Introduction
The biological mechanism underpinning microRNA
(miRNA) biogenesis allows for the understanding of the
incorporation of short hairpin RNAs (shRNA) targeting genes of
interest into natural miRNA backbones (BBmiR). Chimeric miRNA-based
shRNAs (shRNAmiR), driven by RNA polymerase II promoters, mimic
primary miRNA (pri-miRNA) function and are processed via intrinsic
small RNA biogenesis machinery without perturbation of endogenous
miRNA homeostasis (1,2). A variety of shRNAmiRs have been under
preclinical investigation in the context of various diseases
(3).
Multi-target inhibition is a major concern for
shRNAmiR design, especially in the case of infectious settings. To
date, there have been several reports examining multiplexing
approximately 2–7 shRNAmiRs in a single transcript with either the
same BBmiR (homo-BB) or different BBmiRs (hetero-BB). Hetero-BB
multiplexed shRNAmiRs are reconstituted based on authentic
polycistronic miRNA clusters (4–6), as
well as artificial tandem-arrayed miRNAs (7), and typically show additive antiviral
properties compared to their mono-counterparts. However, homo-BB
multiplexed shRNAmiRs commonly adopt the backbone of either miR30
(8–13) or miR155 (14–17)
to accommodate the same or different shRNAs, targeting one or more
genes, yielding inconsistent results regarding efficacy and
stability of the homo-BB scaffold. Little is known regarding the
impact of backbone selection on multi-shRNAmiR construction due to
a lack of comparative studies of both hetero-BB and homo-BB.
The present study reports a paralleled design of
tetra-shRNAmiRs constructed in hetero-BB and homo-BB
simultaneously. Structural composition was based on several
considerations. Firstly, it is known that more than four shRNAmiR
cassettes in tandem result in less efficient knockdown when using
C-terminal firefly luciferase (Fluc)-specific shRNAmiR as a
functional readout (16).
Therefore, a tetrameric approach was chosen. Secondly, in the
tetramer, four shRNAmiRs were designed to target different genes
[programmed cell death protein 1 (PD1), B- and T-lymphocyte
attenuator (BTLA), lymphocyte activation gene 3 (LAG3) and T cell
immunoglobulin mucin 3 (TIM3)] in order to distinguish individual
shRNAmiR-related inhibition separately. Thirdly, miR30a, miR16-1,
miR20a and miR122 were selected to construct the hetero-BB tetramer
based on a previous study of multiplexing seven hetero-BB shRNAmiR
(7). It was reported that when a
shRNAmiR heptamer was processed to produce separate shRNAmiRs, the
four selected miRNAs above exhibited higher processing efficiencies
than the other three. In the present study, we used the modified
miR-E version of miR30a, with enhanced processing and knockdown
abilities, for the construction of both hetero-BB and homo-BB
(18). Importantly, although no
positional effects of shRNAmiRs as components in the multiplex have
been reported (7,10,16),
our study maintained the exact shRNAmiR alignment between homo- and
hetero-BB tetramers, with their multi-target inhibition quantified
and compared in an artificial cell model that co-overexpressed four
target genes.
Materials and methods
Mono- and tetra-shRNAmiR
constructs
Four or five shRNA sequences for each target gene
were inserted into the indicated BBmiRs, generating four sets of
mono-shRNAmiRs (Table I).
Tetra-shRNAmiRs were designed by multiplexing four mono-shRNAmiRs
(Table II). All mono- and
tetra-shRNAmiR genes were synthesized (AuGCT Corp., Beijing, China)
and cloned into pEGFP-C2 vector (Clontech Laboratories, Mountain
View, CA, USA) to generate pEGFP-shRNAmiRs for transient
transfection. Tetra-shRNAmiRs and Enhanced Green Fluorescent
