Introduction
Gliomas are the most common primary brain tumors in
adults (1). Astrocytomas are a
type of glioma that are graded between I and IV according to a
World Health Organization (WHO) grading system, which is determined
following pathological evaluation of the tumor. Low-grade gliomas
(WHO grade I and II) are well differentiated with a good prognosis,
whereas high-grade gliomas (WHO grade III and IV) are malignant
with a poor prognosis (2).
MicroRNAs (miRNAs) are small noncoding RNAs that
function as negative regulators of posttranscriptional gene
expression by binding with the 3′untranslated regions of target
mRNAs to either inhibit their translation or promote degradation
(3,4). Altered miRNA expression has been
associated with cancer development and drug resistance (5). Ectopic miRNA expression may aid in
cancer diagnosis, prognosis and therapy, and may predict possible
responses to treatment (6). miRNA
(miR)-124 is highly conserved in animals and is the most abundantly
expressed miRNA in the embryonic and adult brain, serving a key
role in neurogenesis (7–9). A recent study reported that miR-124
expression is downregulated in a variety of human cancers, and is
therefore considered to be a tumor suppressor (10). miR-124 was also demonstrated to
inhibit the migration and invasion of glioma cells, and reduced
miR-124 expression may be predictive of a poor prognosis in
patients with gliomas (11–13).
miR-124 has numerous mRNA targets that are related to the cell
cycle, oncogenesis and cancer metastasis (14–16),
such as cyclin-dependent kinase (CDK)6, inhibitor of
apoptosis-stimulating protein of p53 (iASPP) and IQ motif
containing GTPase activating protein 1 (IQGAP1) (17–19).
iASPP is involved in cancer metastasis and is a promising
therapeutic target (20,21). Although miR-124 targeting of iASPP
has been implicated in glioma progression (22), the specific role remains to be
elucidated.
In the present study, the expression levels of
miR-124 and iASPP were examined in human glioma tissues and in
human U251 and U87 glioma cell lines. In addition, the methylation
status of miR-124 in U251 and U87 cells was examined and functional
analyses of miR-124 in these cells were conducted. The results
suggested that miR-124 may serve as a molecular biomarker for
glioma tumors of different grades and may represent a novel
therapeutic target for the treatment of gliomas.
Materials and methods
Patients and cell lines
All samples were collected from Beijing Tongren
Hospital (Beijing, China) between 2011 and 2014, and were stored in
liquid nitrogen until use. Patients with glioma (n=66) and normal
control patients (n=14) were examined following receipt of written
informed consent, and this study was approved by The Ethics
Committees of Beijing Tongren Hospital, Capital Medical University
(Beijing, China). Gliomas were diagnosed and graded based on the
WHO classification system, and of the 66 glioma tissues collected,
19 were classified as diffuse astrocytoma (grade II), 25 as
anaplastic astrocytoma (grade III) and 22 as glioblastoma
multiforme (grade IV). The clinicopathological information of all
glioma patients is presented in Table
I. A total of 14 healthy normal brain (NB) tissues were
collected from patients receiving treatment of cerebral injury by
internal decompression and treatment of epilepsy by temporal lobe
resection (9 male and 5 female, 8 <50 years old, 6 ≥50 years
old).
 | Table I.Clinicopathological features of 66
glioma patients. |
Table I.
Clinicopathological features of 66
glioma patients.
| Clinicopathological
feature | WHO II | WHO III | WHO IV |
|---|
| Cases (n) | 19 | 25 | 22 |
| Sex |
|
|
|
| Male | 11 | 15 | 14 |
|
Female | 8 | 10 | 8 |
| Age |
|
|
|
|
<50 | 10 | 13 | 10 |
|
≥50 | 9 | 12 | 12 |
| KPS score |
|
|
|
|
<80 | 4 | 11 | 9 |
|
≥80 | 15 | 14 | 13 |
| Tumor size |
|
|
|
| <5
cm | 12 | 15 | 17 |
| ≥5
cm | 7 | 10 | 5 |
The human glioblastoma cell lines U87 and U251 were
purchased from Peking Union Medical College (Beijing, China) and
maintained in Dulbecco's modified Eagle's medium supplemented with
10% fetal bovine serum (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), 10 U/ml penicillin and 0.1 mg/ml streptomycin
(Sigma-Aldrich; Merck KGaA), at 37°C and 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For RT-qPCR, 0.1 g tissues and 5×106
cells were used to extract total RNA using the mirVana miR
Isolation kit (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocols. The mature form of
miR-124 was detected using the miRNA RT kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and TaqMan Universal PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocols. The primers were used as previously
described (23). Reverse
transcription reactions were incubated for 10 min at 16°C, 30 min
at 37°C and 5 min at 65°C. The real-time PCR protocol was used as
follows: 95°C for 2 min, followed by 40 cycles at 95°C for 15 sec,
60°C for 60 sec and 70°C for 30 sec. U6 was used as an internal
control. qPCR was performed using the CFX Connect Real-Time PCR
Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
and analyzed using CFX Maestro Software, Chinese edition (12005260;
Bio-Rad Laboratories, Inc.). The relative quantitative value for
each target gene was performed using the 2−ΔΔCq method
(24).
Oligonucleotide transfection
miR-124 mimics and miRNA-negative control were
purchased from Life Technologies (Thermo Fisher Scientific, Inc.).
The sequence of the miR-124 mimics was 5′-UAAGGCACGCGGUGAAUGCC-3′
and the miR-negative control sequence was
5′-UUGUACUACACAAAAGUACUG-3′. U251 and U87 cells (3.5×105
cells/well) were seeded into 6-well plates and transfected when the
cells reached 60% confluence; 50 nmol/l miR-124 mimics and
miRNA-negative control were transfected as previously described
(25) using Lipofectamine RNAiMAX
(Thermo Fisher Scientific, Inc.), following the manufacturer's
protocol. The medium was removed and fresh growth medium was added
6 h after transfection at 37°C; cells were incubated for 36 h at
37°C prior to collection for analysis. Three independent repeats
were performed for all experiments.
Bisulfite sequencing PCR
U87 and U251 cells (6×105) and NB
astrocytes were treated with the MethylCode Bisulfite Conversion
kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. Genomic DNA was extracted using the
QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to the
manufacturer's protocols. PCR was performed using TaqMan Universal
PCR Master Mix (Thermo Scientific, Inc.). The primer sequences are
listed in Table II. PCR
amplification was conducted with the following parameters: initial
denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for
30 sec, 60°C for 30 sec and 72°C for 40 sec, followed by 1 cycle of
72°C for 5 min. The amplified fragments were cloned into the
pMD18-T vector (Takara Bio, Inc., Tokyo, Japan) and 8 clones were
randomly selected for sequencing (Invitrogen; Thermo Scientific,
Inc.).
 | Table II.Primer sequences used for bisulfite
genomic sequencing. |
Table II.
