation (37). Indeed, it is this collagen/AGC1
network that gives cartilage its viscoelastic nature with stiff
elastic polymer properties, making it resistant to sudden impact
loading with slow inelastic deformation under sustained load
(38).
The results of the present study indicate that the
expression of the SOX trio is likely to be a good prognostic
marker for cells undergoing chondrogenic differentiation. By
contrast, interpretation of COL2A1 and AGC1 as
markers of chondrogenic differentiation is more complex because, as
other authors have previously demonstrated (36), the expression of these genes
rapidly decreases as the number of passages and the culture period
increases, and these are highly dependent on the expression of
other markers, including the SOX trio genes. Based on the
immunofluoresence and qPCR analyses, earlier literature data
indicating that the production of COL2A1 and AGC1 decreases with
passage number and duration of culture was confirmed (passage 0 vs.
3).
IGF-1 is active during the entire chondrogenic
process. It promotes the synthesis of COL2A1 and proteoglycans and
stabilizes the chondrocyte phenotype in pathological conditions.
IGF-1 and BMP-2 are predominantly present in the proliferative and
hypertrophic layers, however additionally, rarely, in the calcified
chondrocyte zone, in contrast with TGF-β1 (39). Activation of IGF-1 correlates with
the presence of the type 1 IGF receptor, which becomes elevated in
human osteoarthritic chondrocytes as a function of disease
severity. IGF binding proteins regulate the density of IGF-1
receptors on the cell surface and the levels of activated IGF-1
(40). The IGF-1 signaling pathway
is involved in the regulation of growth plate development and cell
size during chondrogenesis. During phosphatidylinositol 3-kinase
(P13K)-protein kinase B (Akt) signaling, IGF-2 and RUNX2 are
dependent on each other to coordinate osteoblast and chondrocyte
differentiation and migration. IGF-1 is suggested to be a major
ligand for the activation of P13K-Akt signaling and RUNX2 (41,42).
Although IGF-1 is associated with chondrogenesis, a previous study
suggested that it may be involved in the maintenance of SC
pluripotency (42). In mouse
spermatogonial SCs, IGF-1 is secreted from Leydig cells as a key
factor in sustaining a pluripotent state. Blockage of endocrine
factor IGF-1 receptor phosphorylation and its downstream P13K/Akt
signaling pathway reduces the activity and expression of
pluripotency genes including Nanog, OCT4 and PR domain zinc
finger protein 1 (43). However,
data concerning human pluripotent SCs are lacking.
IGF-1 expression in cells differentiated in
the presence of TGF-β3 was high compared with IGF-1
expression in human articular chondrocytes (Fig. 8). This phenomenon is associated
with the transition from chondrogenic-like cells into hypertrophic
chondrogenic-like cells, rather than cells from early
chondrogenesis. This observation is confirmed by the high
expression of RUNX2 (16),
which is associated with IGF-1 via the P13K/Akt signaling
pathway.
Hyaluronan (HA) is a linear
polymer-glycosaminoglycan that is distributed throughout the
extracellular space of connective tissues, including articular
cartilage. HA forms the backbone of proteoglycan aggregates, which
primarily consist of AGC1 interacting with COL2A1. HA, together
with proteoglycan aggregates, ensures the load-bearing capacity of
the tissue (44). Takahashi et
al (45) demonstrated that
fragmentation of HA receptor CD44 is a common phenomenon in
dedifferentiated and osteoarthritic chondrocytes, caused by the
secretion and activity of matrix metalloproteinases (MMP), leading
to cleavage of CD44. The disruption of CD44 may cause matrix
turnover and enhanced catabolism, which are hallmarks of early
osteoarthritis. CD44 is highly expressed in human parental
fibroblasts and is gradually lost during the reprogramming process.
It influences the adhesion and motility of fibroblasts throughout
TGF-β activation, and is critical in lesions because, as
fibroblasts migrate to the site of injury, CD44 controls
inflammation and initiates the repair process (46,47).