Protein (EGFP) were also cloned into pLVX-IRES-Puro (Clontech
Laboratories) to produce pLVX-EGFP-tetra-shRNAmiR-IRES-Puro for
lentivirus preparation.
 | Table I.Oligonucleotide sequences of
mono-shRNAmiRs. Target-complementary sequences are underlined. |
Table I.
Oligonucleotide sequences of
mono-shRNAmiRs. Target-complementary sequences are underlined.
| Target | shRNAmiRs | 5′-3′ Sequence |
|---|
| PD1 |
shPD1(257)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgacgtgctaaactggtaccgcattagtgaagccacagatgtaatgcggtaccagtttagcacggtgcctactgcctcggacttcaaggggctagaattc |
|
|
shPD1(458)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgacaaggcgcagatcaaagagagtagtgaagccacagatgtactctctttgatctgcgccttggtgcctactgcctcggacttcaaggggctagaattc |
|
|
shPD1(746)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgaggatttccagtggcgagagaatagtgaagccacagatgtattctctcgccactggaaatccgtgcctactgcctcggacttcaaggggctagaattc |
|
|
shPD1(800)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgagcagacggagtatgccaccattagtgaagccacagatgtaatggtggcatactccgtctgcgtgcctactgcctcggacttcaaggggctagaattc |
|
|
shPD1(818)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgacattgtctttcctagcggaattagtgaagccacagatgtaattccgctaggaaagacaatggtgcctactgcctcggacttcaaggggctagaattc |
|
|
shPD1(NC)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgaatgccgacgccggaatatagctagtgaagccacagatgtagctatattccggcgtcggcatgtgcctactgcctcggacttcaaggggctagaattc |
| BTLA |
shBTLA(085)-BBmiR20a |
ctcgaggtctatctgatgtgacagcttctgtagcacccatgggaaagaatcatgtgattgtttagttatatcacatgattcattcccatgcgtactgctagctgtagaactccagcttcggcgaattc |
|
|
shBTLA(152)-BBmiR20a |
ctcgaggtctatctgatgtgacagcttctgtagcacggagatccctttgaactagaattgtttagttatattctagttcaacgggatctcggtactgctagctgtagaactccagcttcggcgaattc |
|
|
shBTLA(163)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgagagatccctttgaactagaattagtgaagccacagatgtaattctagttcaaagggatctcgtgcctactgcctcggacttcaaggggctagaattc |
|
|
shBTLA(163)-BBmiR20a |
ctcgaggtctatctgatgtgacagcttctgtagcactgaactagaatgccctgtgaaatgtttagttattttcacagggcaatctagttccgtactgctagctgtagaactccagcttcggcgaattc |
|
|
shBTLA(332)-BBmiR20a |
ctcgaggtctatctgatgtgacagcttctgtagcacgtcataccgctgttctgcaaattgtttagttatatttgcagaacaccggtatgaggtactgctagctgtagaactccagcttcggcgaattc |
|
|
shBTLA(NC)-BBmiR20a |
ctcgaggtctatctgatgtgacagcttctgtagcacgattctatcagtcgtgcatacctgtttagttatggtatgcacgacagatagaatggtactgctagctgtagaactccagcttcggcgaattc |
| LAG3 |
shLAG3(651)-BBmiR122 |
ctcgagcgtggctacagagtttccttagcagagctgcccatcaccacttagcggaaagtgtctaaactatctttccgctaagaggtgatggctagctactgctaggcaatccttccctcgataagaattc |
|
|
shLAG3(651)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgaccatcaccacttagcggaaagtagtgaagccacagatgtactttccgctaagtggtgatgggtgcctactgcctcggacttcaaggggctagaattc |
|
|
shLAG3(747)-BBmiR122 |
ctcgagcgtggctacagagtttccttagcagagctgtcaacgtctccatcatgtataatgtctaaactatttatacatgatgcagacgttgctagctactgctaggcaatccttccctcgataagaattc |
|
|
shLAG3(999)-BBmiR122 |
ctcgagcgtggctacagagtttccttagcagagctggccatatccatctgcaggaacatgtctaaactattgttcctgcagaaggatatgggtagctactgctaggcaatccttccctcgataagaattc |
|
|
shLAG3(1102)-BBmiR122 |
ctcgagcgtggctacagagtttccttagcagagctggctttgtgaggtgactccagtatgtctaaactattactggagtcacgtcacaaaggtagctactgctaggcaatccttccctcgataagaattc |
|
|
shLAG3(NC)-BBmiR122 |
ctcgagcgtggctacagagtttccttagcagagctgggaccgctagaacgctcactaatgtctaaactatttagtgagcgttgtagcggtcgtagctactgctaggcaatccttccctcgataagaattc |
| TIM3 |
shTIM3(201)-BBmiR16-1 |
ctcgaggctcttatgatagcaatgtcagcagtgcctgctcaggactgatgaaagggatttaagattctaaaattatatccctttcatccgtcctgaggagtaaggttgaccatactctacagttgtgtgaattc |
|
|
shTIM3(539)-BBmiR16-1 |
ctcgaggctcttatgatagcaatgtcagcagtgcctgccaatgagttacgggactctattaagattctaaaattattagagtcccgtacctcattgggagtaaggttgaccatactctacagttgtgtgaattc |