Primer sequences used for bisulfite
genomic sequencing.
| Primer | Sequence
(5′➝3′) | Size (bp) |
|---|
| P1 | F:
GGGGATTTAGTYGTATTATATA | 297 |
|
| R:
TAAAAACRAAACCCATCTAA |
|
| P2 | F:
ATTATTTGGGATTTGAGYGA | 255 |
|
| R:
CTACTTCRCCCAAAAAACTAAA |
|
| P3 | F:
TTAGTTTTTTGGGYGAAGTAGA | 264 |
|
| R:
CCAAAAACTCTTTATTCTCCCC |
|
| P4 | F:
TTTGTAGGAAAAAGTTTGGAT | 300 |
|
| R:
AAAAAAAAACCTTTCTCCTAAA |
|
| P5 | F:
GGGTTTTTGYGTAAGATTAGTT | 258 |
|
| R:
ACTCCTCAACCAATAACCACA |
|
| P6 | F:
TAGATTGTGGTTATTGGTTGAG | 228 |
|
| R:
TCATCTTTCTTTATAATACRAAAAAA |
|
| P7 | F:
GGGAAGTTTTAGTGAGTAGGTTT | 306 |
|
| R:
AACCATTACCAATCATTTTCTACTA |
|
| P8 | F:
TTTTTAAGGATATTTYGGGGAG | 270 |
|
| R:
ACCCCRAAAAAAAAACCA |
|
| P9 | F:
GTTTGGTTTTTTTTTYGG | 315 |
|
| R:
TACTCAAAAATAAAAACTCRTCTC |
|
| P10 | F:
GAGAYGAGTTTTTATTTTTGAGT | 276 |
|
| R:
AAAAAATAAACCCCCAAAT |
|
| P11 | F:
TTGGGGGTTTATTTTTTGT | 257 |
|
| R:
ACATTAAATCAAAATCCRCTAT |
|
Immunofluorescence staining
Tissues were fixed using 4% Paraformaldehyde for 24
h at room temperature. Immunofluorescence staining was performed on
formalin-fixed/paraffin-embedded sections (4 µm), as previously
described (18). Primary
antibodies include rabbit anti-iASPP polyclonal antibody (ab34898;
1:100; Abcam, Cambridge UK), mouse anti-glial fibrillary acidic
protein (GFAP) monoclonal antibody (MAB360; 1:100; EMD Millipore,
Billerica, MA, USA) or mouse anti-neuronal nuclei (NeuN) monoclonal
antibody (MAB377; 1:100; EMD Millipore). Cy3 donkey anti rabbit IgG
(711-165-152; 1:800; Jackson Immunoresearch, West Grove, PA, USA)
and Alexa Fluor 488 donkey anti mouse IgG (715-545-150; 1:800;
Jackson Immunoresearch) were used as secondary antibodies. Cell
nuclei were stained with DAPI (Invitrogen; Thermo Fisher
Scientific, Inc.). Sections were observed under a fluorescence
microscope (Carl Zeiss AG, Oberkochen, Germany). Four fields were
detected per slide.
Cell viability assay
The effects of miR-124 on cell proliferation were
determined by MTT assay. MTT (Sigma-Aldrich; Merck KGaA) was
dissolved in distilled H2O (5 mg/ml) and sterilized
through a 0.22-µm filter. Briefly, U87 and U251 cells
(3×103 cells/well) were seeded into 96-well plates and
transfected at 24 h after seeding. At 0, 24, 48, 72 and 96 h
following transfection, cells were incubated at 37°C and 20 µl MTT
solution was added to each well and cells were incubated for 2 h at
37°C. Following removal of the medium, the colored formazan product
was then dissolved in 150 µl dimethylsulfoxide. Optical density was
measured at 570 nm using a microplate reader (Bio-Rad Laboratories,
Inc.). Triplicate wells were assayed for each time point.
Cell cycle analysis
At 48 h following miRNA transfection at 37°C,
1×106 U87 and U251 cells were collected and fixed
overnight in 70% ethanol at 4°C and subsequently treated with 0.5
mg/ml RNase in PBS at 37°C for 30 min. Cell nuclei were stained
with 50 µg/ml propidium iodide at 4°C for 30 min. Cells
(1×106) were analyzed for their DNA content using BD
FACSDiva software version 8.0.1 (BD Biosciences, Franklin Lakes,
NJ, USA) in a BD Influx flow cytometer (BD Biosciences).
Western blot analysis
Western blotting was performed as previously
described (26). Briefly, ~0.5 g
of tissue was used to extract the protein. The protein
concentration was determined by a Bicinchoninic Acid Protein Assay
kit (Pierce; Thermo Fisher Scientific, Inc.), using bovine serum
albumin (Pierce; Thermo Fisher Scientific, Inc.) as the standard.
Protein samples (100 µg/lane) were separated by 8 or 10% SDS-PAGE
and subsequently transferred to polyvinylidene fluoride membranes
(EMD Millipore). Following blocking in 5% skimmed milk for 2 h at
room temperature, the membranes were incubated overnight at 4°C
with the following antibodies: rabbit anti-CDK4 polyclonal antibody
(sc-260; 1:200; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
mouse anti-CDK6 polyclonal antibody (3136; 1:1,000; Cell Signaling
Technology, Inc., Danvers, MA, USA), rabbit anti-iASPP polyclonal
antibody (ab34898; 1:1,000; Abcam), rabbit anti-Cyclin D1
polyclonal antibody (sc-717; 1:200; Santa Cruz Biotechnology, Inc.)
and rabbit anti-β-actin polyclonal antibody (sc-7210; 1:5,000;
Santa Cruz Biotechnology, Inc.). β-actin was used as an internal
loading control. Membranes were subsequently incubated with
horseradish peroxidase-conjugated goat anti-rabbit secondary
immunoglobulin G-horseradish peroxidase (HRP) (sc-2004; 1:2,000;
Santa Cruz Biotechnology, Inc.) or goat anti-mouse secondary
immunoglobulin G-HRP (sc-2005; 1:2,000; Santa Cruz Biotechnology,
Inc.) in blocking solution for 1 h at room temperature. Protein
bands were visualized by Western Chemiluminescent HRP Substrate
(EMD Millipore). Protein expression levels were normalized to the
β-actin loading control. The relative optical density of protein
bands was measured following subtraction of the film background
using Image-Pro Plus version 6.0 (Media Cybernetics, Inc.,
Rockville, MD, USA).
Wound-healing assay
Cells were seeded into 6-well plates at a density of
8×105 cells per well. At 24 h after seeding, miR-124
mimics and miR-negative controls were transfected into cells with
at least 70% confluence in 6-well plates. Following 6 h
transfection, cell layers were scratched using a 200 µl sterile
pipette tip to form wound gaps and washed immediately with
serum-free medium twice. Wounds were measured at 0 and 24 h. Images
of the wound width were captured for 6 fields in each well and
analyzed using Image-Pro Plus 6.0 (Media Cybernetics, Inc.).
