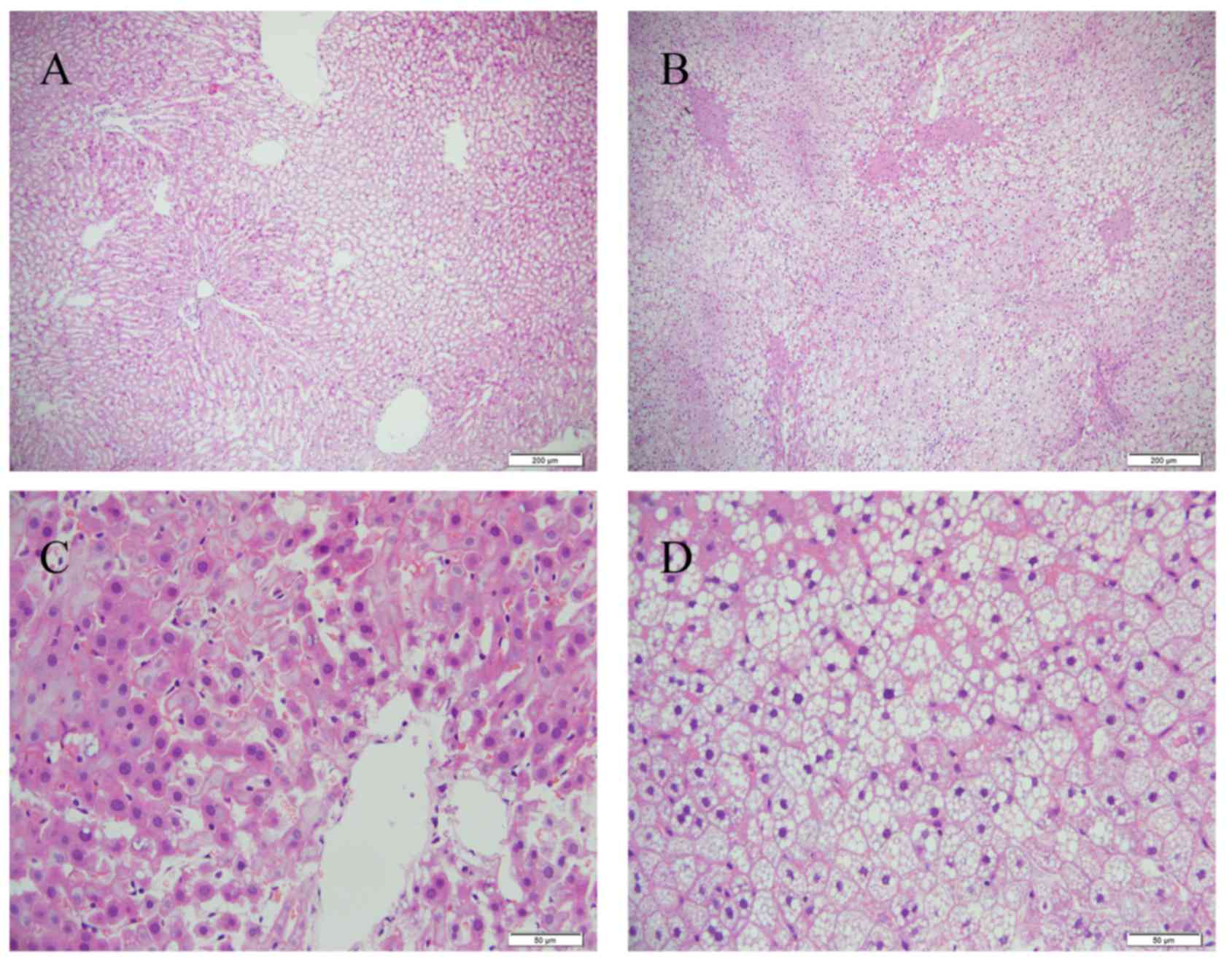
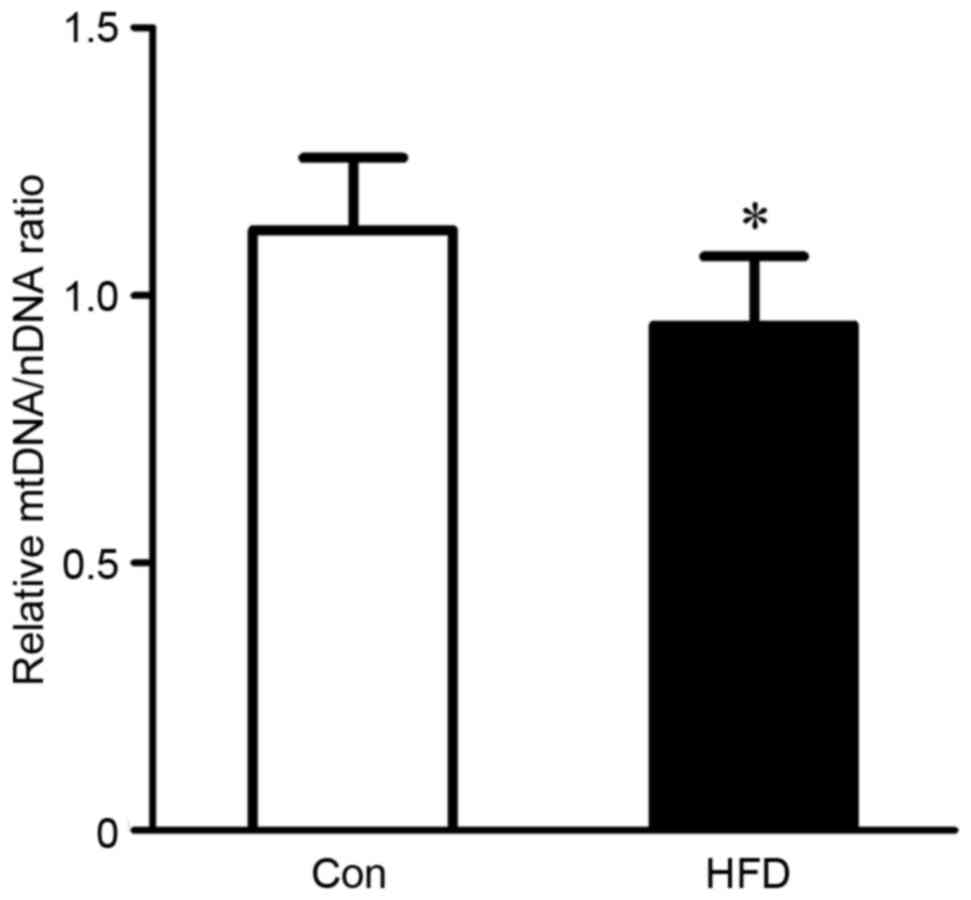
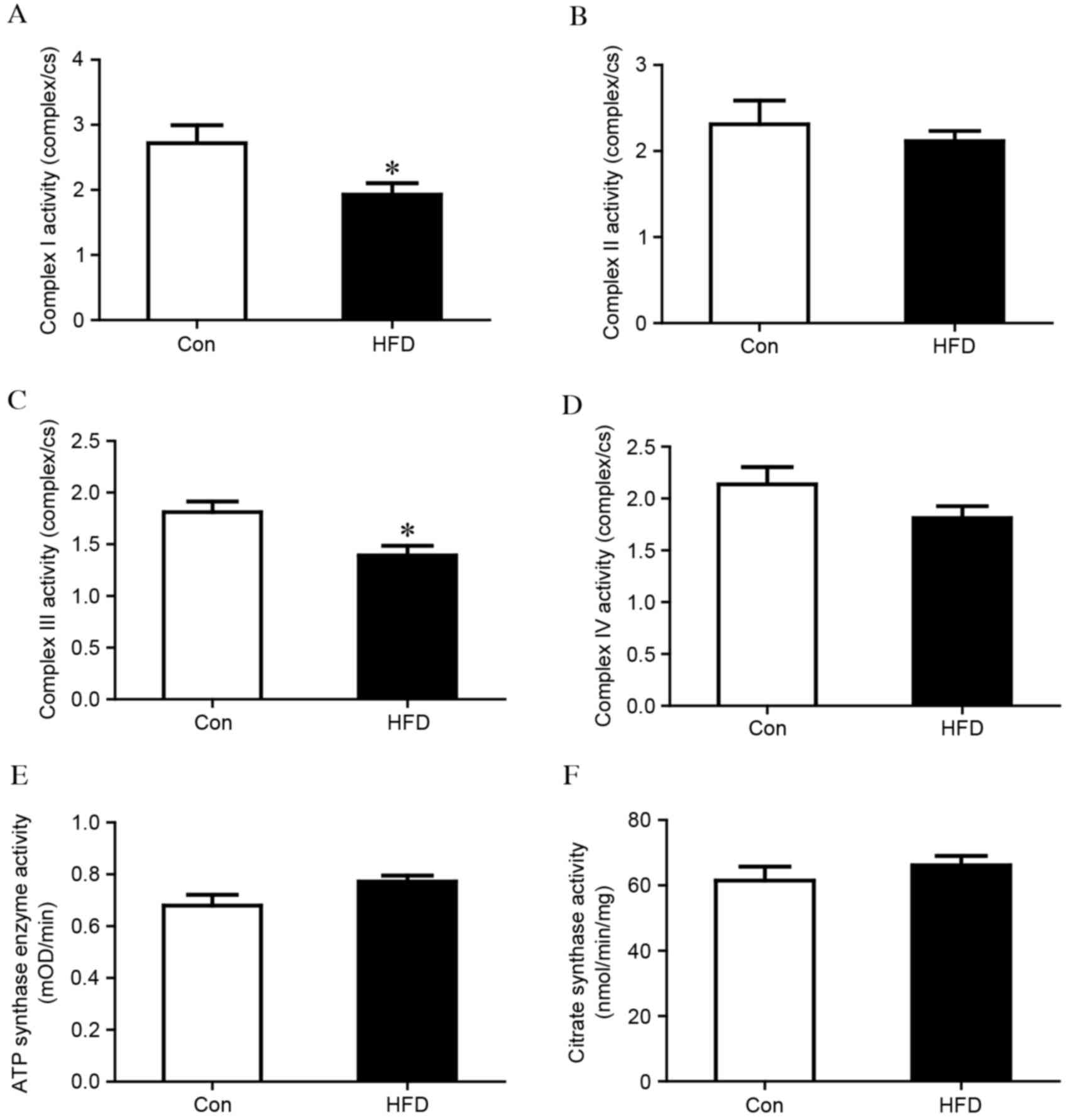
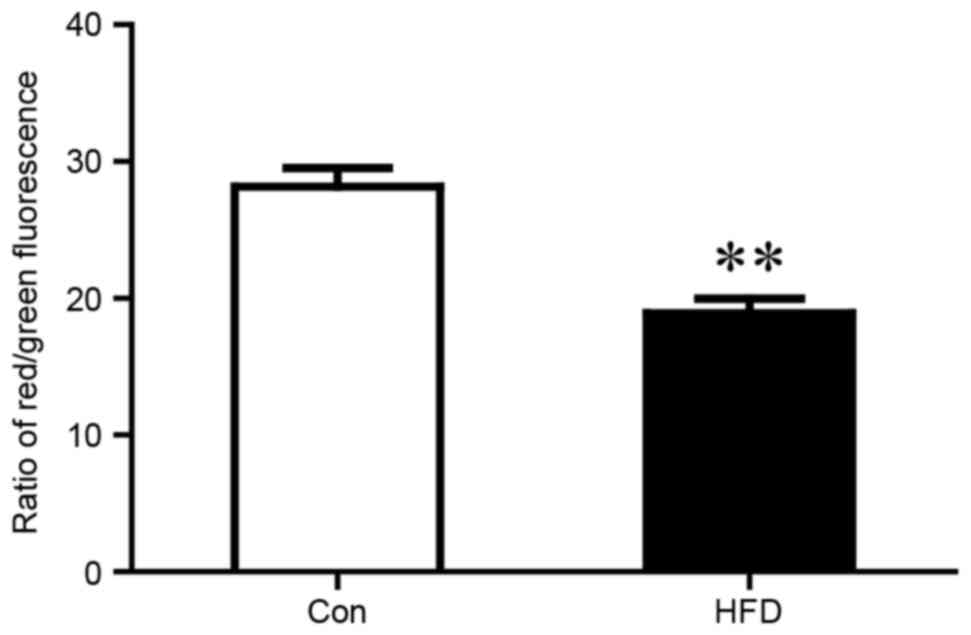
|
1
|
Melin Patrick AJ, Kalinski MI, Kelly KR,
Haus JM, Solomon TP and Kirwan JP: Nonalcoholic fatty liver
disease: Biochemical and therapeutic considerations. Ukr Biokhim Zh
(1999). 81:16–25. 2009.PubMed/NCBI
|
|
2
|
Masuoka HC and Chalasani N: Nonalcoholic
fatty liver disease: An emerging threat to obese and diabetic
individuals. Ann N Y Acad Sci. 1281:106–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Day CP and James OF: Steatohepatitis: A
tale of two ‘hits’? Gastroenterology. 114:842–845. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Day CP: From fat to inflammation.
