Introduction
Sepsis is a life-threatening condition that is
caused by the entry of bacteria, fungi, viruses or parasites to the
bloodstream, which subsequently triggers a systemic uncontrolled
inflammatory response (1). Sepsis
is be divided into three classifications with increasing severity:
Sepsis, severe sepsis and septic shock (2). Severe sepsis is often complicated by
septic shock and multiple organ dysfunction syndrome, and the
incidence and mortality rate of sepsis is higher in elderly
patients (3,4). Additionally, sepsis syndrome and
septic shock are the major cause of mortality in intensive care
units. More than 750,000 cases of sepsis occur in the United States
each year (5), which result in
more than 210,000 deaths per year (6).
At present, the exact mechanism of sepsis
pathophysiology is not clear. A previous study reported that the
pathophysiology includes highly complex interactions between
invading microorganisms, the innate and adaptive immune systems of
the host and multiple downstream events, which lead to organ
dysfunction (7). In addition,
Calvano et al (8)
demonstrated that ~25% of normal human gene expression was altered
in sepsis. Hypermorphic genetic variation in TLR1 is reported to
also have roles in the clinical outcomes of sepsis (9). Despite clinical advances, the
mortality rate of sepsis has not considerably reduced in the last
10 years. (10). Early diagnosis
and treatment is crucial in order to reduce the mortality rate in
patients with sepsis.
The present study aimed to identify differentially
expressed genes (DEGs) in sepsis using integrated analysis.
Functional enrichment analysis using the Gene Ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) was subsequently
performed to investigate the biological functions of the identified
DEGs. A protein-protein interaction (PPI) network was subsequently
constructed from the top 40 DEGs (20 upregulated and 20
downregulated). Furthermore, reverse transcription
quantitative-polymerase chain reaction (RT-qPCR) was performed to
analyze the expression of candidate DEGs in patients with sepsis
and healthy individuals to validate bioinformatics results.
Receiver operating characteristic (ROC) analysis was performed to
analyze the diagnostic ability of the identified DEGs in sepsis.
The DEGs in the present study may have potential as diagnostic
biomarkers or aid the development of novel therapeutics.
Materials and methods
Gene expression profiles
In the current study, relevant datasets from the
Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with the keywords
‘sepsis’ [Medical Subject Headings (MeSH) Terms] OR ‘sepsis’ (All
Fields) AND ‘Homo sapiens’ [Organism name (porgn)] AND ‘gse’
(Filter). The study type was characterized as ‘expression profiling
by array.’ All selected datasets consisted of genome-wide
expression data from blood samples of patients with sepsis and/or
healthy samples. Only standardized or primary datasets were
included in this study. Two datasets were ultimately selected for
screening. The details of datasets GSE69528 (11) and GSE46955 (12) are presented in Table I.
 | Table I.mRNA expression datasets of sepsis
employed in the current study. |
Table I.
mRNA expression datasets of sepsis
employed in the current study.
| GEO accession
no. | Author | Year | Platform | Samples (N:P) | (Refs.) |
|---|
| GSE69528 | Pankla et
al | 2015 | GPL10558 Illumina
HumanHT-12 V4.0 Expression BeadChip | 28:29 | (11) |
| GSE46955 | Wu et
al | 2014 | GPL6104 Illumina
HumanRef-8 v2.0 Expression BeadChip | 12:16 | (12) |
Identification of DEGs
The raw expression data of patients with sepsis was
obtained from the GEO database. The significantly DEGs were
identified by comparison between patients with sepsis and healthy
controls. P-values were combined using the meta-analysis for
microarrays (metaMA version 3.3.3) package (13) in the R software 3.3.3 (https://www.r-project.org/). The false discovery rate
(FDR) was calculated for multiple testing corrections of the raw
P-value through the Benjamin and Hochberg method (14,15).
The threshold for DEG identification was set as FDR <0.05.
Functional and pathway enrichment
analyses of DEGs
To obtain the biological function and signaling
pathways of DEGs, GeneCodis3 (http://genecodis.cnb.csic.es/analysis) was used for
the GO (http://www.geneontology.org/)
annotation and KEGG (http://www.genome.jp/kegg/pathway.html) pathway
enrichment of DEGs. The threshold for GO and KEGG analysis of DEGs
was set at FDR <0.05.
PPI network construction
To gain insight into the interaction between DEGs
and proteins, the BioGRID database (http://thebiogrid.org) was used to obtain the
predicted interactions between the top 40 proteins encoded by DEGs
(20 upregulated and 20 downregulated) and other associated
proteins. The PPI network was subsequently visualized using
Cytoscape software 3.3.0 (http://cytoscape.org/). A node in the PPI network
denotes a protein and an edge denotes interactions between
proteins.
RT-qPCR validation
Blood samples were obtained from 3 male patients
(average, 71) diagnosed with sepsis and 3 male healthy individuals
(average, 71) from Taizhou People's Hospital during November, 2016.
The collected blood was then immediately frozen in liquid nitrogen.
All participating individuals enrolled in the present study
provided informed consent and approval was obtained from the ethics
committee of Taizhou People's Hospital (Taizhou, China). Total RNA
was extracted from the sepsis and control blood samples using
TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's protocol.
cDNA was synthesized using the SuperScript III First-Strand
Synthesis System (Invitrogen; Thermo Fisher Scientific, Inc.) for 5
min at 65°C followed by 60 min at 42°C and 5 min at 72°C. qPCR was
performed using the SYBR Green PCR Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the Applied Biosystems 7500
RT-PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The amplification process was performed under the following
conditions: 15 min at 95°C followed by 40 cycles of 10 sec at 95°C,
30 sec at 55°C, 32 sec at 72°C, and 15 sec at 95°C, 60 sec at 60°C
and 15 sec extension at 95°C. The sequences of reverse and forward
primers for all of the genes analyzed were as follows: IRAK3
forward (F), CAG CCA GTC TGA GGT TAT GTT T and reverse (R), TTG GGA
ACC AAC TTT CTT CAC A; ADM F, CTT ATT CGG CCC CAG GAC ATG, and R,
GCG ACG TTG TCC TTG TCC TTA; ALOX5 F, TGA GCC AGT TCC AGG AAA ACG
and R, ATG GCC ACA CTG TTC GGA ATC; MMP9 F, TGT ACC GCT ATG GTT ACA
CTC G and R, GGC AGG GAC AGT TGC TTC T; S100A8 F, CAG CCC TGC ATG
TCT CTT GTC and R, CCC TGT AGA CGG CAT GGA AAT; SOCS3 F, CAT GGA
GAG GGA CCC AGC ATA and R, GAC ATT CCC AGT GCT CAG CTG; CD14 F, GAC
CTA AAG ATA ACC GGC ACC and R, GCA ATG CTC AGT ACC TTG AGG; ENTPD1
F, AGG TGC CTA TGG CTG GAT TAC and R, CCA AAG CTC CAA AGG TTT CCT;
GAPDH F, GGA GCG AGA TCC CTC CAA AAT and R, GGC TGT TGT CAT ACT TCT
CAT GG.
GAPDH was used as an internal control for gene
detection. The experiment was repeated three times. The relative
expression of genes was calculated using the 2−ΔΔcq
equation (16). Results are
presented as fold change (2−ΔΔcq; patient/control)
values; 2−ΔΔcq >1 indicates an upregulated gene and
2−ΔΔcq <1 indicates a downregulated gene.
ROC analysis
ROC analysis was performed to assess the diagnostic
value of differentially expressed microRNA targets, using the pROC
package 3.3.3 (http://web.expasy.org/pROC/) in the R software 3.3.3
(https://www.r-project.org/). An ROC
curve was generated and a binomial exact confidence interval was
calculated for the area under the curve (AUC).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA).