Relative migration rate=(start distance-end distance)/start
distance.
Statistical analysis
Group distributions were compared parametrically
using the Student's t-test or by one-way analysis of the variance
with Scheffe's post hoc test; group distributions were compared
non-parametrically using the Mann-Whitney U-test. All analyses were
done using GraphPad Prism software version 5.01 (GraphPad Software,
Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-124 in astrocytic
glioma tissues and cell lines
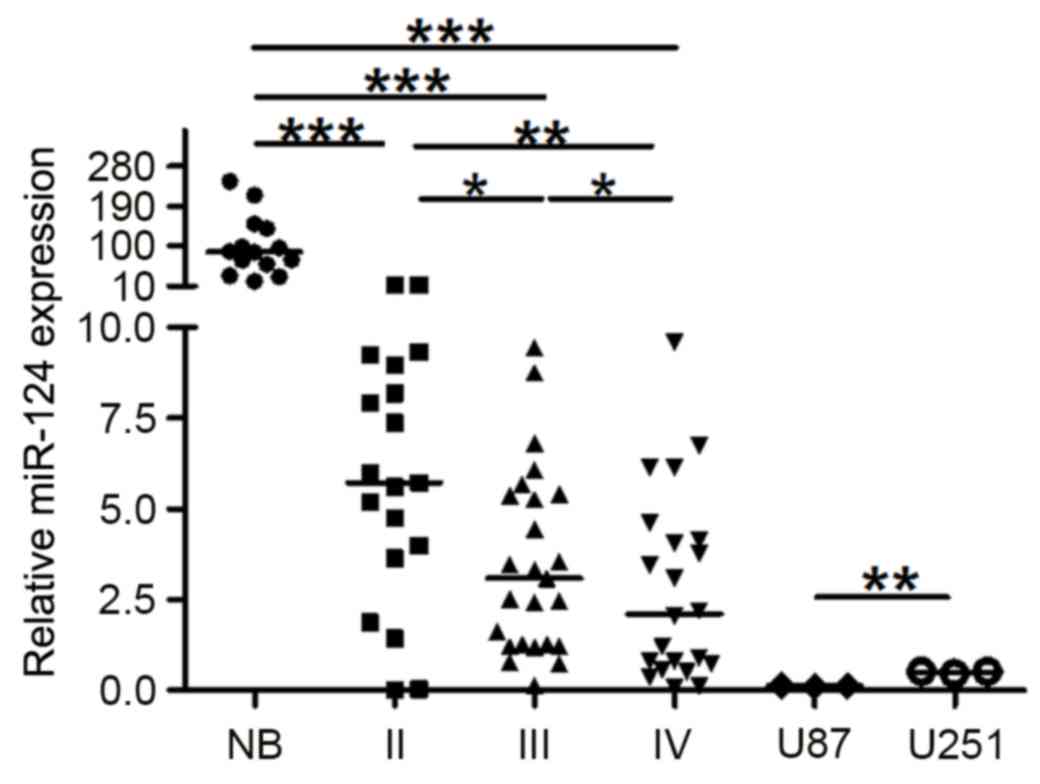
The expression levels of miR-124 were quantified by
RT-qPCR in 66 astrocytoma (grade II, 19; grade III, 25; grade IV,
22), 14 healthy NB control tissues and the glioma cell lines, U87
and U251 (Fig. 1). miR-124
expression was significantly lower in the three astrocytic glioma
tissues compared with NB controls (P<0.001). The expression
level of miR-124 in low-grade gliomas (grade II) was significantly
higher compared with expression in the high-grade gliomas
(P<0.05 vs. grade III; P<0.01 vs. grade IV), whereas miR-124
expression was lower in grade IV gliomas compared with grade III
(P<0.05). miR-124 expression in gliomas is not influenced by
gender or age (data not shown). These results indicated that the
downregulation of miR-124 may be related to the degree of
malignancy of the glioma, and suggested a putative crucial role for
downregulation of miR-124 in the initiation and progression of
glioma. Although U87 and U251 are both human glioblastoma cell
lines and demonstrated low expression level of miR-124, the
expression level of miR-124 in U87 cells was significantly lower
compared with U251 cells (Fig. 1;
P<0.01).
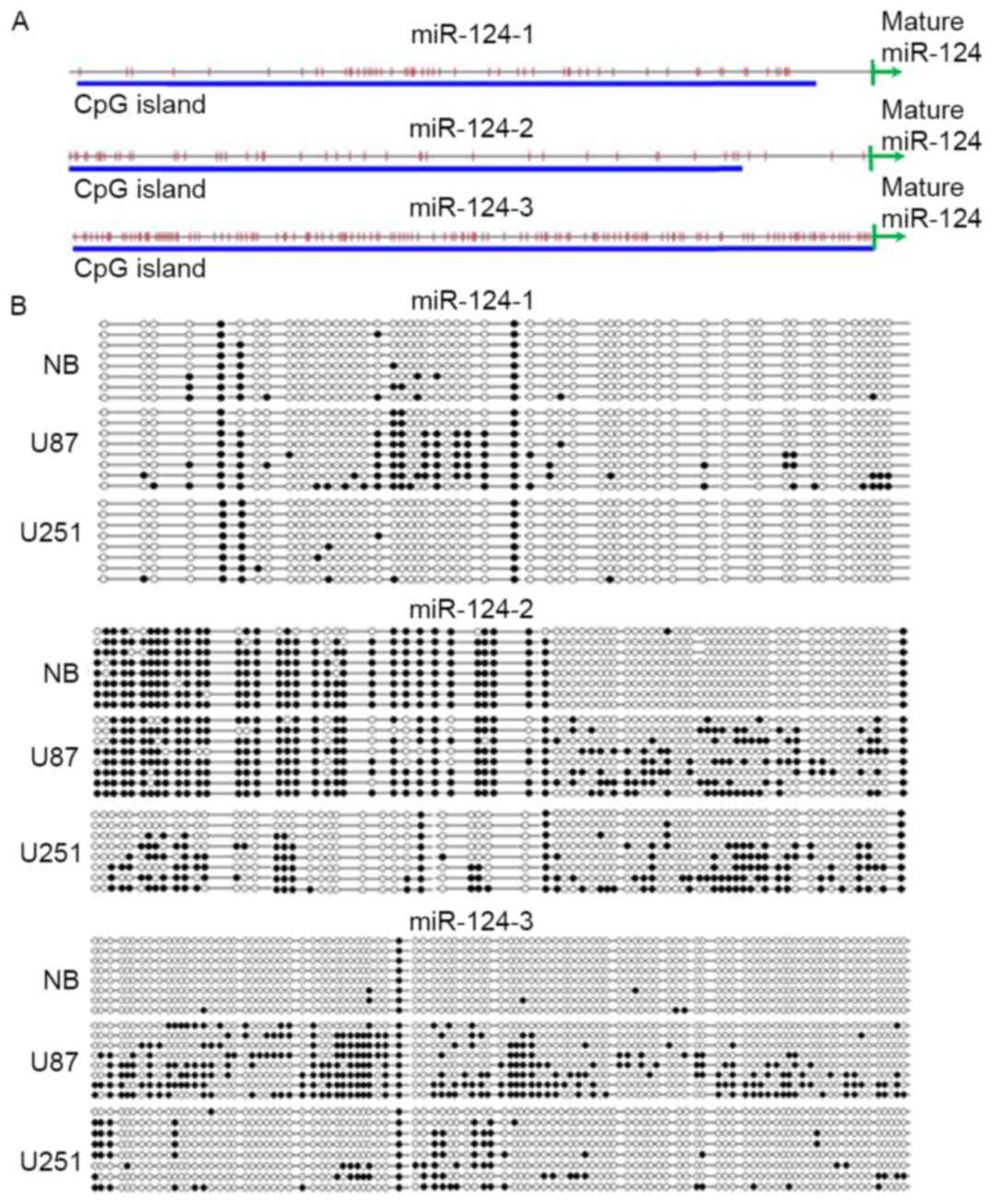
Methylation of the miR-124 promoter in
U87 and U251 cells
The present study also investigated whether the
difference in the expression level of miR-124 between U87 and U251
cells was due to methylation status of the miR-124 promoter. It has
been predicted that more than 90% of the human miRNA promoters are
located 1,000 bp upstream of DNA sequence which can be transcribed
to the mature miRNA. (27).