|
|
shTIM3(750)-BBmiR16-1 |
ctcgaggctcttatgatagcaatgtcagcagtgcctgcaaatgcagtagcagagggaattaagattctaaaattatttccctctgctagtgcatttggagtaaggttgaccatactctacagttgtgtgaattc |
|
|
shTIM3(884)-BBmiR16-1 |
ctcgaggctcttatgatagcaatgtcagcagtgcctggttgtcgctttgcaatgccatttaagattctaaaattatatggcattgcaacgcgacaacgagtaaggttgaccatactctacagttgtgtgaattc |
|
|
shTIM3(884)-BBmiR30a |
ctcgagaaggtatattgctgttgacagtgagcgagttgtcgctttgcaatgccattagtgaagccacagatgtaatggcattgcaaagcgacaacgtgcctactgcctcggacttcaaggggctagaattc |
|
|
shTIM3(NC)-BBmiR16-1 |
ctcgaggctcttatgatagcaatgtcagcagtgcctgacgtctatcgttacggtgttcttaagattctaaaattatgaacaccgtaactatagacgtgagtaaggttgaccatactctacagttgtgtgaattc |
 | Table II.Oligonucleotide sequences of
tetra-shRNAmiRs. Bold motifs in each sequence are recognized by
Xho I, Spe I, Xba I, Not I and
EcoR I in turn. |
Table II.
Oligonucleotide sequences of
tetra-shRNAmiRs. Bold motifs in each sequence are recognized by
Xho I, Spe I, Xba I, Not I and
EcoR I in turn.
|
Oligonucleotides | 5′-3′ Sequence |
|---|
| homo-BB
tetra-shRNAmiR |
ctcgagaaggtatattgctgttgacagtgagcgacgtgctaaactggtaccgcattagtgaagccacagatgtaatgcggtaccagtttagcacggtgcctactgcctcggacttcaaggggctatactagtacaaggtatattgctgttgacagtgagcgagaactagaatgccctgtgaaatagtgaagccacagatgtatttcacagggcattctagttcgtgcctactgcctcggacttcaaggggctaatgtctagaaaacgaaggtatattgctgttgacagtgagcgaccatcaccacttagcggaaagtagtgaagccacagatgtactttccgctaagtggtgatgggtgcctactgcctcggacttcaaggggctattgcggccgcaacaaggtatattgctgttgacagtgagcgagttgtcgctttgcaatgccattagtgaagccacagatgtaatggcattgcaaagcgacaacgtgcctactgcctcggacttcaaggggctagaattc |
| hetero-BB
tetra-shRNAmiR |
ctcgagaaggtatattgctgttgacagtgagcgacgtgctaaactggtaccgcattagtgaagccacagatgtaatgcggtaccagtttagcacggtgcctactgcctcggacttcaaggggctatactagtacgtctatctgatgtgacagcttctgtagcacgtcataccgctgttctgcaaattgtttagttatatttgcagaacaccggtatgaggtactgctagctgtagaactccagcttcggcatgtctagaaaacgcgtggctacagagtttccttagcagagctgcccatcaccacttagcggaaagtgtctaaactatctttccgctaagaggtgatggctagctactgctaggcaatccttccctcgataattgcggccgcaacgctcttatgatagcaatgtcagcagtgcctggttgtcgctttgcaatgccatttaagattctaaaattatatggcattgcaacgcgacaacgagtaaggttgaccatactctacagttgtgtgaattc |
Preparation of lentiviruses carrying
tetra-shRNAmiRs
pLVX-EGFP-tetra-shRNAmiR-IRES-Puro was
co-transfected into 293T cells with the helper plasmids pSPAX2 and
pMD2. G (plasmids 12259 and 12260; Addgene Inc., Cambridge, MA,
USA). At 72 h post-transfection, recombinant lentiviruses were
harvested and adjusted to 1×106 PFU/ml. Lentiviral
transduction was performed at a multiplicity of infection (MOI) of
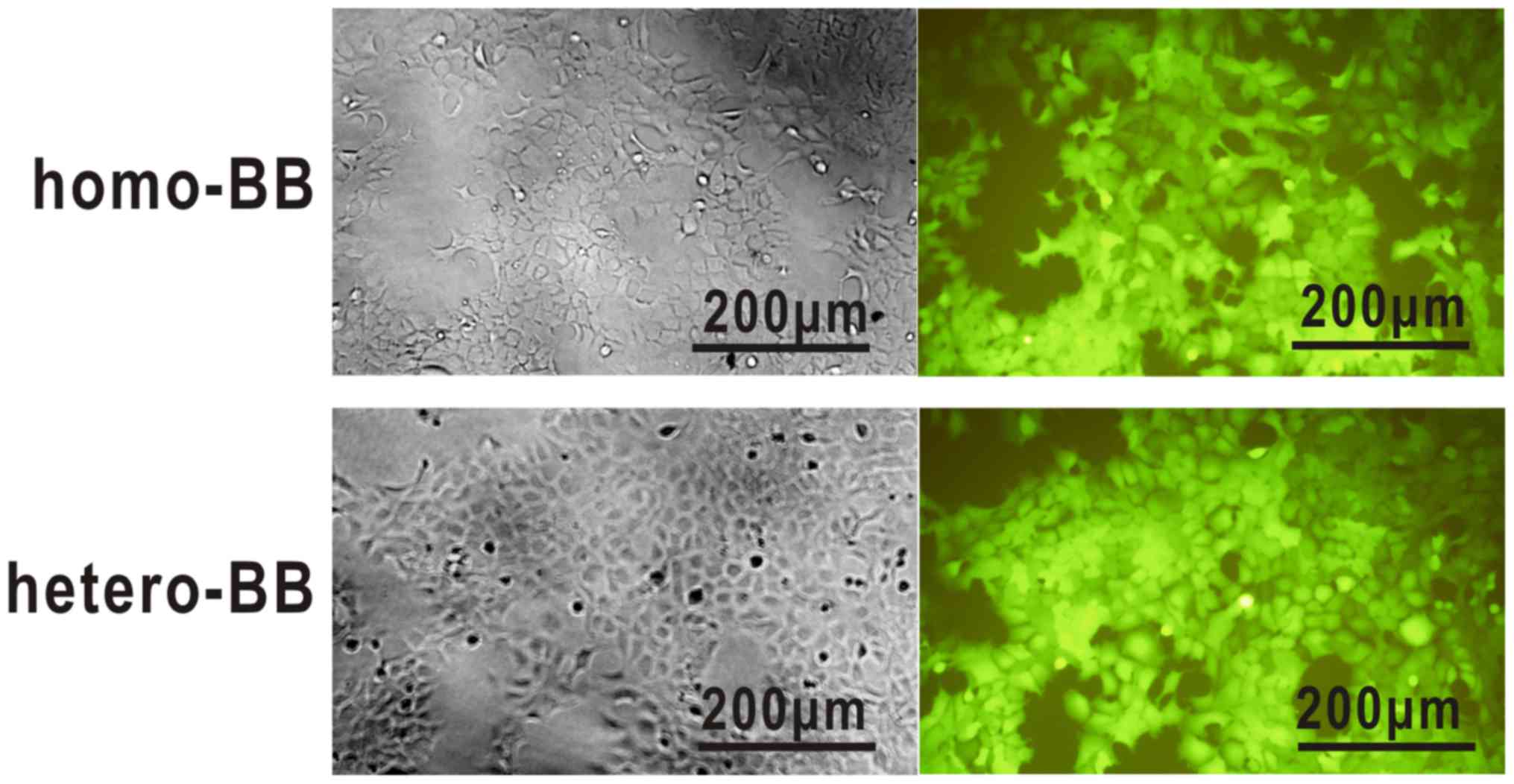
1 in multi-target-overexpressing 293A cells (Fig. 1).
Establishment of
multi-target-overexpressing cells
BTLA, PD1, TIM3 and LAG3 genes were purchased from
Sino Biological Inc. (Beijing, China). Target-encoding lentiviral
vectors were constructed by insertion of BTLA into
pLenti6.3/V5-DEST (Invitrogen, Carlsbad, CA, USA), PD1 into
pLVX-IRES-Puro (Clontech Laboratories), TIM3 into pLVX-IRES-Hyg
(Clontech Laboratories) and LAG3 into pLVX-IRES-Neo (Clontech
Laboratories). Recombinant vectors were co-transfected into 293T
cells with pSPAX2 and pMD2.G for lentivirus production. The
resultant lentiviruses were sequentially transduced into 293A
cells, followed by selection of drug-resistant colonies with 10
µg/ml blasticidin for BTLA expression, 0.25 µg/ml puromycin for PD1
expression, 500 µg/ml hygmycin for TIM3 expression and 1,000 µg/ml
G418 for LAG3 expression. Each selection lasted for 10 days, and
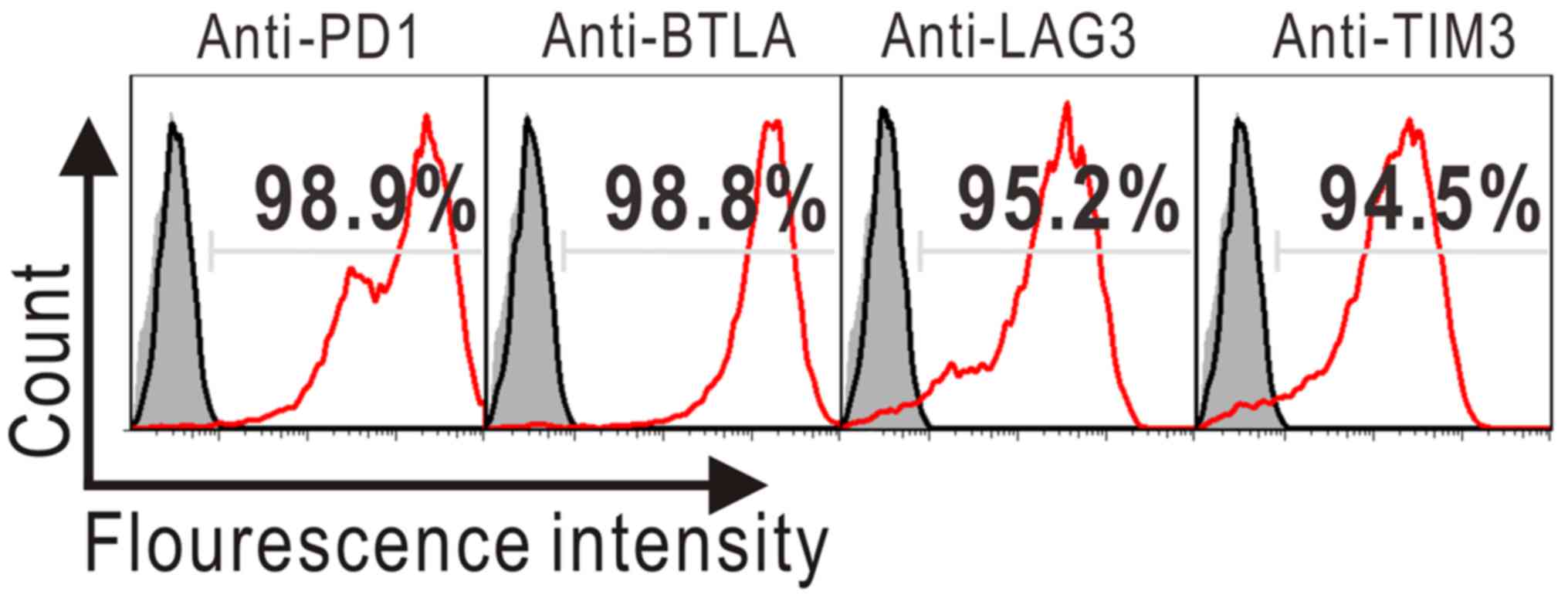
finally multi-target-overexpressing 293A cells were obtained and
identified by flow cytometry.