For the RT-qPCR validation experiment, the difference in mRNA
expression between patients with sepsis and healthy controls was
statistically analyzed by one-way analysis of variance. Data is
presented as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
DEG analysis
A total of 4,402 DEGs were identified with the
threshold of FDR<0.05, including 1,960 upregulated DEGs and
2,442 downregulated DEGs in samples from patients with sepsis
compared with healthy individuals. The top 20 DEGs are listed in
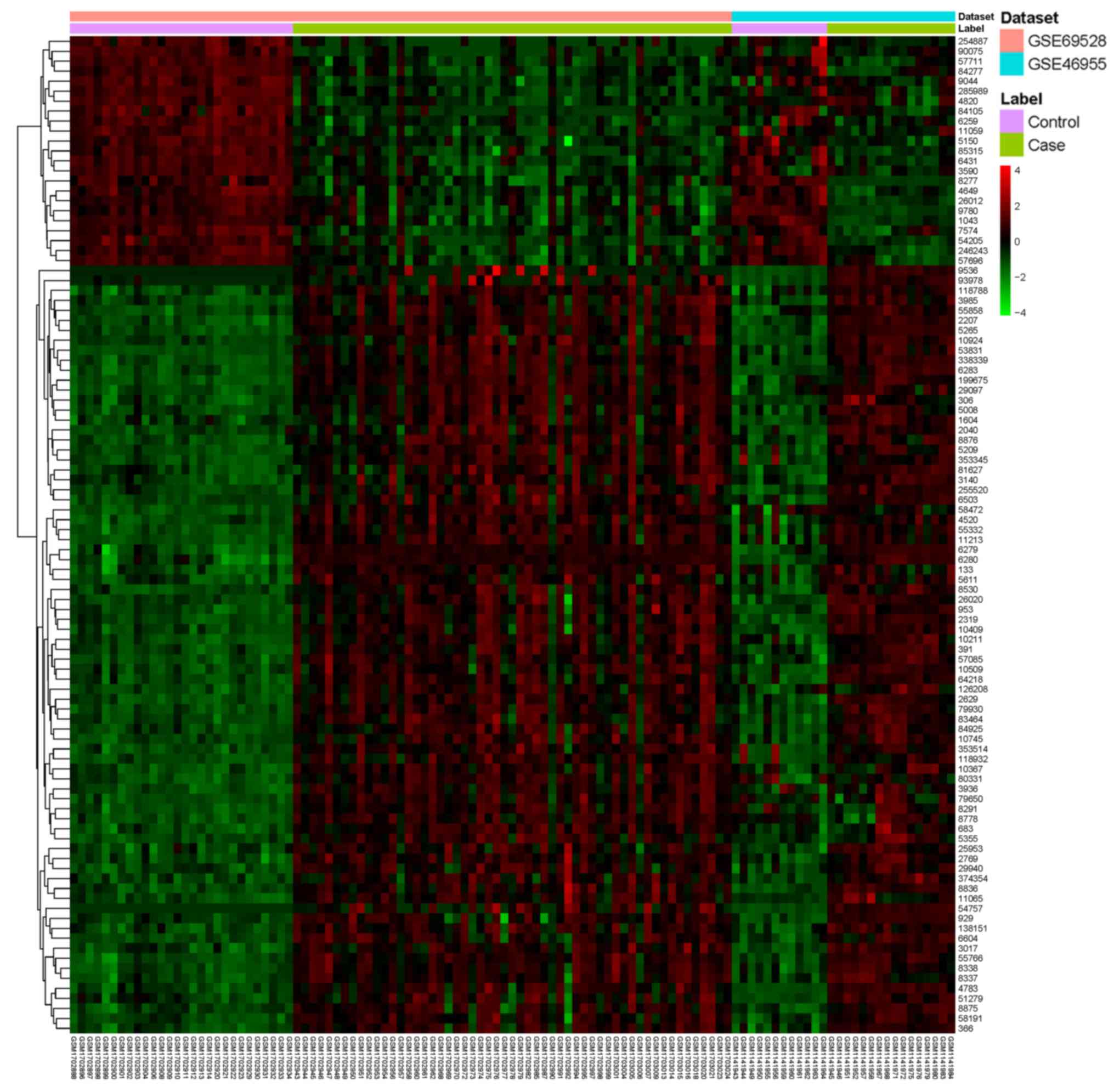
Table II and a heatmap of the top
100 DEGs is presented in Fig.
1.
 | Table II.Top 20 differentially expressed genes
between sepsis and control samples within GSE69528 and GSE46955
datasets. |
Table II.
Top 20 differentially expressed genes
between sepsis and control samples within GSE69528 and GSE46955
datasets.
| NCBI ID | Symbol | Combined ES | P-value | FDR |
Upregulated/downregulated |
|---|
| 6279 | S100A8 | 3.624648 | <0.001 | <0.001 | Upregulated |
| 2207 | FCER1G | 3.597326 | <0.001 | <0.001 | Upregulated |
| 199675 | C19orf59 | 3.516052 | <0.001 | <0.001 | Upregulated |
| 4783 | NFIL3 | 3.510366 | <0.001 | <0.001 | Upregulated |
| 55766 | H2AFJ | 3.477834 | <0.001 | <0.001 | Upregulated |
| 6280 | S100A9 | 3.438282 | <0.001 | <0.001 | Upregulated |
| 338339 | CLEC4D | 3.394467 | <0.001 | <0.001 | Upregulated |
| 5265 | SERPINA1 | 3.378025 | <0.001 | <0.001 | Upregulated |