miR-124 is transcribed from three loci: hsa-miR-124-1,
has-miR-124-2 and has-miR-124-3. Thus, PCR primers were designed in
the 1,000 bp upstream region flanking the miR-124 sequence. These
sequences flanking hsa-miR-124-1, has-miR-124-2 and has-miR-124-3
were located in gi|224589820: c9761982-9760983,
gi|224589820:65290706-65291705 and gi|224589812:61808852-61809851,
respectively (https://www.ncbi.nlm.nih.gov/gene; Fig. 2A). Bisulfite genomic sequencing
results confirmed the hypomethylation of miR-124-1, miR-124-2 and
miR-124-3 CpG islands in astrocytes from human control tissues and
hypermethylation of miR-124-1, miR-124-2 and miR-124-3 in U87 and
U251 cells (Fig. 2B). miR-124 loci
were more highly methylated in U87 cells compared with U251 cells.
These methylation data were consistent with the RT-qPCR result in
these cells, which demonstrated low expression of miR-124 in U87
cells and relatively high expression in U251 cells.
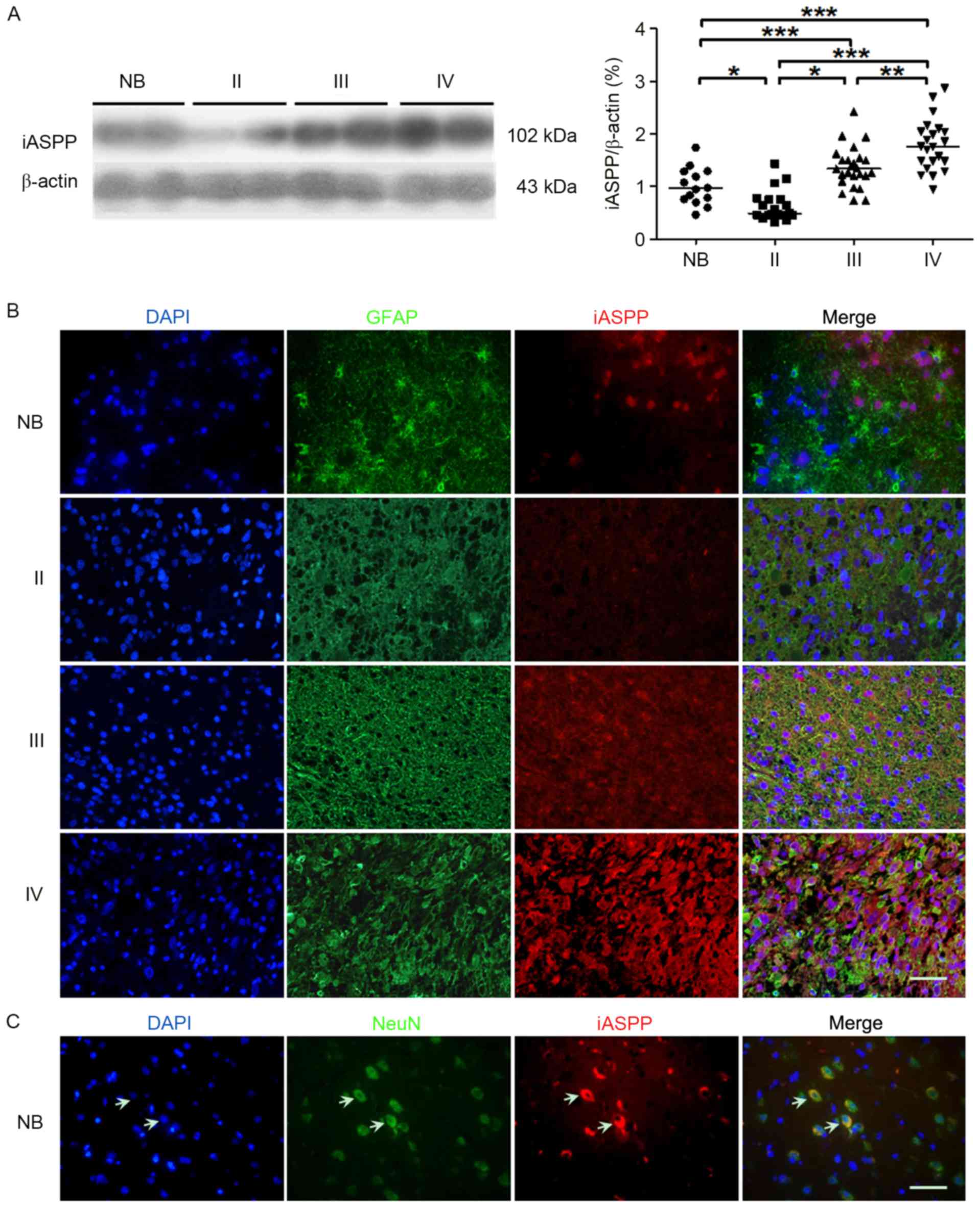
Expression of iASPP protein in
astrocytic glioma tissues
To determine the relationship between the expression
of iASPP and astrocytoma grades, iASPP protein expression levels
were detected by western blotting and immunofluorescence. The
result demonstrated that iASPP protein expression levels
significantly increased with advancing WHO gliomas grade (Fig. 3A). However, the protein expression
level of iASPP was significantly lower in WHO grade I glioma
compared with NB control (P<0.05). The cellular distribution of
iASPP was examined by immunofluorescence staining. iASPP expression
was almost undetected in astrocytes from NB, while gradually
increased in Grad II to Grade IV astrocytomas. Highly positive
staining of iASPP was observed in the neurons of NB (Fig. 3B and C).