Flow cytometry
Target inhibition was quantified by flow cytometry
via measuring downregulation of surface target proteins. Monoclonal
antibodies used herein included phycoerythrin (PE)-conjugated
anti-human PD1 and BTLA (BioLegend, San Diego, CA, USA), and
PE-conjugated anti-human LAG3 and TIM3 (eBioscience, Inc., San
Diego, CA, USA). After incubation on ice with the pertinent
antibodies for 30 min, stained cells were analyzed by FACSCalibur
using Cell-Quest software (both BD Bioscience, San Diego, CA,
USA).
Quantitative real-time PCR
(qRT-PCR)
To detect individual siRNAs generated from
tetra-shRNAmiRs, 500 ng of total RNA was extracted by Trizol
(Invitrogen), followed by qRT-PCR using miScript PCR Starter Kit
(Qiagen, Hilden, Germany). miScript universal primer was used in
combination with the following specific primers:
5′-acgtgctaaactggtaccgcat-3′ (for siPD1-257); 5′-aga act aga atg
ccc tgt gaa a-3′ (for siBTLA-163); 5′-acc atc acc act tag cgg aaa
g-3′ (for siLAG3-651); 5′-agt tgt cgc ttt gca atg cca t-3′ (for
siTIM3-883). The average cycle threshold values of mature siRNAs
were normalized to that of U6 small nuclear RNA (snRNA).
Statistical analysis
Quantitative data are presented as mean ± standard
deviations (SD) from independent experiments in triplicate.
Differences were tested for significance by ANOVA using SPSS 15.0.
P<0.05 was considered to indicate a statistically significant
difference.
Results
BBmiR selection affects the efficacy
of mono-shRNAmiRs
To ensure the efficacy of mono-shRNAmiR design, four
sets of shRNA sequences embedded in the indicated BBmiRs were first
examined for their ability to knockdown their related target genes.
A co-transfection-based cell model was used to directly quantify
shRNA-mediated downregulation of surface target proteins by flow
cytometry analysis. As shown in Fig.
2A, shPD1-257, shBTLA-163, shLAG3-651 and shTIM3-884 were
selected for further experiments.
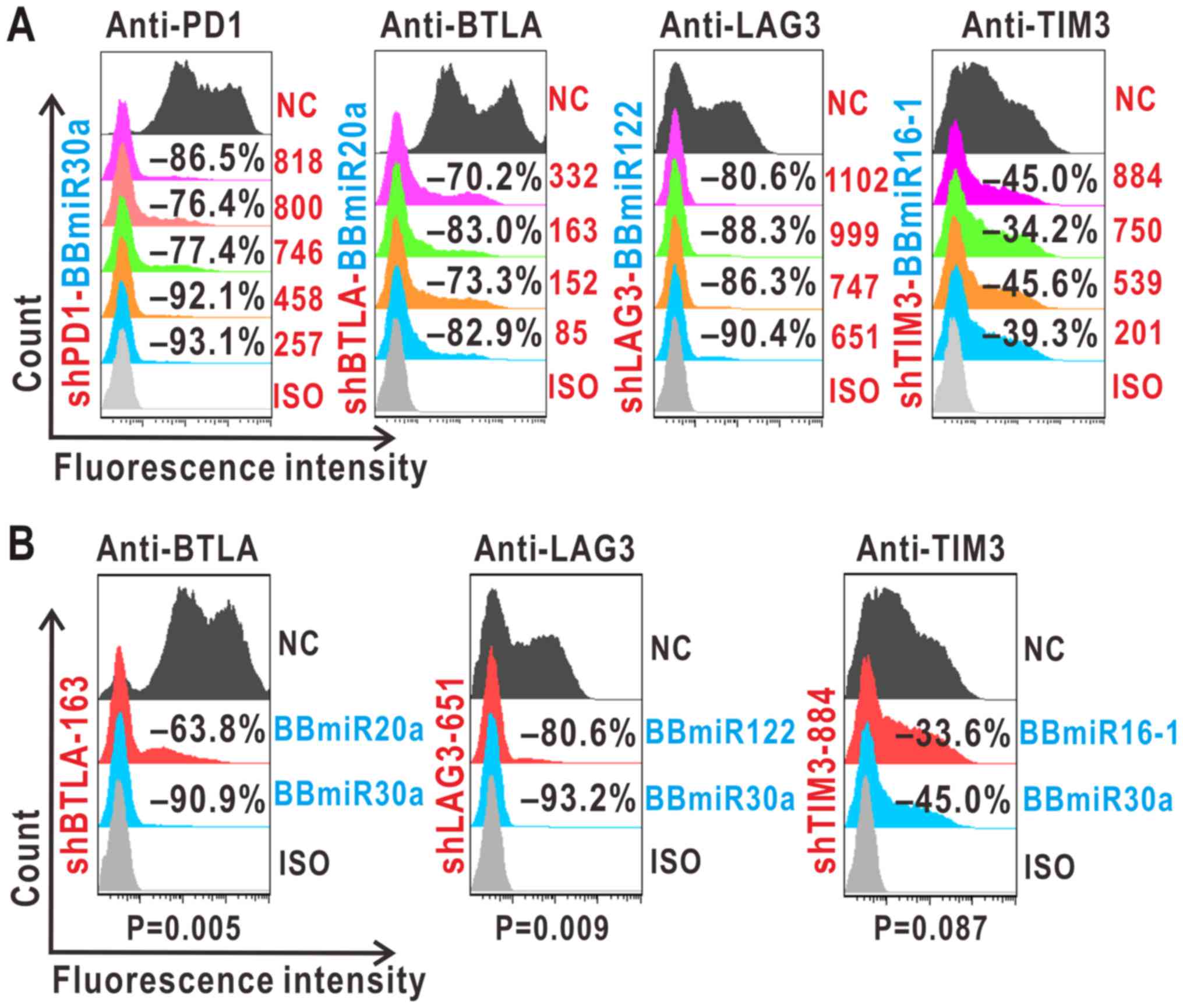 | Figure 2.Identification of mono-shRNAmiRs
targeting PD1, BTLA, LAG3 and TIM3. (A and B) shRNA selection with
the indicated BBmiR. (A) Forty-eight-hour co-transfection of target
genes and pEGFP-shRNAmiRs led to diminished target protein levels
in 293A assayed by flow cytometry. (B) Silencing effects of
different BBmiRs as compared with BBmiR30a at 36 h
post-transfection. Negative percentages denote inhibition against
scrambled shRNA controls (NC). ISO, isotype controls; PD1,
programmed cell death protein 1; BTLA, B- and T-lymphocyte
attenuator; LAG3, lymphocyte activation gene 3; TIM3, T cell
immunoglobulin mucin 3; sh, short hairpin; miR, microRNA; BBmiR,
miRNA backbones. |
Next, several shRNAs were assessed in combination
with different BBmiRs. The resultant mono-shRNAmiRs were examined
as to whether BBmiR substitution could interfere with a particular
shRNA function. As evidenced by Fig.