| 6283 | S100A12 | 3.329135 | <0.001 | <0.001 | Upregulated |
| 54757 | FAM20A | 3.188326 | <0.001 | <0.001 | Upregulated |
| 8836 | GGH | 3.15312 | <0.001 | <0.001 | Upregulated |
| 2629 | GBA | 3.062176 | <0.001 | <0.001 | Upregulated |
| 2319 | FLOT2 | 3.016056 | <0.001 | <0.001 | Upregulated |
| 3017 | HIST1H2BD | 2.983884 | <0.001 | <0.001 | Upregulated |
| 53831 | GPR84 | 2.97356 | <0.001 | <0.001 | Upregulated |
| 55332 | DRAM1 | 2.92859 | <0.001 | <0.001 | Upregulated |
| 8277 | TKTL1 | −2.90765 | <0.001 | <0.001 | Downregulated |
| 57085 | AGTRAP | 2.902596 | <0.001 | <0.001 | Upregulated |
| 11213 | IRAK3 | 2.897167 | <0.001 | <0.001 | Upregulated |
| 10409 | BASP1 | 2.846037 | <0.001 | <0.001 | Upregulated |
Functional and pathway enrichment
analyses of DEGs
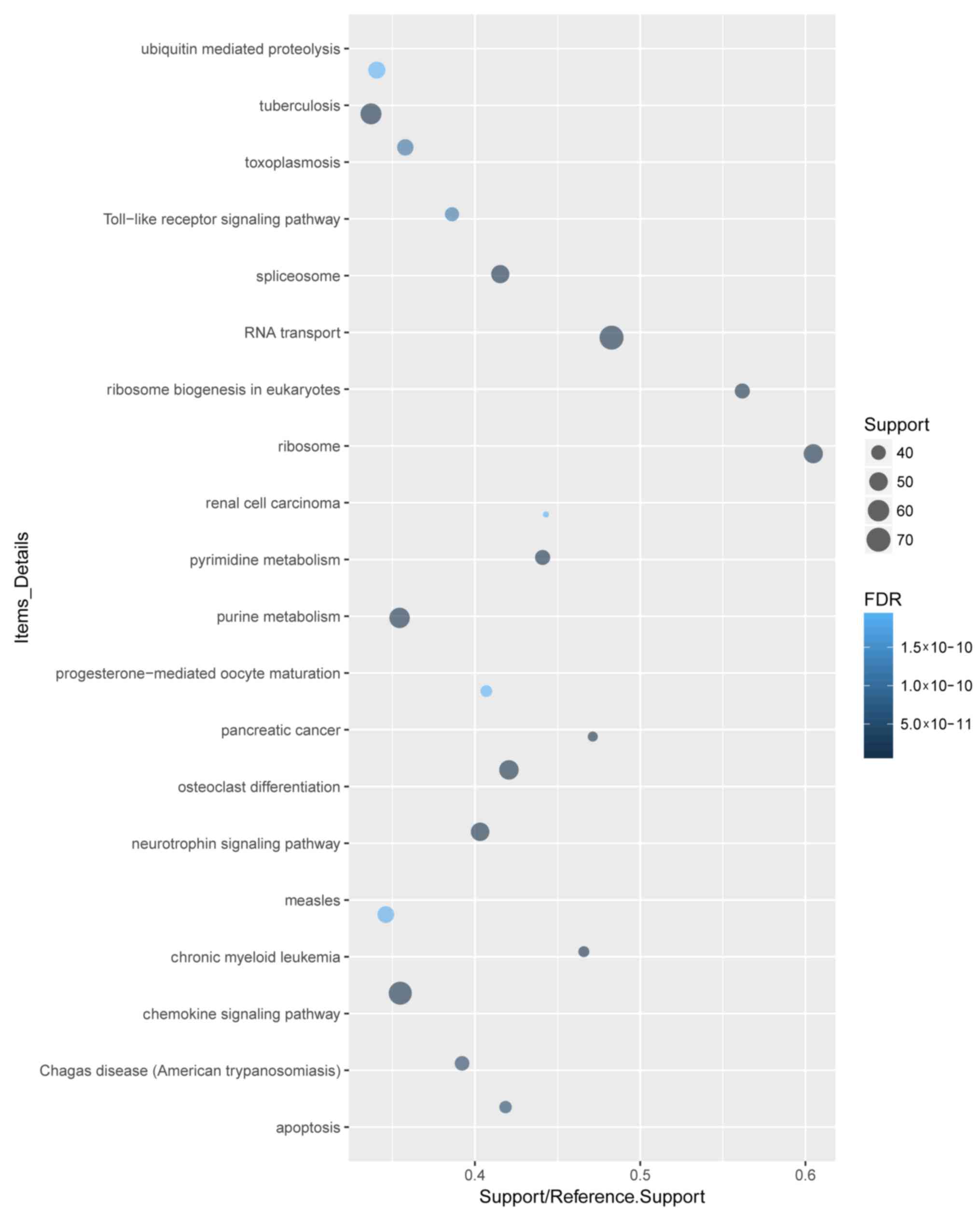
The 4,402 DEGs were analyzed by GO term and KEGG
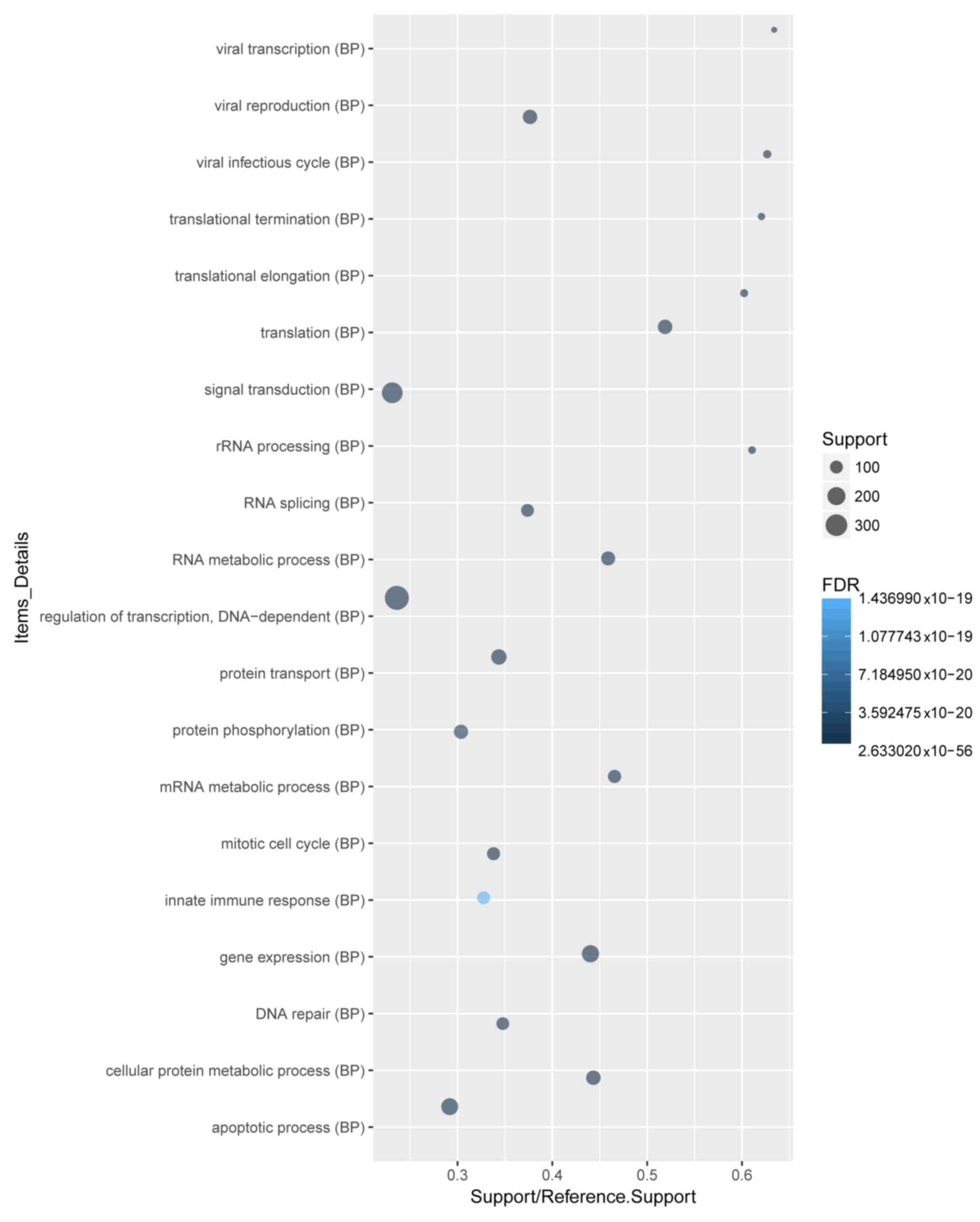
pathway enrichment. A total of 4,102 DEGs were recognized. GO and
KEGG pathway analyses of the top 20 DEGs demonstrated that ‘signal
transduction’, ‘regulation of transcription, DNA-dependent’ and
‘apoptotic process’ were the most enriched biological process (BP)
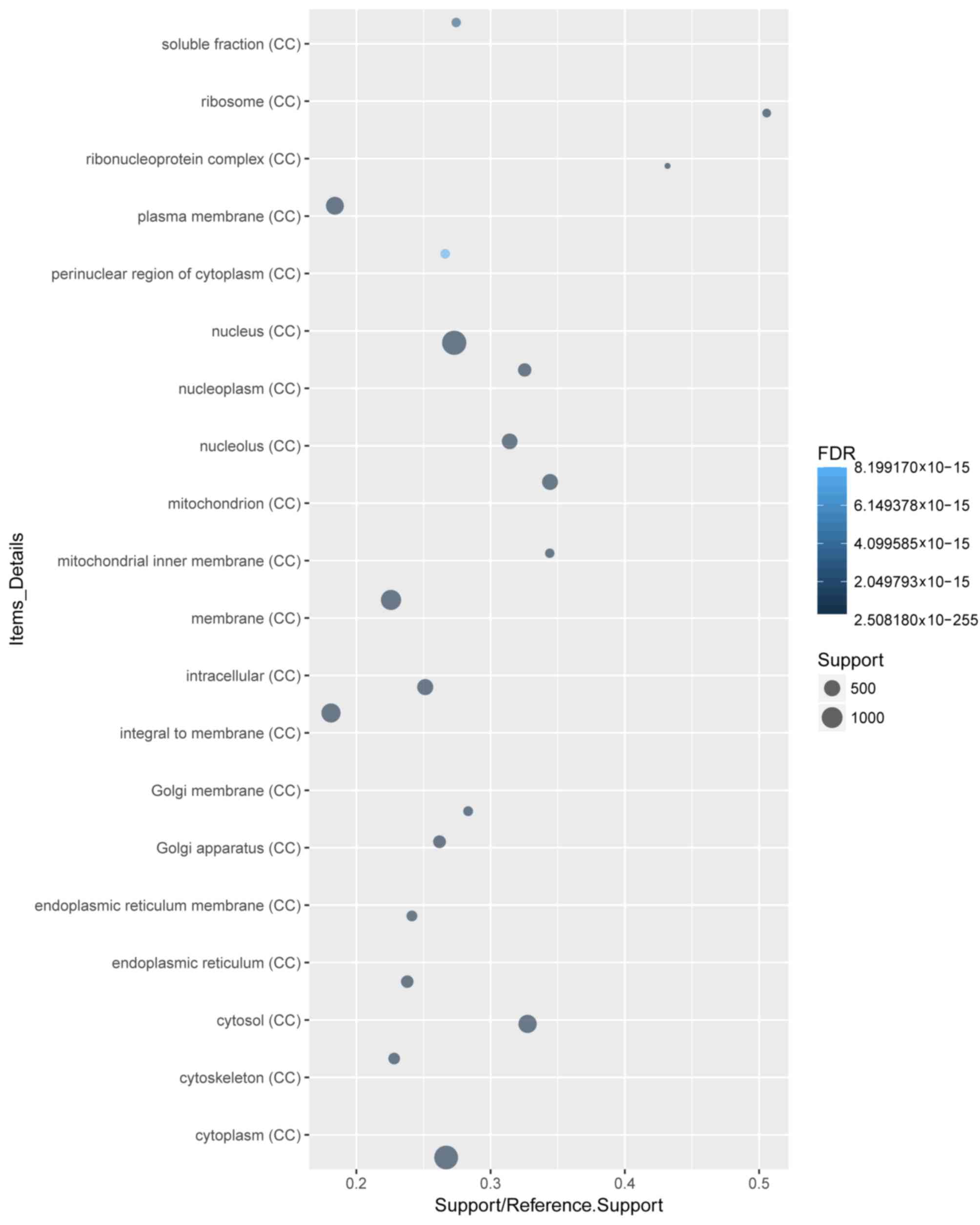
terms (Fig. 2). ‘Cytoplasm’ and
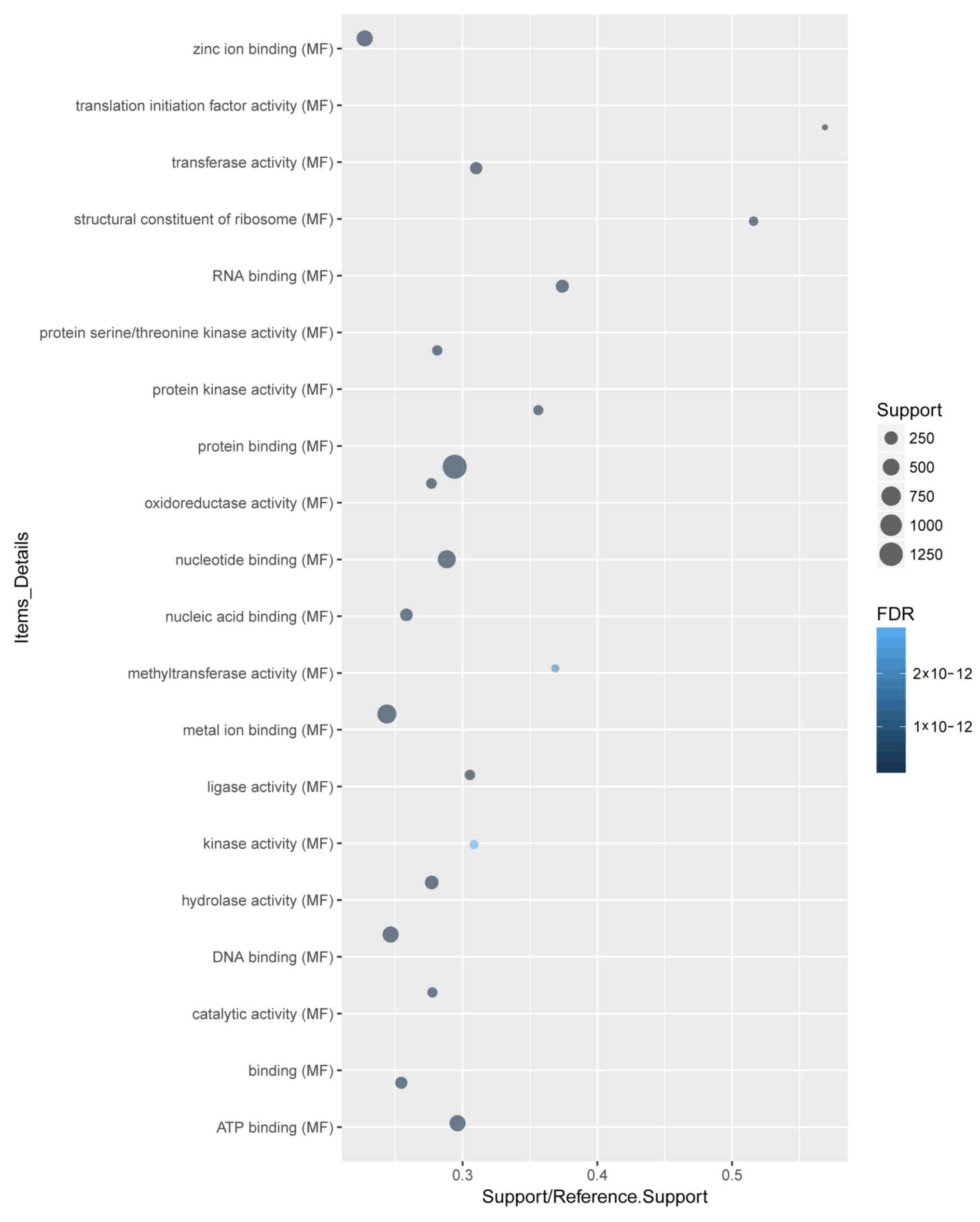
‘protein binding’ were the most enriched cellular component
(Fig. 3) and molecular function
(Fig. 4) terms, respectively.