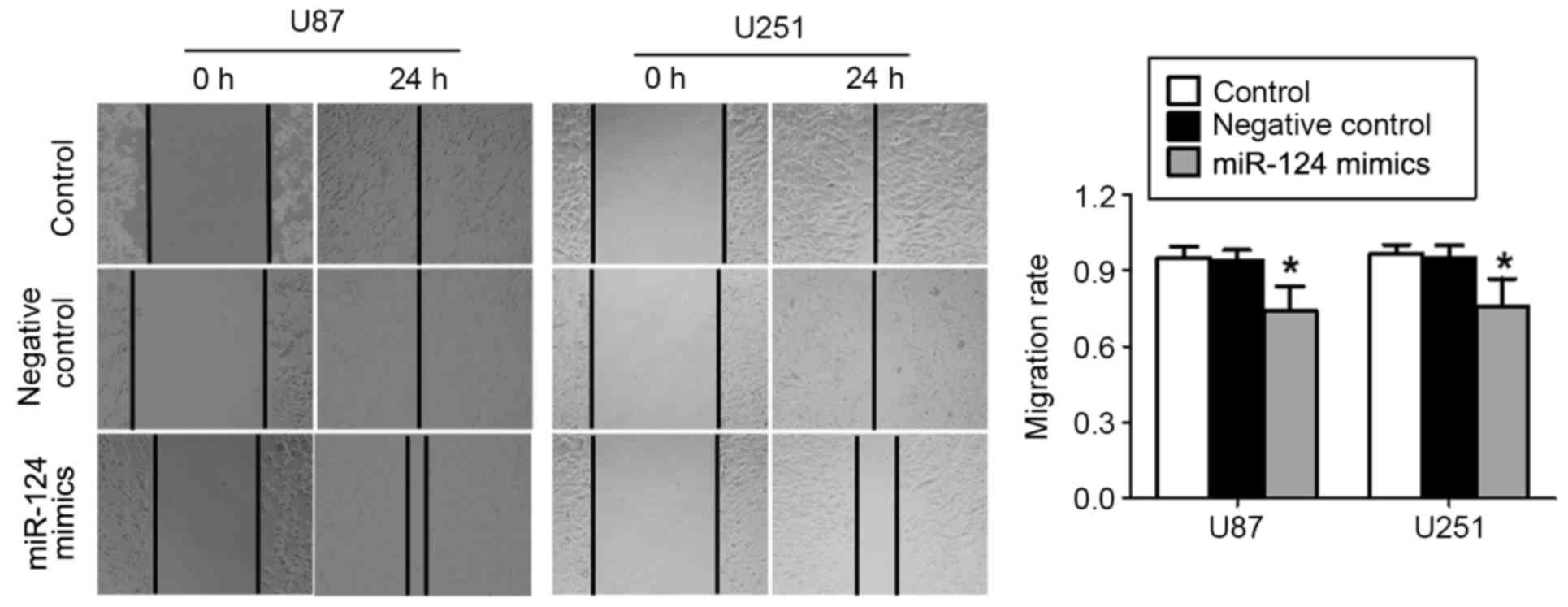
miR-124 inhibits U87 and U251 cell
migration
To confirm the effects of miR-124 on the glioma cell
migration, wound-healing assays were performed in U87 and U251
cells that were transfected with miR-124 mimics or miR-negative
control oligonucleotides. The results revealed that miR-124 mimics
significantly inhibited wound closure in U87 and U251 cells
compared with cells in the miR-negative control groups (P<0.05;
Fig. 4). These results indicated
that the upregulation of miR-124 expression attenuated the
migratory ability of glioma cells.
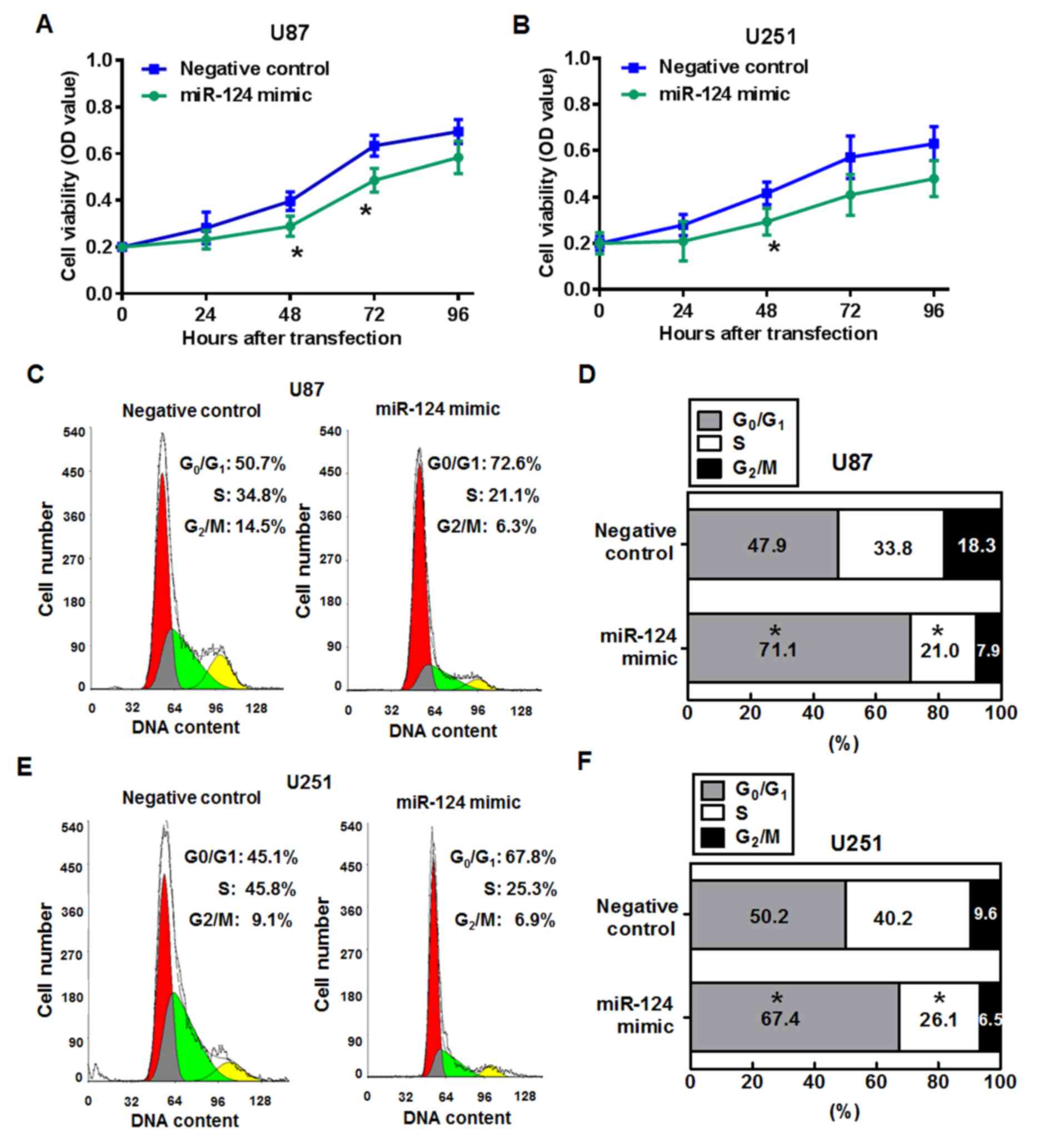
miR-124 inhibits cell viability and
induces cell-cycle arrest in glioma cells
To determine whether miR-124 affects cell growth and
proliferation, MTT assays and cell cycle distribution assays were
performed. Compared with the miR-negative control group, U87 cells
transfected with miR-124 mimics exhibited reduced viability at 48
and 72 h post-transfection (P<0.05; Fig. 5A). miR-124 mimics transfection
significantly inhibited cell viability in U251 cells at 48 h
compared with cells transfected with the miR-negative control
oligonucleotides (P<0.05; Fig.