2B, shBTLA-163-BBmiR30a led to a marked increase in BTLA
inhibition by 27.8±3.71% compared with its BBmiR20a counterpart.
Similarly, in the case of shLAG3-651 and shTIM3-884, their BBmiR30a
chimeras increased LAG3 inhibition by 10.93±5.05% and TIM3
inhibition by 14.20±6.29%, respectively, compared with their
BBmiR122 and BBmiR16-1 chimeras. These results suggest that BBmiR
selection contributes, at least in part, to mono-shRNAmiR
function.
Limited effect of BBmiR substitution
on tetra-shRNAmiR function
Tetra-shRNAmiRs were generated by tandem fusion of
four mono-shRNAmiRs targeting PD1, BTLA, LAG3 and TIM3 (Fig. 3). To determine whether BBmiR
substitution affected the function of multiplexed shRNAmiRs, two
paralleled tetra-shRNAmiRs were designed with exactly the same
targeting alignments but distinct backbones to generate homo-BB
tetra-shRNAmiR with only BBmiR30a and hetero-BB tetra-shRNAmiR with
four different BBmiRs (Fig. 3A).
For functional comparison of these distinct backbone-based
tetra-shRNAmiRs, an artificial cell model overexpressing all the
targets was established and referred to as PBLT+ 293A.
Selection with sequential drugs gave rise to high-level
co-expression of PD1, BTLA, LAG3 and TIM3 (Fig. 4). Of note, when cultured without
drugs for two months, PBLT+ 293A cells stably expressed
BTLA, while the expressions of other three target genes gradually
declined with prolonged culture (Fig.
3B, black lines). Given this slowly declining trend of target
gene expression, non-drug-treated mock-transfected control cells
were monitored at all time points and relative target inhibition
over the mock was calculated to exclude intergroup variations.
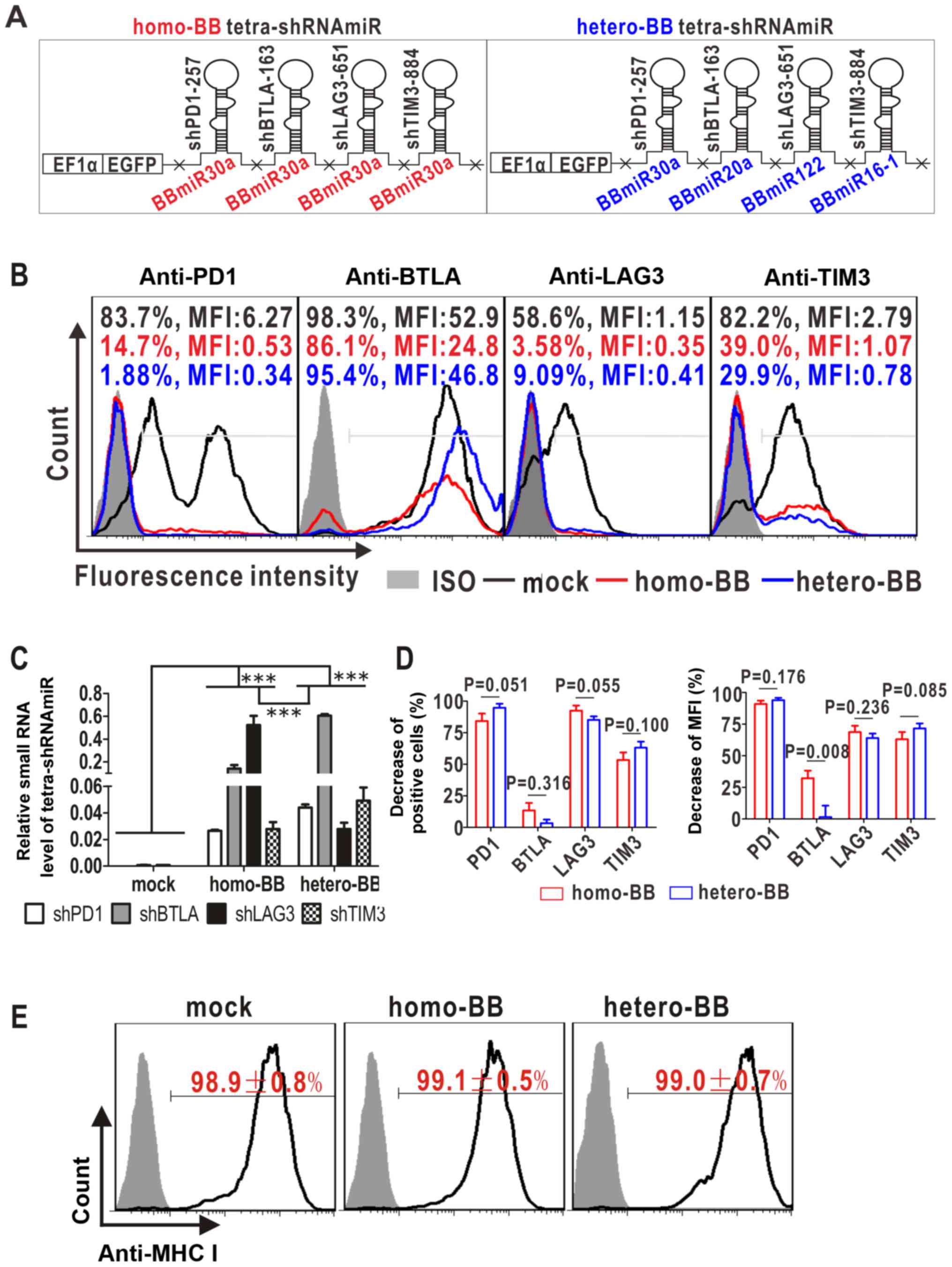 | Figure 3.Multi-target effect of tetra-shRNAmiRs
on the established 293A cells simultaneously overexpressing PD1,
BTLA, LAG3 and TIM3 (designated as PBLT+ 293A). (A)
Schematic diagram of tetra-shRNAmiRs with homo-or hetero-BBmiRs. X,
restriction enzyme sites. (B) Target inhibition by flow cytometry
at 4 days post-infection. A set of representative histographs are
shown, with cell percentages and mean fluorescence intensities
(MFI) of immunostained cells marked. ISO, isotype controls. (C)
Normalized small RNA levels of tetra-shRNAmiRs to U6 at 4 days
post-infection by quantitative RT-PCR. ***P<0.001. (D)
Statistical analyses of the decrease in both cell percentages and
MFI against the void vector control (mock) from three independent
experiments. (E) Undetected off-target effect of tetra-shRNAmiRs on
an irrelevant antigen (MHC I). Isotype controls are filled in grey.