‘Toll-like receptor signaling pathway’ was among the significantly
enriched KEGG pathways associated with sepsis development (Fig. 5).
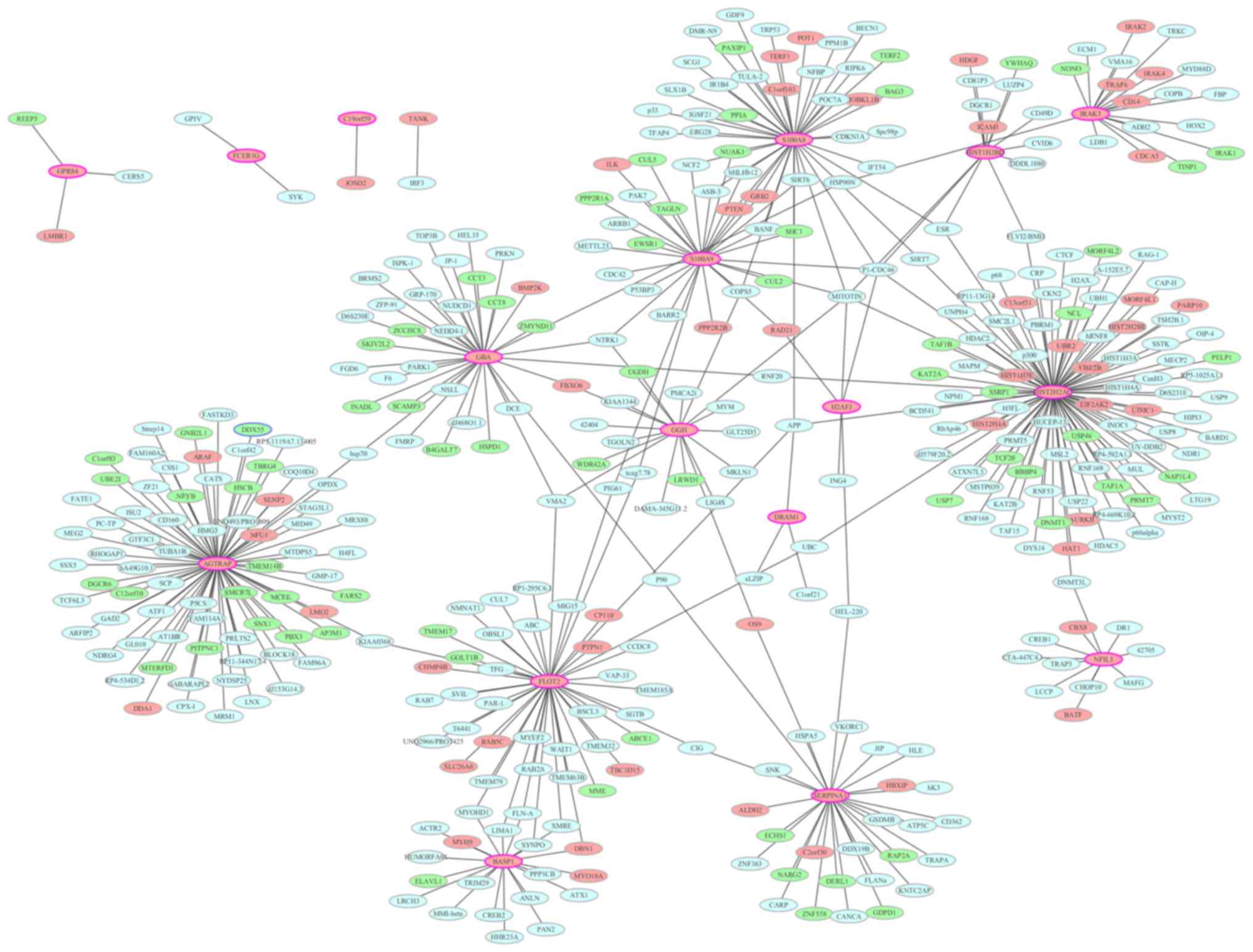
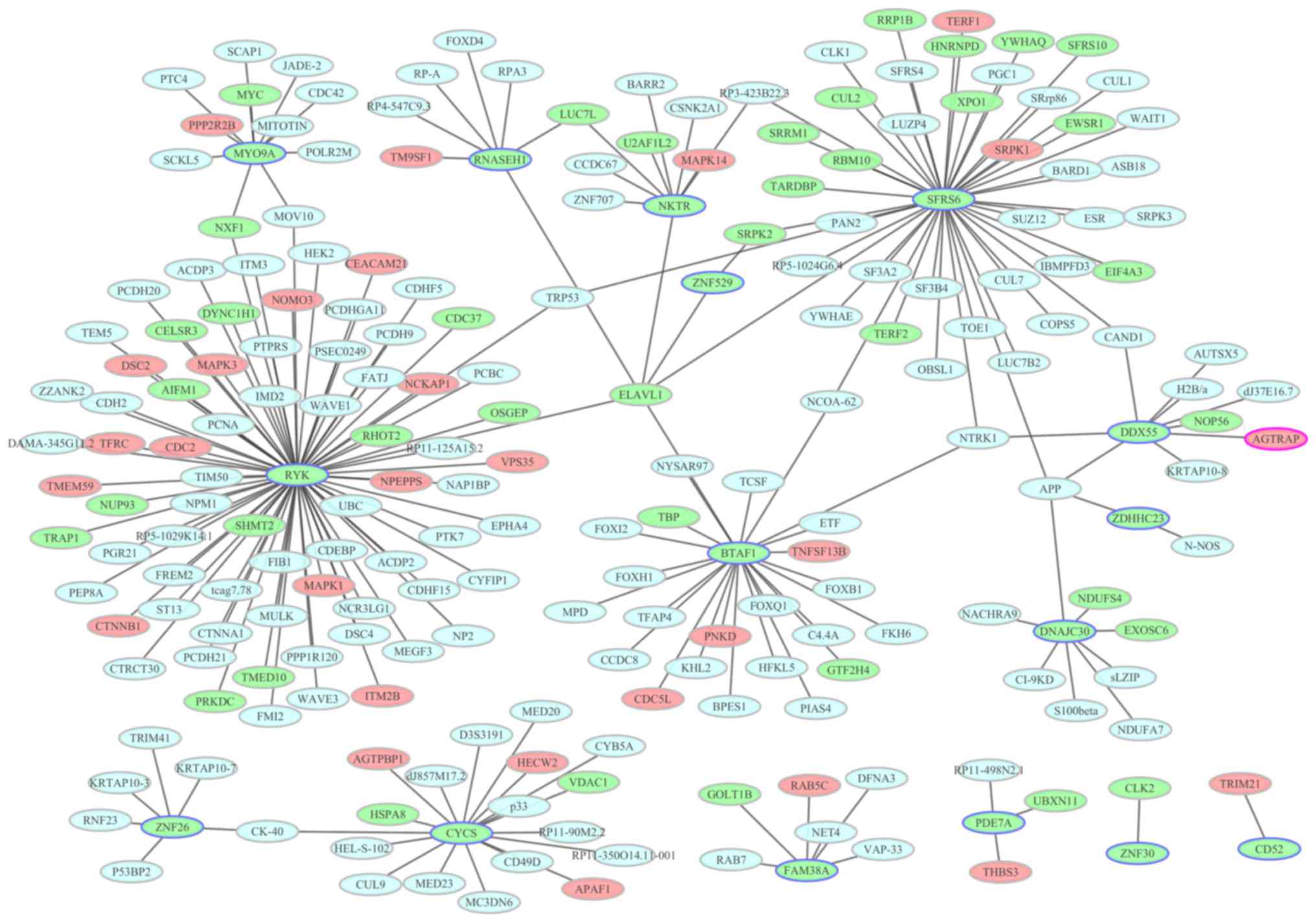
PPI network construction
To identify the interactions between the DEGs, a PPI
network was constructed and visualized using Cytoscape. PPI
networks of the top 20 upregulated and top 20 downregulated DEGs
are presented in Figs. 6 and
7, respectively. Nodes with a high
degree (proteins encoded by DEGs interact with >5 other proteins
encoded by other genes) are described as hub proteins. As
demonstrated in Fig. 6, the
network for upregulated DEGs consisted of 409 nodes and 443 edges.