Gastroenterology. 130:207–210. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pessayre D and Fromenty B: NASH: A
mitochondrial disease. J Hepatol. 42:928–940. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanyal AJ, Sargent Campbell C, Mirshahi F,
Rizzo WB, Contos MJ, Sterling RK, Luketic VA, Shiffman ML and Clore
JN: Nonalcoholic steatohepatitis: Association of insulin resistance
and mitochondrial abnormalities. Gastroenterology. 120:1183–1192.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Begriche K, Igoudjil A, Pessayre D and
Fromenty B: Mitochondrial dysfunction in NASH: Causes, consequences
and possible means to prevent it. Mitochondrion. 6:1–28. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anstee QM and Goldin RD: Mouse models in
non-alcoholic fatty liver disease and steatohepatitis research. Int
J Exp Pathol. 87:1–16. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye H, Sun L, Huang X, Zhang P and Zhao X:
A proteomic approach for plasma biomarker discovery with 8-plex
iTRAQ labeling and SCX-LC-MS/MS. Mol Cell Biochem. 343:91–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pallotti F and Lenaz G: Isolation and
subfractionation of mitochondria from animal cells and tissue
culture lines. Methods Cell Biol. 80:3–44. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wieckowski MR, Giorgi C, Lebiedzinska M,
Duszynski J and Pinton P: Isolation of mitochondria-associated
membranes and mitochondria from animal tissues and cells. Nat
Protoc. 4:1582–1590. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carreras M Pérez, Del Hoyo P, Martin MA,
Rubio JC, Martin A, Castellano G, Colina F, Arenas J and Herruzo
Solis JA: Defective hepatic mitochondrial respiratory chain in
patients with nonalcoholic steatohepatitis. Hepatology.
38:999–1007. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sparks LM, Xie H, Koza RA, Mynatt R,
Hulver MW, Bray GA and Smith SR: A high-fat diet coordinately
downregulates genes required for mitochondrial oxidative
phosphorylation in skeletal muscle. Diabetes. 54:1926–1933. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Anitha A, Nakamura K, Thanseem I,
Matsuzaki H, Miyachi T, Tsujii M, Iwata Y, Suzuki K, Sugiyama T and
Mori N: Downregulation of the expression of mitochondrial electron
transport complex genes in autism brains. Brain Pathol. 23:294–302.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu T, Chen L, Kim E, Tran D, Phinney BS
and Knowlton AA: Mitochondrial proteome remodeling in ischemic
heart failure. Life Sci. 101:27–36. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang PT, Chen CL, Ren P, Guarini G and
Chen YR: BCNU-induced gR2 defect mediates S-glutathionylation of
Complex I and respiratory uncoupling in myocardium. Biochem
Pharmacol. 89:490–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Finsterer J, Rauschka H, Segal L, Kovacs
GG and Rolinski B: Affection of the respiratory muscles in combined
complex I and IV deficiency. Open Neurol J. 11:1–6. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murphy MP: How mitochondria produce
reactive oxygen species. Biochem J. 417:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vial G, Dubouchaud H, Couturier K,
Rousselle Cottet C, Taleux N, Athias A, Galinier A, Casteilla L and
Leverve XM: Effects of a high-fat diet on energy metabolism and ROS
production in rat liver. J Hepatol. 54:348–356. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Paradies G, Petrosillo G, Pistolese M and
Ruggiero FM: Reactive oxygen species affect mitochondrial electron
transport complex I activity through oxidative cardiolipin damage.
Gene. 286:135–141. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gallegos AM, Atshaves BP, Storey SM,
Starodub O, Petrescu AD, Huang H, McIntosh AL, Martin GG, Chao H,
Kier AB and Schroeder F: Gene structure, intracellular
localization, and functional roles of sterol carrier protein-2.
Prog Lipid Res. 40:498–563. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schroeder F, Atshaves BP, McIntosh AL,
Gallegos AM, Storey SM, Parr RD, Jefferson JR, Ball JM and Kier AB:
Sterol carrier protein-2: New roles in regulating lipid rafts and
signaling. Biochim Biophys Acta. 1771:700–718. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mari M, Caballero F, Colell A, Morales A,
Caballeria J, Fernandez A, Enrich C, Checa Fernandez JC and Ruiz
Garcia C: Mitochondrial free cholesterol loading sensitizes to TNF-
and Fas-mediated steatohepatitis. Cell Metab. 4:185–198. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Yin L and Hillgartner FB: SREBP-1
integrates the actions of thyroid hormone, insulin, cAMP, and
medium-chain fatty acids on ACCalpha transcription in hepatocytes.
J Lipid Res. 44:356–368. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martin D, Piulachs MD, Cunillera N, Ferrer
A and Bellés X: Mitochondrial targeting of farnesyl diphosphate
synthase is a widespread phenomenon in eukaryotes. Biochim Biophys
Acta. 1773:419–426. 2007. View Article : Google Scholar : PubMed/NCBI
|