5B).
The cell cycle status was analyzed in U87 and U251
cells using the flow cytometer 48 h post-transfection with miR-124
mimics or miR-negative control (Fig.
5C-F). The percentage of cells at S phase cells was
significantly lower in miR-241 mimics transfected U87 cells
(21.0±5.7%) compared with the percentage of S phase cells in the
miR-negative control group (33.8±4.1%; P<0.05; Fig. 5C and D), whereas the percentage of
G0/G1 phase cells was significantly higher in miR-124 mimics
transfected cells (71.1±6.9%) compared with the miR-negative
controls (47.9±3.8%; P<0.05). Similarly, the proportion of U251
cells in the S-phase fraction was significantly lower following
miR-124 mimics transfection (26.1±5.7%) compared with miR-negative
control transfection (40.2±5.9%; P<0.05; Fig. 5E and F), and the proportion of
cells in the G0/G1 phase was significantly
higher in themiR-124 mimics treated group (50.2±3.8%) compare with
the miR-negative control group (67.4±6.9%; P<0.05). These
results suggested that the reduction in cell viability at 48 h may
occur through cell cycle arrest.
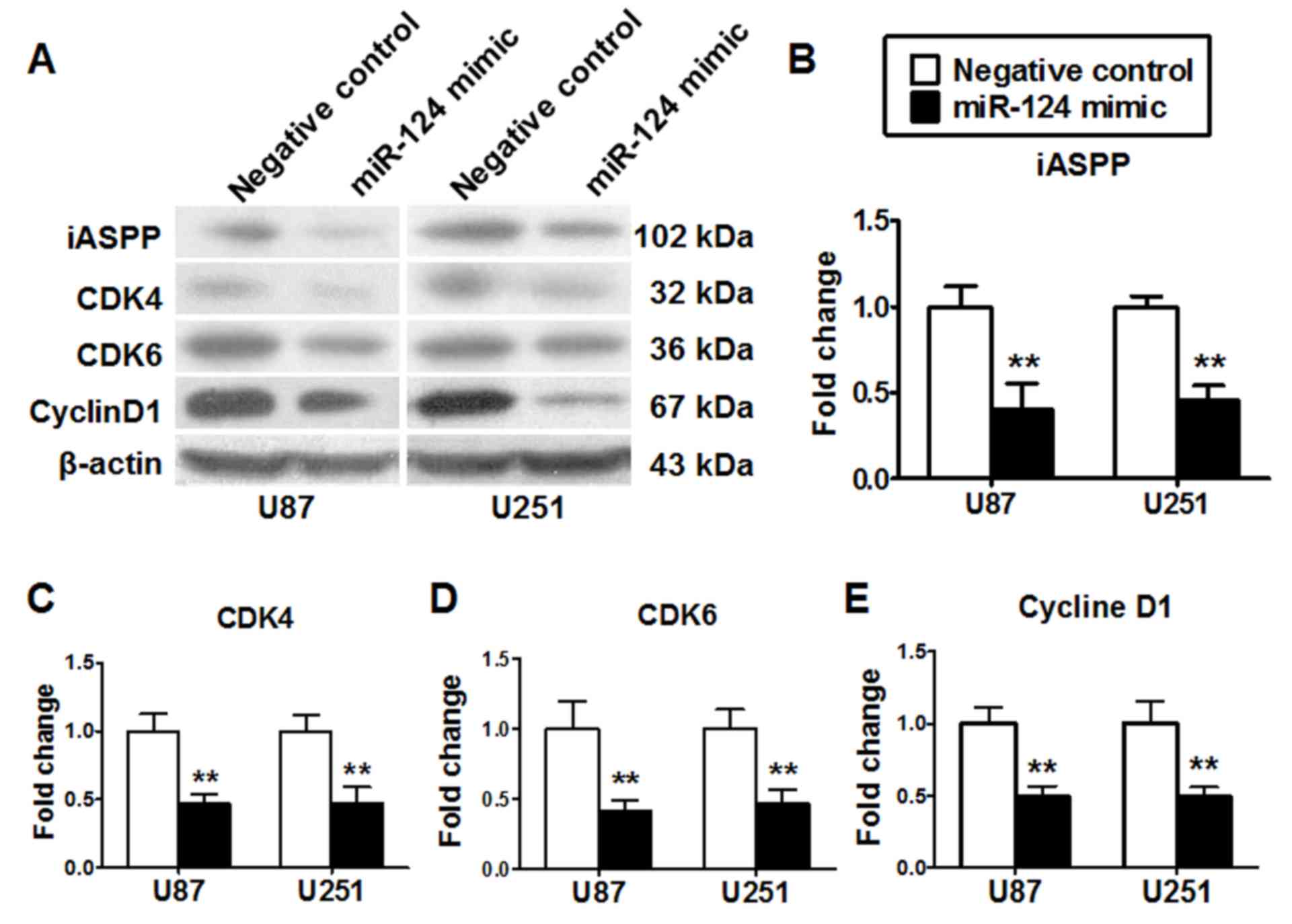
miR-124 inhibits the expression of
target gene associated with cell cycle regulation
The effects of miR-124 on the expression of its
target genes were determined by western blot. The protein
expression levels of the miR-124 target genes CDK4, CDK6 and iASPP,
and cell cycle regulatory protein cyclin D1 were examined in U87
and U251 cells 48 h post-transfection with miR-124 mimics or
miR-negative control. The results demonstrated that miR-124 mimics
transfection led to decreased expression levels of CDK4, CDK6,
iASPP and cyclin D1 in U87 and U251 cells compared with cells
transfected with the miR-negative control oligonucleotides
(P<0.01; Fig. 6).