The most important upregulated hub proteins were histone cluster 2
H2A family member C (degree, 92), angiotensin II
receptor-associated protein (AGTRAP; degree, 74), flotillin 2
(degree, 46), S100 calcium-binding protein (S100)A8 (degree, 43),
glucosylceramidase β (degree, 35), S100A9 (degree, 30), serpin
family A member 1 (degree, 29), brain abundant membrane-attached
signal protein 1 (degree, 20), γ-glutamyl hydrolase (degree, 18),
interleukin 1 receptor-associated kinase 3 (IRAK3; degree, 18),
histone cluster 1 H2B family member D (degree, 13) and nuclear
factor interleukin 3-regulated (degree, 11). In the downregulated
DEG PPI network, there were 224 nodes and 227 edges. In Fig. 7 for the downregulated DEG network,
the most important hub proteins were receptor-like tyrosine kinase
(degree, 75), serine and arginine rich splicing factor 6 (degree,
45), B-TFIID TATA-box binding protein-associated factor 1 (degree,
24), cytochrome C, somatic (degree, 18) and myosin IXA (degree,
11).
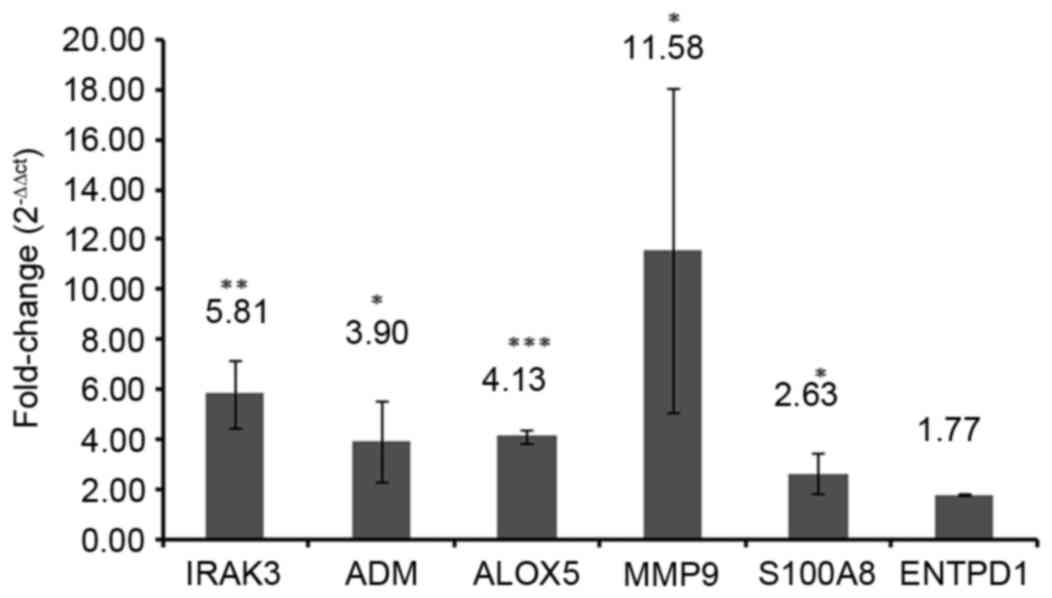
RT-qPCR validation
To verify the results of the bioinformatics
analyses, the expression levels of selected DEGs were quantified by
RT-qPCR in three sepsis and three healthy blood samples. A total of
6 DEGs, including IRAK3, adrenomedullin (ADM), arachidonate
5-lipoxygenase (ALOX5), matrix metallopeptidase 9 (MMP9), S100A8
and ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1) were
selected as candidate genes based on the previous literature
reviews (1,17–21).
IRAK3 (P<0.01), ADM (P<0.05), ALOX5 (P<0.001), MMP9
(P<0.05) and S100A8 (P<0.05) were significantly upregulated
in the blood samples of patients with sepsis compared with those of
healthy controls (Fig. 8). No
significant difference was identified between the ENTPD1 mRNA
expression in blood samples from patients with sepsis and healthy
controls in the present study (Fig.
8).
ROC curve analysis
ROC curve analyses were performed and the AUC was
calculated to assess the discriminatory ability of several DEGs in
the GEO dataset. The AUC of 7 DEGs, including AGTRAP (1.000), IRAK3
(1.000), ADM (0.994), ALOX5 (0.916), MMP9 (0.967), S100A8 (1.000)
and ENTPD1 (0.984) was >0.9 (Fig.
9). AGTRAP, IRAK3 and S100A8 had the largest AUC among these 7
DEGs. For sepsis diagnosis, the 1-specificity (proportion of false
positive results) and sensitivity (proportion of true positive
results) values, respectively, were as follows: AGTRAP, 100 and
100%; IRAK3, 100 and 100%; ADM, 100 and 96.6%; ALOX5, 100 and
82.8%; MMP9, 100 and 89.7%; S100A8, 100 and 100%; and ENTPD1, 100
and 93.1%
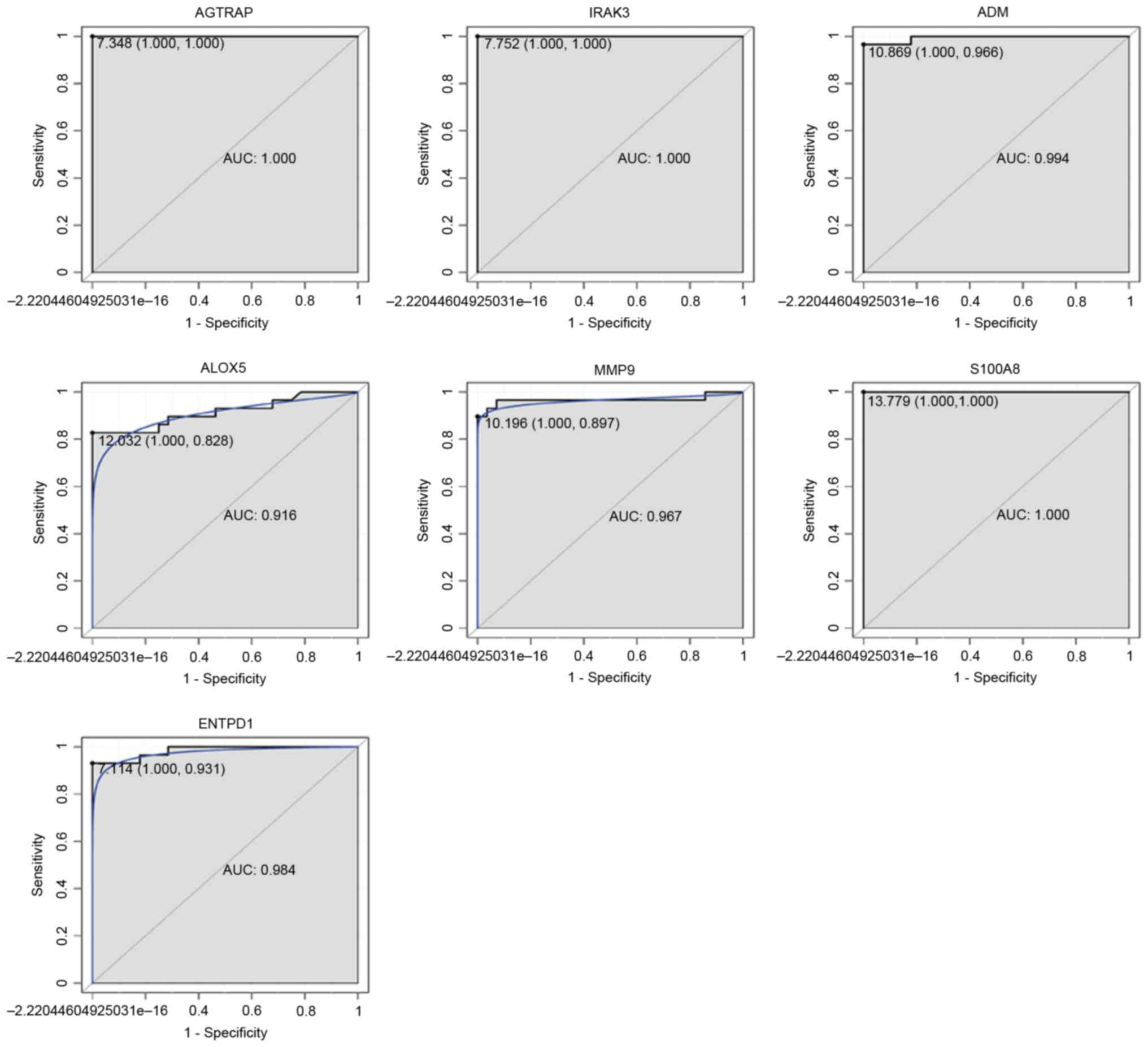 | Figure 9.ROC curves of selected DEGs between
patients with sepsis and healthy controls. The ROC curves were
utilized to indicate the diagnostic ability of these selected DEGs
with 1-Specificity (the proportion of false positive results;
x-axis) and sensitivity (the proportion of true positive results;
y-axis). ROC, receiver operating curve; DEG, differentially
expressed gene; AGTRAP, angiotensin II receptor-associated protein;
IRAK3, interleukin 1 receptor-associated kinase 3; ADM,
adrenomedullin; ALOX5, arachidonate 5-lipoxygenase; MMP9, matrix
metallopeptidase 9; S100A8, S100 calcium-binding protein A8;
ENTPD1, ectonucleoside triphosphate diphosphohydrolase 1; AUC, area
under the curve. |
Discussion
Sepsis is a prevalent condition that is one of the
major causes of mortality in hospitalized patients. An
understanding of the mechanisms underlying sepsis pathophysiology
is vital in order to develop effective, novel therapeutics and
diagnostic tools. In the present study, a total of 4,402 DEGs,
including 1,960 upregulated and 2,442 downregulated DEGs, were
identified to be associated with sepsis. These DEGs were
significantly enriched in the TLR signaling pathways. Several hub
genes with high degrees, including IRAK3, S100A8, AGTRAP and
S100A9, were subsequently identified in the PPI networks consisting
of the top 20 upregulated and top 20 downregulated DEGs. Candidate
genes IRAK3, ADM, ALOX5, MMP9, S100A8 and ENTPD1 were then
validated by RT-qPCR to confirm conclusions drawn from the
bioinformatics analysis. In addition, the DEGs AGTRAP, IRAK3, ADM,
ALOX5, MMP9, S100A8 and ENTPD1 were also identified to have
diagnostic value in sepsis.