The cells differentiated in the conditioned medium
presented CD44 levels similar to those observed in human
articular chondrocytes while, by contrast, cells obtained from the
TGF-β3 medium did not express CD44 (Fig. 8). This lack of CD44
expression may be due to the activity of MMPs, which were highly
expressed during the present study and may have presented
undesirable features of dedifferentiated and/or osteoarthritic
chondrocytes (16). It is
necessary to be cautious when considering CD44 as a marker for the
chondrogenic process in vitro due to the fact that, as the
published data indicate (44,45),
CD44 is present in fibroblasts and at the vestigial level in human
pluripotent SCs.
COMP is an important component of the cartilage ECM.
It has the ability to interact with COL2A1 and AGC1 as well as
other ECM components. COMP has a large impact on cartilage
phenotype development, and on the matrix organization and load
support function of cartilage. A COMP deficiency in the joints is
correlated with arthritis. This protein holds promise as a
diagnostic and prognostic factor as a marker of disease progression
and the effect of treatment (48,49).
In the present study, the conditioned medium
stimulated expression of COMP. Furthermore, the level of
expression was similar to that observed in human articular
chondrocytes, suggesting that cells cultured in the conditioned
medium have chondrogenic properties. In contrast, COMP was
not observed in cells cultured in the presence of TGF-β. PHDFs and
hiPSCs expressed this marker at low levels, thus it is possible to
use this marker to assess the gene profile expression of
differentiating cells (Fig.
8).
TNC is an oligomeric glycoprotein of ECM expressed
during various processes, including neural development, tissue
remodeling, wound healing, angiogenesis and tumorigenesis. This
marker was suggested to be tissue-specific due to its high
concentration in articular cartilage. However, compared with human
articular chondrocytes, malignant cells produce TNC in higher
quantities (50). TNC has
proliferative and anti-adhesive properties and is considered,
therefore, to have metastatic potential (50). Data indicate that TNC is highly
active during early chondrogenic differentiation, for example
during mesenchymal condensation, and is turned off in cartilage
with progressive chondrocyte maturation. Although the
fibrinogen-like domain of TNC is indispensable, it is not
sufficient by itself for the induction of chondrogenesis (51). TNC is involved in fibroblast
migration and infiltration into the provisional matrix in response
to injury. This suggests that TNC expression and degradation
is tightly controlled to ensure efficient tissue rebuilding
(52).
TNC was expressed by all the cells evaluated
in the present study. The highest levels of expression were
detected in PHDFs vs. the positive controls (human articular
chondrocytes) (Fig. 9). Although
TNC is expressed in the articular cartilage, it is also
involved in multiple cellular processes, thus reducing its value as
a marker of chondrogenesis.
Control of the WNT/β-catenin signaling pathway helps
to make the reprogramming process more efficient. Augmented
reprogramming is observed as a result of interaction between
WNT/β-catenin and reprogramming factors (OCT4, SOX2
and Kruppel-like factor 4) and other endogenous core pluripotency
genes, although it does not affect v-myc myelocytomatosis viral
oncogene homolog expression. This signaling pathway is critical for
the reprogramming process and its influence is most apparent during
the initial stage where interaction with the T-cell factor is
important. Nevertheless, WNT/β-catenin is not required to maintain
cell pluripotency (53). Qiu et
al (54) demonstrated that the
self-renewal-promoting WNT/β-catenin effect is predominantly
triggered by two of its downstream targets, KLF2 and
transcription factor CP2-like 1 (TCFP2L1). The
downregulation of these two genes impairs mouse embryonic stem cell
self-renewal mediated by WNT/β-catenin, and conversely the
overexpression of KLF2 and TCFP2L1 recapitulates the
self-renewal-promoting effect (54). Furthermore, Nanog and β-catenin
(coded by CTNNB1) cooperate in establishing pluripotency
during the reprogramming process. Nanog inhibits Dickkopf-related
protein 1, which leads to β-catenin activation and accumulation,
which, in turn, is essential for Nanog-dependent conversion of
pre-miPSCs into miPSCs. Thus, the crosstalk between Nanog and the
WNT/β-catenin signaling pathway is relevant for ESC physiology, as
it results in a synergistic effect (55). In human PSCs, the contribution of
the WNT/β-catenin signaling pathway in promoting self-renewal of
hESCs is unclear. Certain data suggest that this signaling pathway
is involved in hESC proliferation and self-renewal, but following
multiple passages of hESCs the effect disappears. Davidson et
al (56) demonstrated that the
WNT/β-catenin signaling pathway is assigned to differentiation
towards mesodermal lineages rather than self-renewal. They also
demonstrated that OCT3/4, a key pluripotency factor, represses
endogenous WNT/β-catenin signaling in hESCs.