Discussion
miR-124 is a tumor suppressor, and its expression is
significantly decreased in many types of cancer (28,29).
miR-124 expression has been reported to decrease tumor initiation,
promotion and progression in different cell culture systems and in
animal models (11–13). In the present study, reduced
expression of miR-124 was observed in astrocytoma tissue, with a
more pronounced reduction in high-grade gliomas, which was
consistent with previous reports (11,12).
These findings indicated that the low expression levels of miR-124
may be involved in the pathological progression of astrocytoma.
Epigenetic silencing of miRNAs together with their
target tumor suppressors is becoming a common hallmark of human
tumors (30). DNA
methylation-based silencing is functionally involved in
carcinogenesis (31). Epigenetic
silencing of miR-124 by hypermethylation of CpG islands frequently
occurs in different tumors, including leukemia, lung cancer and
colon cancer (28). The present
study demonstrated that the expression level of miR-124 was lower
in U87 cells compared with U251 cells, which may be due to a higher
level of methylation in the miR-124 promoter region in U87 cells. A
recent study demonstrated that miR-124 expression is upregulated in
glioblastoma cell lines following treatment with a combination of
5-aza-2′-deoxycytidine (decitabine), a DNA demethylating agent, and
trichostatin A (TSA), a histone deacetylase inhibitor, whereas no
significant difference in expression was observed in cells treated
with decitabine or TSA alone (31). These data suggested that the
expression of miR-124 was not regulated by DNA methylation alone,
but by a variety of epigenetic modifications. Further
investigations are required to define the full extent of DNA
methylation that affects miR-124 expression in glioma cell lines
and tumors.
As a direct target of miR-124, iASPP is involved in
cellular processes, including cell cycle, apoptosis and autophagy
(32–34). iASPP is upregulated in a number of
tumors, such as leukemia, lung cancer and hepatocellular carcinoma
(35–37). In the present study, iASPP was
expressed at low levels in the astrocytes of NB tissue, whereas
expression was high in NB neurons, which may be the reason that the
expression level of iASPP is higher in NB tissues compared with
astrocytoma Grade II. The present study did not detect the
expression of iASPP in astrocytes of NB tissue by western blotting;
therefore, thorough investigations need to be conducted to explore
the expression and role of iASPP both in astrocytoma cells and in
all kinds of cell types of NB.
Results from the present study demonstrated that
miR-124 may be involved in regulating cell viability and migration
in human glioblastoma cells. Overexpression of miR-124 resulted in
significant changes in cell migration and viability, and miR-124
may regulate glioblastoma cell viability by arresting the cell
cycle at the G0/G1 phase. The cell cycle is tightly regulated by a
number of molecular pathways and checkpoints, which may be altered
in tumors. miR-124 has a variety of mRNA targets that are
associated with the cell cycle, such as CDK6, iASPP and CDK4
(17,18,38).
In this study, the results of these previously published reports
were confirmed; further suggesting that miR-124 may influence the
cell cycle in glioma cells. CDK4, CDK6 and iASPP have recently been
identified as direct targets of miR-124 (17,18,38).
iASPP was reported to downregulate the expression of cyclin D1
(32), which controls the
transition from the G1 phase to the S phase. Another direct target
of miR-124 is IQGAP1 (not examined by the present study), which is
suppressed by miR-124 leading to a downregulation of β-catenin and
downstream cyclin D1 expression (19).
In conclusion, the present results indicated that
miR-124 is downregulated in glioma cells and this reduced
expression level may promote tumor growth and progression. In
addition, miR-124 may serve as a potential biomarker for
pathological diagnosis and represents a promising therapeutic
target for the treatment of glioma.
Acknowledgements
The authors thank Yanglong Li and Kaiyuan Song
(Beijing Tongren Hospital, Capital Medical University, Beijing,
China) for collecting tissue samples. This work was supported by
The National Natural Science Foundation of China (grant nos.
81000504, 81471209 and 81641055) and The Beijing Natural Science
Foundation (grant no. 7132112).
References
|
1
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012. View Article : Google Scholar
|
|
2
|
Dunbar E and Yachnis AT: Glioma diagnosis:
Immunohistochemistry and beyond. Adv Anat Pathol. 17:187–201. 2010.
View Article : Google Scholar
|
|
3
|
Shenoy A and Blelloch RH: Regulation of
microRNA function in somatic stem cell proliferation and
differentiation. Nat Rev Mol Cell Biol. 15:565–576. 2014.
View Article : Google Scholar :
|
|
4
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar :
|
|
5
|
Garofalo M and Croce CM: Role of microRNAs
in maintaining cancer stem cells. Adv Drug Deliv Rev. 81:53–61.
2015. View Article : Google Scholar
|
|
6
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar
|
|
7
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar
|
|
8
|
Kloosterman WP, Wienholds E, de Bruijn E,
Kauppinen S and Plasterk RH: In situ detection of miRNAs in animal
embryos using LNA-modified oligonucleotide probes. Nat Methods.
3:27–29. 2006. View
Article : Google Scholar
|
|
9
|
Cao X, Pfaff SL and Gage FH: A functional
study of miR-124 in the developing neural tube. Genes Dev.
21:531–536. 2007. View Article : Google Scholar :
|
|
10
|
Cao Q, Li YY, He WF, Zhang ZZ, Zhou Q, Liu
X, Shen Y and Huang TT: Interplay between microRNAs and the STAT3
signaling pathway in human cancers. Physiol Genomics. 45:1206–1214.
2013. View Article : Google Scholar
|
|
11
|
Chen T, Wang XY, Li C and Xu SJ:
Downregulation of microRNA-124 predicts poor prognosis in glioma
patients. Neurol Sci. 36:131–135. 2015. View Article : Google Scholar
|
|
12
|
Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang
C, Liu X, Wang X, Li H, Kang C, et al: MiR-124 governs glioma
growth and angiogenesis and enhances chemosensitivity by targeting
R-Ras and N-Ras. Neuro Oncol. 16:1341–1353. 2014. View Article : Google Scholar :
|
|
13
|
An L, Liu Y, Wu A and Guan Y: microRNA-124
inhibits migration and invasion by down-regulating ROCK1 in glioma.