IRAK3 is an immune-associated enzyme. Notably, IRAK3
expression was elevated in blood monocytes from patients with
sepsis, and expression levels in pediatric patients were directly
associated with adverse clinical outcomes (22,23).
In addition, increased expression was also significantly increased
following septic shock (24).
Cazalis et al (24)
identified four variants of the IRAK3 gene that were associated
with acute lung injury development during severe sepsis. Targeting
IRAK3 in sepsis may have an impact on the progression of this
condition (25). The present study
also identified a significant upregulation of IRAK3 expression in
the blood samples of patients with sepsis compared with healthy
controls. Furthermore, IRAK3 was identified as a hub gene (degree,
18) in the PPI network for upregulated DEGs. Notably, it had a
large value (AUC, 1) for sepsis diagnosis, confirming that IRAK3
may be critically involved in the development of sepsis and may be
employed as a diagnostic marker and potential drug target in sepsis
therapy.
ADM expression is induced by hypoxia, oxidative
stress and proinflammatory cytokines. It exerts various effects,
including the regulation of inflammation, infection and
angiogenesis (26–29). ADM is also implicated in the
vasodilation and hypotension that is associated with septic shock
(30). Hirata et al
(27) demonstrated that serum
levels of ADM were increased in sepsis and the highest plasma
concentrations of ADM were detected in patients with septic shock
(26). It has been reported that
ADM expression increased with increasing sepsis severity and was
associated with increasingly adverse outcomes (17,31).
In the current study, the expression of ADM was demonstrated to be
significantly increased in the blood sample data from patients with
sepsis compared with healthy controls, which is consistent with a
previous report (27).
Furthermore, RT-qPCR analysis confirmed this increase in ADM
expression, and diagnoses were also markedly associated with levels
of ADM expression in patients with sepsis. The results of the
present study highlighted the pivotal involvement of ADM in sepsis,
which may aid the evaluation of sepsis diagnosis and prognosis.
ALOX5 expression is reported to be increased during
inflammatory and immune responses (32,33).
It is typically expressed by leukocytes and is associated with the
biosynthesis of leukotrienes. Various leukotriene mediators are
thought to be pathogenic in inflammatory diseases such as sepsis
(34). Monteiro et al
(35) demonstrated that the
products of ALOX5 induced lung injury during sepsis. Furthermore,
ALOX5 expression in a mouse model exacerbated sepsis-induced
multiple organ injury (36) and
mice with an increased susceptibility to sepsis exhibited increased
ALOX5 expression (37). In the
present study, ALOX5 expression was significantly upregulated in
blood samples from patients with sepsis compared with controls, and
ALOX5 was determined to have a high diagnostic value, indicating
that ALOX5 may have a crucial role in the inflammatory and immune
processes of sepsis and has potential as a diagnostic marker.
It has been established that MMP9 is associated with
inflammation and inhibits platelet aggregation (38–41).
Previous studies demonstrated that the mRNA and protein expression
levels of MMP9 in patients with sepsis were significantly higher
compared with healthy individuals, and its expression levels have
been investigated as prognostic biomarkers of sepsis (42–45).
In the present study, RT-qPCR demonstrated that the expression of
MMP9 was significantly upregulated in the blood samples from
patients with sepsis compared with healthy controls, which was
concordant with the bioinformatics analysis results. Furthermore,
MMP9 was significantly associated with sepsis diagnosis, further
demonstrating the function of MMP9 in the pathophysiology of
sepsis.
S100A8 is an important inflammatory mediator
(46), and a previous study has
demonstrated that the expression levels of S100A8 is upregulated in
sepsis (21). The peripheral blood
level of S100A8 was elevated in patients with sepsis-associated
encephalopathy (SAE) and may be associated with SAE severity
(21). Furthermore, the presence
of S100A8 in amniotic fluid was reported to be an important
predictor of early-onset neonatal sepsis incidence (47–49).
In the current study, S100A8 expression was significantly increased
in blood samples of patients with sepsis compared with healthy
controls and was also identified as a hub gene (degree, 43) in the
PPI network of upregulated DEGs in sepsis. Notably, S100A8 had the
largest AUC in sepsis diagnosis, indicating that S100A8 may have
potential as a drug target and diagnostic marker in sepsis
therapy.
ENTPD1 has been previously reported to reduce
sepsis-associated mortality and improve the survival rate in
microbial sepsis cases through the attenuation of systemic
inflammation (50,51). Additionally, numerous studies have
demonstrated that ENTPD1 protected organs from hypoxic and
non-infectious inflammation (52–55),
which are implicated in the development of sepsis (7,56).
The present study identified that ENTPD1 expression was increased
in sepsis compared with healthy controls in the GEO data; however,
the increased expression was not significant in the RT-qPCR
results. Therefore, further research is required to investigate the
involvement of ENTPD1 in sepsis pathophysiology. Furthermore, for
ENTPD1, a large diagnosis value for sepsis was calculated, which
indicates that ENTPD1 may be employed as a diagnostic marker.
In addition to the above genes that were validated
by RT-qPCR in the present study, AGTRAP and S100A9 were also
identified to be associated with sepsis in the PPI network. AGTRAP
has been reported to be implicated in septic shock caused by
bacterial or viral infections (24). Nakada et al (57) also demonstrated that AGTRAP may be
a potential pharmacogenetic biomarker in sepsis. S100A9 is a
damage-associated molecule that has been reported to be a mediator
of severe sepsis (58). In
addition, S100A9 mRNA expression was increased in circulating cells
in septic shock (59).
Furthermore, S100A9 and S100A8 form a heterodimer that is
implicated in multiple organ failure in septic shock (60). van Zoelen et al (61) demonstrated that the expression of
S100A8/S100A9 was significantly elevated in the plasma of patients
with severe sepsis. In the present study, the expression of AGTRAP
and S100A9 was demonstrated to be upregulated in blood samples from
patients with sepsis and to be implicated in sepsis development.
AGTRAP was associated with a high diagnostic value with the highest
AUC, indicating that AGTRAP may be a potential diagnostic marker
for sepsis.
According to KEGG enrichment analysis, DEGs were
demonstrated to be significantly enriched in the TLR signaling
pathway. In humans, at least ten different TLRs recognize specific
microbial patterns to initiate inflammatory signaling pathways
(62). TLRs are major contributors
to sepsis development, particularly in the early stages where
pro-inflammatory cytokines are produced (63). Savva and Roger (64) demonstrated that regulating the
TLR/lymphocyte antigen 96-mediated inflammatory response may be a
potential approach in the treatment and prevention of septic shock.