The WNT/β-catenin signaling pathway is also
involved in chondrogenesis. N-cadherin, required for temporal
mitogen-activated protein kinase (MAPK) 1/2, p38 MAPK and
BMP-2-mediated regulation of chondrogenic genes including
SOX9, AGC1 and COL2A1, modulates the potential
WNT-induced nuclear activity of β-catenin. N-cadherin-mediated
redistribution of β-catenin appears to be a mechanism by which
WNT-mediated chondrogenesis is kept under control (57). The WNT/β-catenin pathway is
involved in the fracture repair process and bone healing through
early cartilage callus formation, endochondral ossification,
induction of vascularization, late stage remodeling and recovery of
mechanical strength. Inhibition of this pathway results in
decreased expression of the following chondrogenic and osteogenic
genes: Type I, II and X collagen, MMP-13, alkaline
phosphatase, osteocalcin, SOX9, RUNX2 and vascular
endothelial growth factor (58).
WNT/β-catenin signaling is activated by TGF-β-mediated SMAD family
member 3 (SMAD3), which increases β-catenin signaling and its
nuclear translocation. The cooperation between TGF-β members and
β-catenin results in increased expression of cyclin D1
in the chondrocytes (59).
The results of the present study confirm that the
β-catenin signaling pathway may be involved in the self-renewal of
pluripotent SCs and in the chondrogenic process. Expression of
β-catenin was evident in cells differentiated in the presence of
TGF-β3, those differentiated in the HC-402-05a-conditioned medium
and also in all control cells: Human articular chondrocytes, human
primary fibroblasts and hiPSCs (Fig.
9). This level of expression in differentiated cells may be
associated with N-cadherin-mediated redistribution of β-catenin
during chondrogenesis. In turn, the high level of β-catenin
mRNA in hiPSCs may be associated with interactions between
WNT/β-catenin and pluripotency factors including Nanog. The
presence of the activated β-catenin signaling pathway may explain
the high level of SMAD3 expression (16), as a result of SMAD3-mediated
activation of β-catenin. Because the WNT/β-catenin signaling
pathway is engaged in multiple processes, from self-renewal to
differentiation, it is difficult to use as a marker for iPSC
differentiation.
In the present study, two protocols to obtain
chondrocyte-like cells from hiPSCs via embryoid bodies have been
described, either with the addition of TGF-β3 to the chondrogenic
medium, or using a chondrogenic medium conditioned with HC-402-05a
cells. The chondrocyte-like cells obtained in the present study
expressed genes that are present during early chondrogenesis.
Furthermore, the value of several of these genes as markers of
chondrogenic progression was demonstrated: PAX9,
SOX5, SOX6, SOX9 and COMP were all good
markers of hiPSC differentiation. In contrast, other markers
including IGF-1, TNC and β-catenin were less
valuable. Notably, because certain markers are also expressed by
PHDFs, these must be used with caution, taking into account the
dedifferentiation process or transcriptional memory of their
parental somatic cells. Thus, the origin of hiPSCs has an impact on
their further differentiation toward chondrocyte-like cells
deriving from the same germ layer as the parental cells of hiPSCs;
reprogrammed fibroblasts.
The present study contributes to an improved
understanding of the chondrogenic process. In addition, the
obtained hiPSC-derived chondrocytes were demonstrated to be quite
unstable and the chondrogenic features varied among the number of
passages and duration of culture. Therefore, current protocols
based on hiPSC differentiation require further improvements,
particularly with regard to future scaled-up culture of
differentiated hiPSCs and their subsequent application in clinical
practice. The present study provides a method to more efficiently
assess forced differentiation towards chondrocytes. Nevertheless,
given the preliminary nature of the present study, more research is
required to reach definitive conclusions.