PLoS One. 8:e694782013. View Article : Google Scholar :
|
|
14
|
Shkumatava A, Stark A, Sive H and Bartel
DP: Coherent but overlapping expression of microRNAs and their
targets during vertebrate development. Genes Dev. 23:466–481. 2009.
View Article : Google Scholar :
|
|
15
|
Clark AM, Goldstein LD, Tevlin M, Tavaré
S, Shaham S and Miska EA: The microRNA miR-124 controls gene
expression in the sensory nervous system of Caenorhabditis elegans.
Nucleic Acids Res. 38:3780–3793. 2010. View Article : Google Scholar :
|
|
16
|
Chi SW, Zang JB, Mele A and Darnell RB:
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature.
460:479–486. 2009.
|
|
17
|
Pierson J, Hostager B, Fan R and Vibhakar
R: Regulation of cyclin dependent kinase 6 by microRNA 124 in
medulloblastoma. J Neurooncol. 90:1–7. 2008. View Article : Google Scholar
|
|
18
|
Liu X, Li F, Zhao S, Luo Y, Kang J, Zhao
H, Yan F, Li S and Ji X: MicroRNA-124-mediated regulation of
inhibitory member of apoptosis-stimulating protein of p53 family in
experimental stroke. Stroke. 44:1973–1980. 2013. View Article : Google Scholar
|
|
19
|
Lu SH, Jiang XJ, Xiao GL, Liu DY and Yuan
XR: miR-124a restoration inhibits glioma cell proliferation and
invasion by suppressing IQGAP1 and β-catenin. Oncol Rep.
32:2104–2110. 2014. View Article : Google Scholar
|
|
20
|
Liu Z, Zhang X, Huang D, Liu Y, Zhang X,
Liu L, Li G, Dai Y, Tan H, Xiao J and Tian Y: Elevated expression
of iASPP in head and neck squamous cell carcinoma and its clinical
significance. Med Oncol. 29:3381–3388. 2012. View Article : Google Scholar
|
|
21
|
Wang LL, Xu Z, Peng Y, Li LC and Wu XL:
Downregulation of inhibitor of apoptosis-stimulating protein of p53
inhibits proliferation and promotes apoptosis of gastric cancer
cells. Mol Med Rep. 12:1653–1658. 2015. View Article : Google Scholar :
|
|
22
|
Zhao WH, Wu SQ and Zhang YD:
Downregulation of miR-124 promotes the growth and invasiveness of
glioblastoma cells involving upregulation of PPP1R13L. Int J Mol
Med. 32:101–107. 2013. View Article : Google Scholar
|
|
23
|
Zhang Y, Zheng L, Huang J, Gao F, Lin X,
He L, Li D, Li Z, Ding Y and Chen L: MiR-124 Radiosensitizes human
colorectal cancer cells by targeting PRRX1. PLoS One. 9:e939172014.
View Article : Google Scholar :
|
|
24
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
25
|
Wang D, Zhang H, Li M, Frid MG, Flockton
AR, McKeon BA, Yeager ME, Fini MA, Morrell NW, Pullamsetti SS, et
al: MicroRNA-124 controls the proliferative, migratory and
inflammatory phenotype of pulmonary vascular fibroblasts. Circ Res.
114:67–78. 2014. View Article : Google Scholar
|
|
26
|
Liu X, Wang L, Zhao S, Ji X, Luo Y and
Ling F: β-catenin overexpression in malignant glioma and its role
in proliferation and apoptosis in glioblastoma cells. Med Oncol.
28:608–614. 2011. View Article : Google Scholar
|
|
27
|
Zhou X, Ruan J, Wang G and Zhang W:
Characterization and identification of microRNA core promoters in
four model species. PLoS Comput Biol. 3:e372007. View Article : Google Scholar :
|
|
28
|
Sato F, Tsuchiya S, Meltzer SJ and Shimizu
K: MicroRNAs and epigenetics. FEBS J. 278:1598–1609. 2011.
View Article : Google Scholar
|
|
29
|
Papagiannakopoulos T and Kosik KS:
MicroRNAs: Regulators of oncogenesis and stemness. BMC Med.
6:152008. View Article : Google Scholar :
|
|
30
|
Toiyama Y, Okugawa Y and Goel A: DNA
methylation and microRNA biomarkers for noninvasive detection of
gastric and colorectal cancer. Biochem Biophys Res Commun.
455:43–57. 2014. View Article : Google Scholar :
|
|
31
|
Silber J, Lim DA, Petritsch C, Persson AI,
Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello
JF, et al: miR-124 and miR-137 inhibit proliferation of
glioblastoma multiforme cells and induce differentiation of brain
tumor stem cells. BMC Med. 6:142008. View Article : Google Scholar :
|
|
32
|
Li G, Wang R, Gao J, Deng K, Wei J and Wei
Y: RNA interference-mediated silencing of iASPP induces cell
proliferation inhibition and G0/G1 cell cycle arrest in U251 human
glioblastoma cells. Mol Cell Biochem. 350:193–200. 2011. View Article : Google Scholar
|
|
33
|
Cai Y, Qiu S, Gao X, Gu SZ and Liu ZJ:
iASPP inhibits p53-independent apoptosis by inhibiting
transcriptional activity of p63/p73 on promoters of proapoptotic
genes. Apoptosis. 17:777–783. 2012. View Article : Google Scholar
|
|
34
|
Chikh A, Sanzà P, Raimondi C, Akinduro O,
Warnes G, Chiorino G, Byrne C, Harwood CA and Bergamaschi D: iASPP
is a novel autophagy inhibitor in keratinocytes. J Cell Sci.
127:3079–3093. 2014. View Article : Google Scholar
|
|
35
|
Deng Q, Sheng L, Su D, Zhang L, Liu P, Lu
K and Ma S: Genetic polymorphisms in ATM, ERCC1, APE1 and iASPP
genes and lung cancer risk in a population of southeast China. Med
Oncol. 28:667–672. 2011. View Article : Google Scholar
|
|
36
|
Zhang X, Wang M, Zhou C, Chen S and Wang
J: The expression of iASPP in acute leukemias. Leuk Res.
29:179–183. 2005. View Article : Google Scholar
|
|
37
|
Lin BL, Xie DY, Xie SB, Xie JQ, Zhang XH,
Zhang YF and Gao ZL: Down-regulation of iASPP in human
hepatocellular carcinoma cells inhibits cell proliferation and
tumor growth. Neoplasma. 58:205–210. 2011. View Article : Google Scholar
|
|
38
|
Deng X, Ma L, Wu M, Zhang G, Jin C, Guo Y
and Liu R: miR-124 radiosensitizes human glioma cells by targeting
CDK4. J Neurooncol. 114:263–274. 2013. View Article : Google Scholar
|