Previous research has also demonstrated that TLR4 rs11536889 may be
a marker of organ failure in sepsis (65).
Certain limitations are associated with the present
study. The expression patterns in sepsis of two identified DEGs,
ENTPD1 and AGTRAP, is unknown; therefore, future in vivo and
in vitro experiments are required to investigate the
expression and function of these genes in sepsis pathology.
Additionally, studies with larger cohorts of patients with sepsis
are required to confirm the diagnostic and therapeutic value of the
identified genes. Larger numbers of blood samples are also required
for further investigation to validate the RT-qPCR results
obtained.
In conclusion, the results of the present study
demonstrated that IRAK3, ADM, ALOX5, MMP9 and S100A8 expression was
significantly upregulated in patients with sepsis compared with
healthy controls, and they may therefore significantly contribute
to the pathophysiology of sepsis. The identification of these genes
may contribute to the development of early diagnostic tools,
prognostic markers or therapeutic targets in sepsis.
Acknowledgements
The present study was funded by the Project Funds of
Taizhou People's Hospital (grant no. ZL201716).
Glossary
Abbreviations
Abbreviations:
|
ADM
|
adrenomedullin
|
|
AGTRAP
|
angiotensin II receptor-associated
protein
|
|
ALOX5
|
arachidonate 5-lipoxygenase
|
|
DEGs
|
differentially expressed genes
|
|
ENTPD1
|
ectonucleoside triphosphate
diphosphohydrolase 1
|
|
FDR
|
false discovery rate
|
|
GEO
|
Gene Expression Omnibus
|
|
GO
|
Gene Ontology
|
|
IRAK3
|
interleukin 1 receptor-associated
kinase 3
|
|
KEGG
|
Kyoto Encyclopedia of Genes
Genomes
|
|
MMP9
|
matrix metallopeptidase 9
|
|
PPI
|
protein-protein interaction
|
|
RT-qPCR
|
reverse transcription
quantitative-polymerase chain reaction
|
|
S100A8
|
S100 calcium-binding protein A8
|
|
S100A9
|
S100 calcium-binding protein A9
|
|
SAE
|
sepsis-associated encephalopathy
|
|
TLR
|
toll-like receptor
|
References
|
1
|
Bao R, Shui X, Hou J, Li J, Deng X, Zhu X
and Yang T: Adenosine and the adenosine A2A receptor agonist,
CGS21680, upregulate CD39 and CD73 expression through E2F-1 and
CREB in regulatory T cells isolated from septic mice. Int J Mol
Med. 38:969–975. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jawad I, Lukšić I and Rafnsson SB:
Assessing available information on the burden of sepsis: Global
estimates of incidence, prevalence and mortality. J Glob Health.
2:0104042012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martin GS, Mannino DM and Moss M: The
effect of age on the development and outcome of adult sepsis. Criti
Care Med. 34:15–21. 2006. View Article : Google Scholar
|
|
4
|
Strehlow MC, Emond SD, Shapiro NI,
Pelletier AJ and Camargo CA Jr: National study of emergency
department visits for sepsis, 1992 to 2001. Ann Emerg Med.
48:326–331, 331.e1-e3. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin GS, Mannino DM, Eaton S and Moss M:
The epidemiology of sepsis in the United States from 1979 through
2000. N Engl J Med. 348:1546–1554. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murphy SL: Deaths: Final data for 1998.
Natl Vital Stat Rep. 48:1–105. 2000.PubMed/NCBI
|
|
7
|
Hotchkiss RS and Karl IE: The
pathophysiology and treatment of sepsis. N Engl J Med. 348:138–150.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calvano SE, Xiao W, Richards DR, Felciano
RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK,
et al: A network-based analysis of systemic inflammation in humans.
Nature. 437:1032–1037. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wurfel MM, Gordon AC, Holden TD, Radella
F, Strout J, Kajikawa O, Ruzinski JT, Rona G, Black RA, Stratton S,
et al: Toll-like receptor 1 polymorphisms affect innate immune
responses and outcomes in sepsis. Am J Respir Crit Care Med.
178:710–720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Angus DC, Linde-Zwirble WT, Lidicker J,
Clermont G, Carcillo J and Pinsky MR: Epidemiology of severe sepsis
in the United States: Analysis of incidence, outcome, and
associated costs of care. Crit Care Med. 29:1303–1310. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pankla R, Buddhisa S, Berry M, Blankenship
DM, Bancroft GJ, Banchereau J, Lertmemongkolchai G and Chaussabel
D: Genomic transcriptional profiling identifies a candidate blood
biomarker signature for the diagnosis of septicemic melioidosis.
Genome Biol. 10:R1272009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu JQ, Sassé TR, Wolkenstein G, Conceicao
V, Saksena MM, Soedjono M, Perera SS, Wang B, Dwyer DE and Saksena
NK: Transcriptome analysis of primary monocytes shows global
down-regulation of genetic networks in HIV viremic patients versus
long-term non-progressors. Virology. 435:308–319. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reiner-Benaim A: FDR control by the BH
procedure for two-sided correlated tests with implications to gene
expression data analysis. Biom J. 49:107–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate-a practical and powerful approach to
multiple testing. J Royal Stat Soc. 57:289–300. 1995.
|
|
16
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pino-Yanes M, Ma SF, Sun X, Tejera P,
Corrales A, Blanco J, Pérez-Méndez L, Espinosa E, Muriel A, Blanch
L, et al: Interleukin-1 receptor-associated kinase 3 gene
associates with susceptibility to acute lung injury. Am J Respir
Cell Mol Biol. 45:740–745. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kesik V, Ataş E, Gülcan Kurt Y, Aydın FN,
Babacan O, Gülgün M and Korkmazer N: Adrenomedullin predicts high
risk and culture positivity in children with solid tumors suffering
from neutropenic fever. J Infect Chemother. 22:617–621. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pavanelli WR, Gutierrez FR, Mariano FS,
Prado CM, Ferreira BR, Teixeira MM, Canetti C, Rossi MA, Cunha FQ
and Silva JS: 5-lipoxygenase is a key determinant of acute
myocardial inflammation and mortality during Trypanosoma cruzi
infection. Microbes Infect. 12:587–597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shogan BD, Belogortseva N, Luong PM,
Zaborin A, Lax S, Bethel C, Ward M, Muldoon JP, Singer M, An G, et
al: Collagen degradation and MMP9 activation by Enterococcus
faecalis contribute to intestinal anastomotic leak. Sci Transl Med.
7:286ra2682015. View Article : Google Scholar
|
|
21
|
Zhang LN, Wang XH, Wu L, Huang L, Zhao CG,
Peng QY and Ai YH: Diagnostic and predictive levels of
calcium-binding protein A8 and tumor necrosis factor
receptor-associated factor 6 in sepsis-associated encephalopathy: A
prospective observational study. Chin Med J (Engl). 129:1674–1681.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hall MW, Gavrilin MA, Knatz NL, Duncan MD,
Fernandez SA and Wewers MD: Monocyte mRNA phenotype and adverse
outcomes from pediatric multiple organ dysfunction syndrome.
Pediatr Res. 62:597–603. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Escoll P, del Fresno C, García L, Vallés
G, Lendínez MJ, Arnalich F and López-Collazo E: Rapid up-regulation
of IRAK-M expression following a second endotoxin challenge in
human monocytes and in monocytes isolated from septic patients.
Biochem Biophys Res Commun. 311:465–472. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cazalis MA, Lepape A, Venet F, Frager F,
Mougin B, Vallin H, Paye M, Pachot A and Monneret G: Early and
dynamic changes in gene expression in septic shock patients: A
genome-wide approach. Intensive Care Med Exp. 2:202014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng JC, Cheng G, Newstead MW, Zeng X,
Kobayashi K, Flavell RA and Standiford TJ: Sepsis-induced
suppression of lung innate immunity is mediated by IRAK-M. The J
Clin Invest. 116:2532–2542. 2006.PubMed/NCBI
|
|
26
|
Ueda S, Nishio K, Minamino N, Kubo A, Akai
Y, Kangawa K, Matsuo H, Fujimura Y, Yoshioka A, Masui K, et al:
Increased plasma levels of adrenomedullin in patients with systemic
inflammatory response syndrome. Am J Respir Crit Care Med.
160:132–136. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hirata Y, Mitaka C, Sato K, Nagura T,
Tsunoda Y, Amaha K and Marumo F: Increased circulating
adrenomedullin, a novel vasodilatory peptide, in sepsis. J Clin
Endocrinol Metabol. 81:1449–1453. 1996. View Article : Google Scholar
|
|
28
|
Sugo S, Minamino N, Shoji H, Kangawa K,
Kitamura K, Eto T and Matsuo H: Interleukin-1, tumor necrosis
factor and lipopolysaccharide additively stimulate production of
adrenomedullin in vascular smooth muscle cells. Biochem Biophys Res
Commun. 207:25–32. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Musson DS, McLachlan JL, Sloan AJ, Smith
AJ and Cooper PR: Adrenomedullin is expressed during rodent dental
tissue development and promotes cell growth and mineralization.