Acknowledgements
The authors would like to thank Mr. Bradley Londres
for his invaluable assistance in editing the manuscript. The
present study was supported by the National Science Centre (grant
no. 2012/07/E/NZ3/01819).
Glossary
Abbreviations
Abbreviations:
|
AGC1
|
aggrecan
|
|
Akt
|
protein kinase B
|
|
BMP-2
|
bone morphogenetic protein-2
|
|
COL2A1
|
type II collagen
|
|
COMP
|
cartilage oligomeric matrix
protein
|
|
CTNNB1
|
β-catenin; EBs-embryoid bodies
|
|
ECM
|
extracellular matrix
|
|
EGF
|
epidermal growth factor
|
|
FGF-2
|
fibroblast growth factor 2
|
|
FGFR3
|
fibroblast growth factor receptor
3
|
|
HC-402-05a
|
human primary chondrocyte cell
line
|
|
hESCs
|
human embryonic stem cells
|
|
hiPSCs
|
human induced pluripotent stem
cells
|
|
IGF-1
|
insulin-like growth factor 1
|
|
IHH
|
indian hedgehog
|
|
MMP
|
matrix metalloproteinase
|
|
NCAM
|
neural cell adhesion molecule
|
|
NKX3.2
|
NK-related homeodomain protein
|
|
PAX9
|
paired box 9
|
|
P13K
|
phosphatidylinositol 3-kinase
|
|
PHDFs
|
primary human dermal fibroblasts
|
|
RUNX2
|
runt-related transcription factor
2
|
|
SCs
|
stem cells
|
|
SOX2
|
5, 6, 9, sex determining region Y-box
2, 6, 9
|
|
Tcf-Lef
|
T-cell factor/lymphoid enhancer
factor
|
|
TCFP2L1
|
transcription factor CP2-like 1
|
|
TGF-β3
|
transforming growth factor β3
|
References
|
1
|
Liu T, Li Q, Wang S, Chen C and Zheng J:
Transplantation of ovarian granulosa-like cells derived from human
induced pluripotent stem cells for the treatment of murine
premature ovarian failure. Mol Med Rep. 13:5053–5058.
2016.PubMed/NCBI
|
|
2
|
Cao B, Li Z, Peng R and Ding J: Effects of
cell-cell contact and oxygen tension on chondrogenic
differentiation of stem cells. Biomaterials. 64:21–32. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lach M, Trzeciak T, Richter M, Pawlicz J
and Suchorska WM: Directeddifferentiation of induced pluripotent
stem cells into chondrogenic lineages for articular cartilage
treatment. J Tissue Eng. 5:20417314145527012014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ren Y, Deng CL, Wan WD, Zheng JH, Mao GY
and Yang SL: Suppresive effects of induced pluripotent stem
cell-conditioned medium in in vitro hypertrophic scarring
fibroblast activation. Mol Med Rep. 11:2471–2476. 2015.PubMed/NCBI
|
|
5
|
Chen FH and Tuan RS: Mesenchymal stem
cells in arthritic diseases. Arthritic Res Ther. 10:2232008.
View Article : Google Scholar
|
|
6
|
Kulcenty K, Wróblewska J, Mazurek S,
Liszewska E and Jaworski J: Molecular mechanisms of induced
pluripotency. Contemp Oncol (Pozn). 19:A22–A29. 2015.PubMed/NCBI
|
|
7
|
Suchorska WM, Augustyniak E and Łukjanow
M: Genetic stability of pluripotent stem cells during anti-cancer
therapies. Exp Ther Med. 11:695–702. 2016.PubMed/NCBI
|
|
8
|
Lee J, Taylor SE, Smeriglio P, Lai J,
Maloney WJ, Yang F and Bhutani N: Early induction of a
prechondrogenic population allows efficient generation of stable
chondrocytes from human induced pluripotent stem cells. FASEB J.