PD1, programmed cell death protein 1; BTLA, B- and T-lymphocyte
attenuator; LAG3, lymphocyte activation gene 3; TIM3, T cell
immunoglobulin mucin 3; sh, short hairpin; miR, microRNA. |
Transduction of tetra-shRNAmiRs resulted in
processing and production of individual mature siRNAs in
PBLT+ 293A cells, as detected by quantitative RT-PCR
(Fig. 3C). Construction with the
hetero-BB contributed to more abundant generation of small RNAs
targeting PD1, BTLA and TIM3, whereas the homo-BB design was
associated with lower processing efficiencies, presumably due to
competition for small RNA biogenesis machinery. Unexpectedly,
LAG3-targeted siRNA production predominated in the homo-BB group
rather than the hetero-BB group, indicating a more potentiated
processing capability of shLAG3-BBmiR30a than that of
shLAG3-BBmiR122.
Multi-target effects of the two tetra-shRNAmiRs were
further compared. As shown in Fig. 3B
and D, tetramer-transduced PBLT+ 293A cells
exhibited potent inhibition on PD1, LAG3 and TIM3 in both groups,
as evidenced by the remarkable parallel decreases in cell
percentages and mean fluorescence intensities (MFI), and no
significant difference resulted from BBmiR substitution. In
contrast, BTLA inhibition was relatively weak in both tetramers
(Fig. 3B and D). This may be in
part attributed to the significant expression of BTLA in
non-drug-treated mock controls, which exhibited extremely high MFI
of 56.4±5.9 in 97.6±1.1% BTLA-positive cells, in contrast to the
much weaker expression of the other three targets with MFI below 10
(Fig. 3B, black lines and black
numbers). Thus there is a possibility that tetra-shRNAmiRs were
insufficient to markedly downregulate highly expressed BTLA.
Interestingly, homo-BB-tetramer transductants exhibited stronger
BTLA inhibition than hetero-BB counterparts, with significant
differences in MFI decrease (Fig.
3D). No off-target effects were observed as evidenced by
unaltered expression of an irrelevant antigen (MHC I) after
infection of tetra-shRNAmiRs (Fig.
3E). Taken together, in this particularly tested tetra-shRNAmiR
formulation, BBmiR substitution had limited effects on overall
functionality.
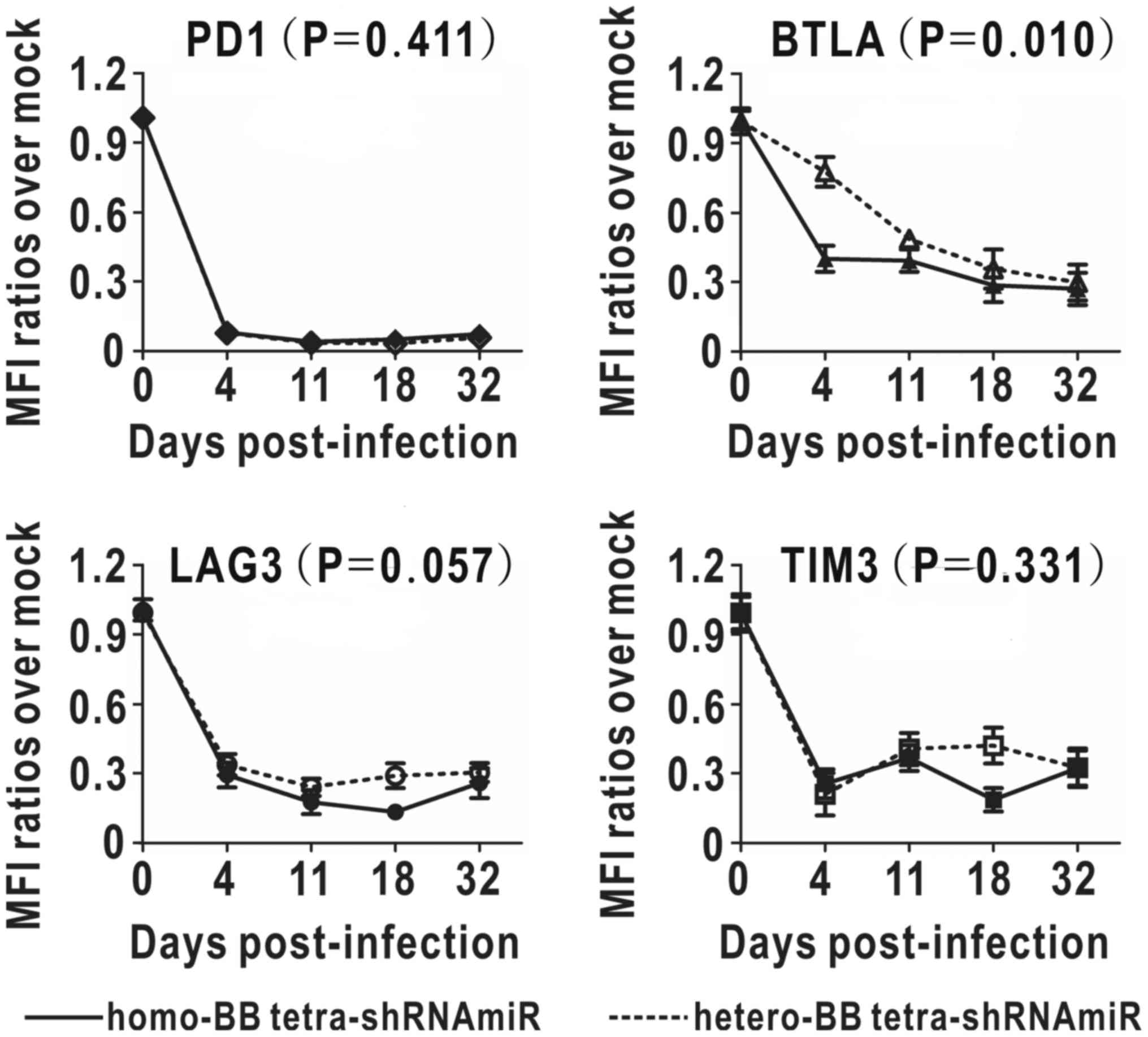
Sustained functionality of homo- and
hetero-BB tetra-shRNAmiRs
To compare long-term inhibitory effects of the two
tetramers, MFI decreases of multi-targets were monitored for one
month (Fig. 5). Simultaneous
downregulation of four targets were observed in both groups.