Biol Cell. 102:145–157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
So S, Hattori Y, Kasai K, Shimoda S and
Gross SS: Up-regulation of rat adrenomedullin gene expression by
endotoxin: Relation to nitric oxide synthesis. Life sciences.
58:Pl309–P1315. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simon TP, Martin L, Doemming S, Humbs A,
Bruells C, Kopp R, Hartmann O, Struck J, Bergmann A, Marx G and
Schuerholz T: Plasma adrenomedullin in critically ill patients with
sepsis after major surgery: A pilot study. J Crit Care. 38:68–72.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Talwar S, Munson PJ, Barb J, Fiuza C,
Cintron AP, Logun C, Tropea M, Khan S, Reda D, Shelhamer JH, et al:
Gene expression profiles of peripheral blood leukocytes after
endotoxin challenge in humans. Physiol Genomics. 25:203–215. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Prabhakar U, Conway TM, Murdock P, Mooney
JL, Clark S, Hedge P, Bond BC, Jazwinska EC, Barnes MR, Tobin F, et
al: Correlation of protein and gene expression profiles of
inflammatory proteins after endotoxin challenge in human subjects.
DNA Cell Biol. 24:410–431. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Peters-Golden M and Henderson WR Jr:
Leukotrienes. N Engl J Med. 357:1841–1854. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Monteiro AP, Soledade E, Pinheiro CS,
Dellatorre-Teixeira L, Oliveira GP, Oliveira MG, Peters-Golden M,
Rocco PR, Benjamim CF and Canetti C: Pivotal role of the
5-lipoxygenase pathway in lung injury after experimental sepsis. Am
J Respir Cell Mol Biol. 50:87–95. 2014.PubMed/NCBI
|
|
36
|
Collin M, Rossi A, Cuzzocrea S, Patel NS,
Di Paola R, Hadley J, Collino M, Sautebin L and Thiemermann C:
Reduction of the multiple organ injury and dysfunction caused by
endotoxemia in 5-lipoxygenase knockout mice and by the
5-lipoxygenase inhibitor zileuton. J Leukoc Biol. 76:961–970. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Das UN: Combination of aspirin with
essential fatty acids is superior to aspirin alone to prevent or
ameliorate sepsis or ARDS. Lipids Health Dis. 15:2062016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stamenkovic I: Extracellular matrix
remodelling: The role of matrix metalloproteinases. J Pathol.
200:448–464. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chakrabarti S and Patel KD: Regulation of
matrix metalloproteinase-9 release from IL-8-stimulated human
neutrophils. J Leukoc Biol. 78:279–288. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sheu JR, Fong TH, Liu CM, Shen MY, Chen
TL, Chang Y, Lu MS and Hsiao G: Expression of matrix
metalloproteinase-9 in human platelets: Regulation of platelet
activation in in vitro and in vivo studies. Br J Pharmacol.
143:193–201. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee YM, Lee JJ, Shen MY, Hsiao G and Sheu
JR: Inhibitory mechanisms of activated matrix metalloproteinase-9
on platelet activation. Eur J Pharmacol. 537:52–58. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin LY, Li CF, Zhu GF, Wu CT, Wang J and
Yan SF: Effect of siRNA against NF-κB on sepsisinduced acute lung
injury in a mouse model. Mol Med Rep. 10:631–637. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cizmeci MN, Kara S, Kanburoglu MK, Simavli
S, Duvan CI and Tatli MM: Detection of cord blood hepcidin levels
as a biomarker for early-onset neonatal sepsis. Med Hypotheses.
82:310–312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hoffmann U, Bertsch T, Dvortsak E,
Liebetrau C, Lang S, Liebe V, Huhle G, Borggrefe M and Brueckmann
M: Matrix-metalloproteinases and their inhibitors are elevated in
severe sepsis: Prognostic value of TIMP-1 in severe sepsis. Scand J
Infect Dis. 38:867–872. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Edgar JD, Gabriel V, Gallimore JR,
McMillan SA and Grant J: A prospective study of the sensitivity,
specificity and diagnostic performance of soluble intercellular
adhesion molecule 1, highly sensitive C-reactive protein, soluble
E-selectin and serum amyloid A in the diagnosis of neonatal
infection. BMC Pediatr. 10:222010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gebhardt C, Nemeth J, Angel P and Hess J:
S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol.
72:1622–1631. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Buhimschi IA and Buhimschi CS: The role of
proteomics in the diagnosis of chorioamnionitis and early-onset
neonatal sepsis. Clin Perinatol. 37:355–374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Buhimschi CS, Buhimschi IA, Abdel-Razeq S,
Rosenberg VA, Thung SF, Zhao G, Wang E and Bhandari V: Proteomic
biomarkers of intra-amniotic inflammation: Relationship with
funisitis and early-onset sepsis in the premature neonate. Pediatr
Res. 61:318–324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Buhimschi CS, Bhandari V, Dulay AT, Nayeri
UA, Abdel-Razeq SS, Pettker CM, Thung S, Zhao G, Han YW, Bizzarro M
and Buhimschi IA: Proteomics mapping of cord blood identifies
haptoglobin ‘switch-on’ pattern as biomarker of early-onset
neonatal sepsis in preterm newborns. PLoS One. 6:e261112011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Csóka B, Németh ZH, Törő G, Koscsó B,
Kókai E, Robson SC, Enjyoji K, Rolandelli RH, Erdélyi K, Pacher P
and Haskó G: CD39 improves survival in microbial sepsis by
attenuating systemic inflammation. FASEB J. 29:25–36. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Haskó G, Csóka B, Koscsó B, Chandra R,
Pacher P, Thompson LF, Deitch EA, Spolarics Z, Virág L, Gergely P,
et al: Ecto-5′-nucleotidase (CD73) decreases mortality and organ
injury in sepsis. J Immunol. 187:4256–4267. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Eltzschig HK, Thompson LF, Karhausen J,
Cotta RJ, Ibla JC, Robson SC and Colgan SP: Endogenous adenosine
produced during hypoxia attenuates neutrophil accumulation:
Coordination by extracellular nucleotide metabolism. Blood.
104:3986–3992. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Köhler D, Eckle T, Faigle M, Grenz A,
Mittelbronn M, Laucher S, Hart ML, Robson SC, Müller CE and
Eltzschig HK: CD39/ectonucleoside triphosphate diphosphohydrolase 1
provides myocardial protection during cardiac ischemia/reperfusion
injury. Circulation. 116:1784–1794. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Grenz A, Zhang H, Hermes M, Eckle T,
Klingel K, Huang DY, Müller CE, Robson SC, Osswald H and Eltzschig
HK: Contribution of E-NTPDase1 (CD39) to renal protection from
ischemia-reperfusion injury. FASEB J. 21:2863–2873. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hart ML, Gorzolla IC, Schittenhelm J,
Robson SC and Eltzschig HK: SP1-dependent induction of CD39
facilitates hepatic ischemic preconditioning. J Immunol.
184:4017–4024. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Riedemann NC, Guo RF and Ward PA: The
enigma of sepsis. J Clin Invest. 112:460–467. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nakada TA, Russell JA, Boyd JH, McLaughlin
L, Nakada E, Thair SA, Hirasawa H, Oda S and Walley KR: Association
of angiotensin II type 1 receptor-associated protein gene
polymorphism with increased mortality in septic shock. Crit Care
Med. 39:1641–1648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mares CA, Ojeda SS, Morris EG, Li Q and
Teale JM: Initial delay in the immune response to Francisella
tularensis is followed by hypercytokinemia characteristic of severe
sepsis and correlating with upregulation and release of
damage-associated molecular patterns. Infect Immun. 76:3001–3010.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fontaine M, Pachot A, Larue A, Mougin B,
Landelle C, Venet F, Allombert C, Cazalis MA, Monneret G and Lepape
A: Delayed increase of S100A9 messenger RNA predicts
hospital-acquired infection after septic shock. Crit Care Med.
39:2684–2690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Feuerstein GZ: Cardiac RAGE in sepsis:
Call TOLL free for anti-RAGE. Circ Res. 102:1153–1154. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
van Zoelen MA, Vogl T, Foell D, Van Veen
SQ, van Till JW, Florquin S, Tanck MW, Wittebole X, Laterre PF,
Boermeester MA, et al: Expression and role of myeloid-related
protein-14 in clinical and experimental sepsis. Am J Respir Crit
Care Med. 180:1098–1106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
West AP, Koblansky AA and Ghosh S:
Recognition and signaling by toll-like receptors. Ann Rev Cell Dev
Biol. 22:409–437. 2006. View Article : Google Scholar
|
|
63
|
Buchholz BM and Bauer AJ: Membrane TLR
signaling mechanisms in the gastrointestinal tract during sepsis.
Neurogastroenterol Motil. 22:232–245. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Savva A and Roger T: Targeting toll-like
receptors: Promising therapeutic strategies for the management of
sepsis-associated pathology and infectious diseases. Front Immunol.
4:3872013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mansur A, von Gruben L, Popov AF, Steinau
M, Bergmann I, Ross D, Ghadimi M, Beissbarth T, Bauer M and Hinz J:
The regulatory toll-like receptor 4 genetic polymorphism rs11536889
is associated with renal, coagulation and hepatic organ failure in
sepsis patients. J Transl Med. 12:1772014. View Article : Google Scholar : PubMed/NCBI
|