29:3399–3410. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye J, Hong J and Ye F: Reprogramming rat
embryonic fibroblasts into induced pluripotent stem cells using
transposon vectors and their chondrogenic differentiation in
vitro. Mol Med Rep. 11:989–994. 2015.PubMed/NCBI
|
|
10
|
Fu C, Yan Z, Xu H, Zhang C, Zhang Q, Wei
A, Yang X and Wang Y: Isolation, identification and differentiation
of human embryonic cartilage stem cells. Cell Biot Int. 39:777–787.
2015. View Article : Google Scholar
|
|
11
|
Oldershaw RA, Baxter MA, Lowe ET, Bates N,
Grady LM, Soncin F, Brison DR, Hardingham TE and Kimber SJ:
Directed differentiation of human embryonic stem cells toward
chondrocytes. Nat Biotechnol. 28:1187–1194. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toh WS and Cao T: Derivation of
chondrogenic cells from human embryonic stem cells for cartilage
tissue engineering. Methods Mol Biol. Jul 12–2014.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mardani M, Hashemibeni B, Ansar MM,
ZarkeshEsfahani SH, Kazemi M, Goharian V, Esmaeili N and Esfandiary
E: Comparison between chondrogenic markers of differentiated
chondrocytes from adipose deived stem cells and articular
chondrocytes in vitro. Iran J Basic Med Sci. 16:763–773.
2013.PubMed/NCBI
|
|
14
|
Lee HJ, Choi BH, Min BH and Park SR:
Changes in Surface markers of human mesenchymal stem cells during
the chondrogenic differentiation and dedifferentiation processes in
vitro. Arthritis Rheum. 60:2325–2332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Augustyniak E, Trzeciak T, Richter M,
Kaczmarczyk J and Suchorska W: The role of growth factors in stem
cell-directed chondrogenesis: A real hope for damaged cartilage
regeneration. Int Orthop. 39:995–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Augustyniak E, Suchorska WM, Trzeciak T
and Richter M: Gene expression profile in human induced pluripotent
stem cells: Chondrogenic differentiation in vitro, part B.
Mol Med Rep. 15:2402–2414. 2017.
|
|
17
|
Wróblewska J: A new method to generate
human induced pluripotent stem cells (iPS) and the role of the
protein KAP1 in epigenetic regulation of self-renewal. PhD
dissertation. Poznan University of Medical Sciences. http://www.wbc.poznan.pl/Content/373798/index.pdf2015.
|
|
18
|
Suchorska WM, Lach MS, Richter M,
Kaczmarczyk J and Trzeciak T: Bioimaging: An useful tool to monitor
differentiation of human embryonic stem cells into chondrocytes.
Ann Biomed Eng. 44:1845–1859. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nejadnik H, Diecke S, Lenkov OD, Chapelin
F, Doing J, Tong X, Derugin N, Chan RC, Gaur A, Yang F, et al:
Improved approach for chondrogenic differentiation of human induced
pluripotent stem cells. Stem Cell Rev. 11:242–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta (CT)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rodrigo I, Hill RE, Balling R, Münsterberg
A and Imai K: Pax1 and Pax9 activate Bapx1 to induce chondrogenic
differentiation in the sclerotome. Development. 130:473–482. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blake JA and Ziman MR: Pax genes:
Regulators of lineage specification and progenitor cell
maintenance. Development. 141:737–751. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Singh P and Schwarzbauer JE: Fibronectin
and stem cel differentiation-lessons from chondrogenesis. J Cell
Sci. 125:3703–3712. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gigout A, Jolicoeur M, Nelea M, Raynal N,
Farndale R and Buschmann MD: Chondrocyte aggregation in suspension
culture is GFOGER-GPP- and beta1 integrin-dependent. J Biol Chem.
283:31522–31530. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang J and Hall BK: Differential
expression of neural cell adhesion molecule (NCAM) during
osteogenesis and secondary chondrogenesis in the embryonic chick.