Inhibition rate of PD1 was over 90%, and those of BTLA, LAG3 and
TIM3 ranged from 67 to 74% at the final endpoint. There was no
intergroup difference shown in persistent inhibition of PD1, LAG3
and TIM3, in contrast to BTLA inhibition, which was significantly
enhanced with homo-BB structure. The enhancement of BTLA inhibition
in the homo-BB group peaked on day four post-infection when
comparing with the hetero-BB group. On the whole, long-term
inhibitory effects of tetra-shRNAmiRs with substituted BBmiRs were
almost comparable at the end of one month.
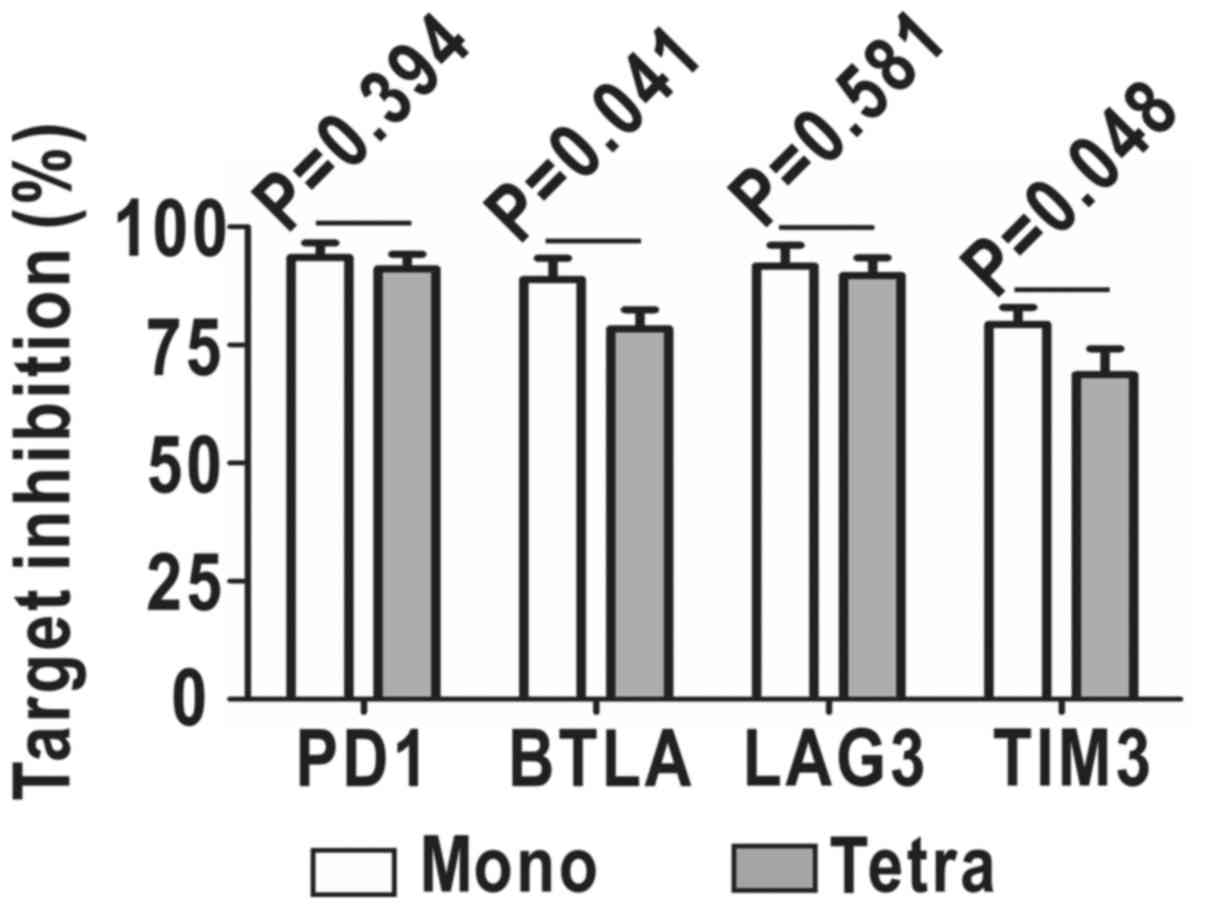
Inhibition efficiency of homo-BB
tetra-shRNAmiR
In order to verify whether homo-BB tetra-shRNAmiR
would reduce the efficiency of each shRNA, we co-transfected target
genes together with mono-shRNAmiRs or homo-BB tetra-shRNAmiR into
293A cells, and directly quantified shRNA-mediated downregulation
of surface target proteins by flow cytometry. As shown in Fig. 6, knockdown efficiencies on PD1 and
LAG3 were comparable between mono-shRNAmiRs and the tetra-shRNAmiR.
Despite a slight decrease in downregulation of BTLA and TIM3 by
~10%, the tetra-shRNAmiR still exhibited potent inhibition ~70%.
Therefore, the BBmiR-shared strategy of multiplexing shRNAmiR may
likely result in a little lower efficiencies of multi-target
inhibition, but is functionally competent and conveniently
manipulated.
Discussion
Recent insights into tandem shRNAmiR strategies have
accumulated out of separate studies with either homo-BB or
hetero-BB alone. Our design of BBmiR-substituted tetra-shRNAmiRs,
to the best of our knowledge, is the first attempt to comparatively
explore the contribution of BBmiR scaffolds to multiplexed shRNAmiR
functions. To date, there has been only one report on comparison of
homo- and hetero-dimers of natural miRNAs (19), which differs from our study in
several ways. Firstly, no artificial shRNA sequences were exchanged
into dimerized miRNAs, failing to offer a structure-function
relationship of shRNAmiR multiplexes. Secondly, several cell models
were developed by introducing natural miRNA-complementary motifs
into 3′-UTR of GFP reporter, hardly affording simultaneous
assessment of both miRNA functions. In contrast, our study is
characterized by combining shRNA-adapted artificial miRNA tetramer
with strictly paralleled BBmiRs, and by targeting four different
genes to validate individual knockdown effect at the same time.
Intriguingly, two seemingly paradoxical findings
were highlighted by our study. The first was that despite superior
functionalities of miR30a-backboned mono-shRNAmiR, the homo-BB
tetramer exerted multi-inhibitory effects almost similar to the
hetero-BB tetramer. One of possible interpretations was a lowered
individual threshold of expression and processing due to shRNAmiR
multiplexing. As evidenced by a previous study on seven-chained
hetero-BB shRNAmiRs, the four BBmiRs that we chose for our study
decreased individual processing efficiencies by 70–80% compared
with their mono-counterparts (7).
These sharply narrowed assay windows could reduce functional
differences between the two tetramers. Another underlying
possibility was potent target inhibition beyond a linear range.
This was indicated by loss of early-phased difference in BTLA
downregulation when reaching late-phase inhibition as high as 70%
(Fig. 5).