Int J Dev Biol. 39:519–528. 1995.PubMed/NCBI
|
|
26
|
Nakatani K, Tanaka H, Ikeda K, Sakabe M,
Kadoya H, Seki S, Kaneda K and Nakajima Y: Expression of NCAM in
activated portal fibroblasts during regeneration of the rat liver
after partial hepatectomy. Arch Histol Cytol. 69:61–72. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Francavilla C, Loeffler S, Piccini D, Kren
A, Christofori G and Cavallaro U: Neural cel adhesion molecule
regulates the cellular response to fibroblast growth factor. J Cell
Sci. 120:4388–4394. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rainbow RS, Kwon H and Zeng L: The role of
Nkx3. 2 in chondrogenesis. Front Biol (Beijing). 9:376–381. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kawato Y, Hirao M, Ebina K, Shi K,
Hashimoto J, Honjo Y, Yoshikawa H and Myoui A: Nkx3.2 promotes
primary chondrogenic differentiation by upregulating
Col2a1transcription. PLoS One. 7:e347032012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Choi SW, Jeong DU, Kim JA, Lee B, Joeng
KS, Long F and Kim DW: Indian Hedgehog signaling triggers Nkx3.2
protein degradation during chondrocyte maturation. Biochem J.
443:789–798. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu CF and Lefebvre V: The transcription
factors SOX9 and SOX5/SOX6 cooperate genome-wide through
super-enhancer to drive chondrogenesis. Nucleic Acids Res.
43:8183–8203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han Y and Lefebvre V: L-Sox5 and Sox6
drive expression of the aggrecan gene in cartilage by securing
binding of Sox9 to a far-upstream enhancer. Mol Cell Biol.
28:4999–5013. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamamizu K, Schlessinger D and Ko MS: SOX9
accelerates ESC differentiation to three germ layer lineages by
repressing SOX2 expression through P21 (WAF1/CIP1). Development.
141:4254–4266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akiyama H, Lyons JP, Mori-Akiyama Y, Yang
X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR,
et al: Interactions between Sox9 and beta-catenin control
chondrocyte differentiation. Genes Dev. 18:1072–1087. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao J, Hu N, Zhou N, Lin L, Zhao C, Yi S,
Fan T, Bao W, Liang X, Chen H, et al: Sox9 potentiates BMP-2
induced chondrogenic differentiation and inhibits BMP-induced
osteogenic differentiation. PLoS One. 9:e890252014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hamda T, Sakai T, Hiraiwa H, Nakashima M,
Ono Y, Mitsuyama H and Ishiguro N: Surface markers and gene
expression to characterize the differentiation of monolayer
expanded human articular chondrocytes. Nagoya J Med Sci.
75:101–111. 2013.PubMed/NCBI
|
|
37
|
Sivan SS, Wachtel E and Roughley P:
Structure, function, aging and turnover of aggrecan in the
invertebral disc. Biochim Biophys Acta. 1840:3181–3189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kiani C, Chen L, Wu YJ, Yee AJ and Yang
BB: Structure and function of aggrecan. Cell Res. 12:19–32. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Meng Q, Long X, Deng M, Cai H and Li J:
The expressions of IGF-1, BMP-2 and TGF-β1 in cartilage of condylar
hyperplasia. J Oral Rehabil. 38:34–40. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agriogiannis GD, Sifakis S, Patsouris ES
and Konstantinidou AE: Insulin-like growth factors in embryonic
fetal growth and skeletal development (Review). Mol Med Rep.
10:579–584. 2014.PubMed/NCBI
|
|
41
|
Fujita T, Azuma Y, Fukuyama R, Hattori Y,
Yoshida C, Koida M, Ogita K and Komori T: Runx2 induces osteoblast
and chondrocyte differentiation and enhances their migration by
coupling with P13K-Akt signaling. J Cell Biol. 166:85–95. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guntur AR and Rosen CJ: IGF-1 regulation
of key signaling pathway in bone. Bonekey Rep. 2:4372013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang YH, Chin CC, Ho HN, Chou CK, Shen
CN, Kuo HC, Wu TJ, Wu YC, Hung YC, Chang CC and Ling TY:
Pluripotency of mouse spermatogonial stem cells maintained by
IGF-dependent pathway. FASEB J. 23:2076–2087. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nicoll SB, Barak O, Csóka AB, Bhatnagar RS
and Stern R: Hyaluronidases and CD44 undergo differential
modulating during chondrogenesis. Biochem Biophys Res Commun.