The second confounding issue was inconsistency
between individual mature siRNA levels derived from tetra-shRNAmiRs
and consequent knockdown efficiencies. Technically, we used the
same primers in qRT-PCR for each pair of backbone-grafted shRNAmiRs
targeting the same genes, thus amplified products of exactly the
same mature siRNAs would be quantitatively analyzable for
comparison of tetramer processing. In this study, processing
preference was generally observed in the hetero-BB tetramer, as
evidenced by higher production of siPD1, siBTLA and siTIM3. Of
note, the homo-BB tetramer, although challenged by resource
competition, produced higher amounts of siLAG3, which might be
attributed to an unknown processing bias. However, such differences
in mature siRNA levels did not functionally correlate, given almost
comparable inhibitory activities between the two tetramers.
Explanations behind this might involve PCR-based limitations of
magnifying intergroup variations and failing to discern incorrectly
processed products with minor alterations. To overcome this
problem, deep sequencing may be a more reliable approach to confirm
processing accuracy (7), and more
efforts are needed to underpin the mechanisms behind this
discrepancy.
Lentiviral risks of recombination-mediated deletion
have been documented sporadically for both homo-BB-polymerized
shRNAmiRs (12) and homo-dimerized
natural miRNAs (19). However,
extensive studies still confirm the validity of the sharing of the
same BBmiR30 for shRNAmiR multiplexing, which functions as a
co-inhibitor of different targets (8–10),
as well as additive inhibition against different sites of one
target (11), and dosage-dependent
inhibition against an identical site of one target (10). These findings are further supported
by our current data on the one-month prolonged efficacy of the
homo-BB tetra-shRNAmiR, which also provides the first evidence of
feasibility of repetitively utilizing the modified backbone of the
miR30a derivative miR-E. This shared backbone-based tetramerization
did not substantially attenuate joint target inhibition, as shown
by functional comparison with mono-shRNAmiRs in the co-transfection
assay (Fig. 6).
In summary, this comparative study on
BBmiR-paralleled shRNAmiR tetramers, by establishing a cell model
to dissect target inhibition simultaneously, offered preliminary
clues regarding scaffold influence on tetramer function. In our
assay system, BBmiR substitution was not a major contributing
factor, with nearly no marked functional difference between homo-
and hetero-tetramers. Further efforts remain necessary with regards
to the detailed validation of extended repertoires of
scaffold-grafted shRNAmiR polymers in more physiologically relevant
models.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372459).
References
|
1
|
Bofill-De Ros X and Gu S: Guidelines for
the optimal design of miRNA-based shRNAs. Methods. 103:157–166.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Davidson BL and McCray PB Jr: Current
prospects for RNA interference-based therapies. Nat Rev Genet.
12:329–340. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calloni R and Bonatto D: Scaffolds for
artificial miRNA expression in animal cells. Hum Gene Ther Methods.
26:162–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu YP, Haasnoot J, ter Brake O, Berkhout
B and Konstantinova P: Inhibition of HIV-1 by multiple siRNAs
expressed from a single microRNA polycistron. Nucleic Acids Res.
36:2811–2824. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chung J, Zhang J, Li H, Ouellet DL,
DiGiusto DL and Rossi JJ: Endogenous MCM7 microRNA cluster as a
novel platform to multiplex small interfering and nucleolar RNAs
for combinational HIV-1 gene therapy. Hum Gene Ther. 23:1200–1208.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aagaard LA, Zhang J, von Eije KJ, Li H,
Saetrom P, Amarzguioui M and Rossi JJ: Engineering and optimization
of the miR-106b cluster for ectopic expression of multiplexed
anti-HIV RNAs. Gene therapy. 15:1536–1549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Choi JG, Bharaj P, Abraham S, Ma H, Yi G,
Ye C, Dang Y, Manjunath N, Wu H and Shankar P: Multiplexing seven
miRNA-Based shRNAs to suppress HIV replication. Mol Ther.
23:310–320. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu X, Santat LA, Chang MS, Liu J,
Zavzavadjian JR, Wall EA, Kivork C, Simon MI and Fraser ID: A
versatile approach to multiple gene RNA interference using
microRNA-based short hairpin RNAs. BMC Mol Biol. 8:982007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shin KJ, Wall EA, Zavzavadjian JR, Santat
LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI and
Fraser ID: A single lentiviral vector platform for microRNA-based
conditional RNA interference and coordinated transgene expression.
Proc Natl Acad Sci USA. 103:13759–13764. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun D, Melegari M, Sridhar S, Rogler CE
and Zhu L: Multi-miRNA hairpin method that improves gene knockdown
efficiency and provides linked multi-gene knockdown. Biotechniques.
41:59–63. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Snyder LL, Esser JM, Pachuk CJ and Steel
LF: Vector design for liver-specific expression of multiple
interfering RNAs that target hepatitis B virus transcripts.
Antiviral Res. 80:36–44. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Osório L, Gijsbers R, Oliveras-Salvá M,
Michiels A, Debyser Z, Van den Haute C and Baekelandt V: Viral
vectors expressing a single microRNA-based short-hairpin RNA result
in potent gene silencing in vitro and in vivo. J Biotechnol.
169:71–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou H, Xia XG and Xu Z: An RNA polymerase
II construct synthesizes short-hairpin RNA with a quantitative
indicator and mediates highly efficient RNAi. Nucleic Acids Res.
33:e622005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fowler DK, Williams C, Gerritsen AT and
Washbourne P: Improved knockdown from artificial microRNAs in an
enhanced miR-155 backbone: A designer's guide to potent
multi-target RNAi. Nucleic Acids Res. 44:e482016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chung KH, Hart CC, Al-Bassam S, Avery A,
Taylor J, Patel PD, Vojtek AB and Turner DL: Polycistronic RNA
polymerase II expression vectors for RNA interference based on
BIC/miR-155. Nucleic Acids Res. 34:e532006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu T, Chen P, Fu Q, Liu Y, Ishaq M, Li J,
Ma L and Guo D: Comparative studies of various artificial microRNA
expression vectors for RNAi in mammalian cells. Mol Biotechnol.
46:34–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shan ZX, Lin QX, Yang M, Deng CY, Kuang
SJ, Zhou ZL, Xiao DZ, Liu XY, Lin SG and Yu XY: A quick and
efficient approach for gene silencing by using triple putative
microRNA-based short hairpin RNAs. Mol Cell Biochem. 323:81–89.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fellmann C, Hoffmann T, Sridhar V,
Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan
Y, et al: An optimized microRNA backbone for effective single-copy
RNAi. Cell Rep. 5:1704–1713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amendola M, Passerini L, Pucci F, Gentner
B, Bacchetta R and Naldini L: Regulated and multiple miRNA and
siRNA delivery into primary cells by a lentiviral platform. Mol
Ther. 17:1039–1052. 2009. View Article : Google Scholar : PubMed/NCBI
|