292:819–825. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Takahashi N, Knudson CB, Thankamony S,
Ariyoshi W, Mellor L, Im HJ and Knudson W: Induction of CD44
cleavage in articular chondrocytes. Arthritis Rheum. 62:1338–1348.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Quintanilla RH Jr, Asprer JC, Vaz C,
Tanavde V and Lakshmipathy U: CD44 is a negative cell surface
marker for pluripotent stem cell identification during human
fibroblast reprogramming. PLoS One. 9:e854192014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Acharya PS, Majumdar S, Jacob M, Hayden J,
Mrass P, Weninger W, Assoian RK and Puré E: Fibroblast migration is
mediated by CD44-dependent TGF beta activation. J Cell Sci.
121:1393–1402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Halem-Smith H, Calderon R, Song Y, Tuan RS
and Chen FH: Cartilage oligomerix matrix protein enhances matrix
assembly during chondrogenesis of human mesenchymal stem cells. J
Cell Biochem. 113:1245–1252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tseng S, Reddi AH and Di Cesare PE:
Cartilage oligomeric matrix protein (COMP): A biomeraker of
arthritis. Biomark Insights. 4:33–44. 2009.PubMed/NCBI
|
|
50
|
Ghert MA, Qi WN, Erickson HP, Block JA and
Scully SP: Tenascin-C expression and distribution in cultured human
chondrocytes and chondrosarcoma cells. J Orthop Res. 20:834–841.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Murphy LI, Fischer D, Chiquet-Ehrismann R
and Mackie EJ: Tenascin-C induced stimulation of chondrogenesis is
dependent on the presence of the C-terminal fibrinogen-like
globular domain. FEBS Lett. 480:189–192. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Trebaul A, Chan EK and Midwood KS:
Regulation of fibroblast migration by tenascin-C. BiochemSoc Trans.
35:695–697. 2007. View Article : Google Scholar
|
|
53
|
Tomizawa M, Shinozaki F, Motoyoshi Y,
Sugiyama T, Yamamoto S and Ishige N: Involvement of the Wnt
signaling pathway in feeder-free culture of human induced
pluripotent stem cells. Mol Med Rep. 12:6797–6800. 2015.PubMed/NCBI
|
|
54
|
Qiu D, Ye S, Ruiz B, Zhou X, Liu D, Zhang
Q and Ying QL: Klf2 and Tfcp2l1, Two Wnt/β-catenin targets, act
synergistically to induce and maintain naive pluripotency. Stem
Cell Reports. 5:314–322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Marucci L, Pedone E, Di Vicino U,
Sanuy-Escribano B, Isalan M and Cosma MP: β-catenin fluctuates in
mouse ESCs and is essential for Nanog-mediated reprogramming of
somatic cells to pluripotency. Cell Rep. 8:1686–1696. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Davidson KC, Adams AM, Goodson JM,
McDonald CE, Potter JC, Berndt JD, Biechele TL, Taylor RJ and Moon
RT: Wnt/β-catenin signaling promotes differentiation, not
self-renewal, of human embryonic stem cells and is repressed by
Oct4. Proc Natl Acad Sci USA. 109:4485–4490. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Modarresi R, Lafond T, Roman-Blas JA,
Danielson KG, Tuan RS and Seghatoleslami MR: N-cadherin mediated
distribution of beta-catenin alters MAP kinase and BMP-2 signaling
on chondrogenesis-related gene expression. J Cell Biochem.
95:53–63. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang Y, Zhang X, Du K, Yang F, Shi Y,
Huang J, Tang T, Chen D and Dai K: Inhibition of β-catenin
signaling in chondrocytes induces delayed fracture healing in mice.
J Orthop Res. 30:304–310. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li TF, Chen D, Wu Q, Chen M, Sheu TJ,
Schwarz EM, Drissi H, Zuscik M and O'Keefe RJ: Transforming growth
factor-beta stimulates cyclin D1 expression through activation of
beta-catenin signaling in chondrocytes. J BiolChem.
281:21296–21304. 2006.
|