Introduction
Lower back pain is a common disorder that has
attracted much attention due to its effects on human health. Up to
80% of individuals worldwide may experience lower back pain at some
point in their lives, of whom a large proportion receive surgery,
resulting in heavy burdens on families and societies.
Intervertebral disc degeneration (IDD) is regarded as a primary
contributor to the pathological process of lower back pain
(1). As human IDD is a complex and
multifactorial process with contributions from genes, mechanical
stresses, cellular senescence and decreased nutrition (1,2),
more studies are required to further investigate the mechanism of
IDD and to identify specific factors that may serve important roles
in this condition.
Cartilage intermediate layer protein (CILP) is
specifically expressed in intervertebral discs (IVDs) (3), and its expression is increased with
ageing and disc degeneration (4,5).
Previous studies have identified a genetic correlation of CILP with
IDD among the upregulated genes in IDD (6–9), and
a functional single nucleotide polymorphism (SNP) of the CILP gene
has been demonstrated to increase the direct interaction of CILP
with transforming growth factor-β (TGF-β) and to prevent TGF-β from
activating the transcription of cartilage matrix genes in rabbit
nucleus pulposus (NP) cells (3).
In addition, increased CILP expression via N-terminal
domain-mediated inhibitory effects on chondrocyte insulin-like
growth factor (IGF)-1 responsiveness has been demonstrated to
impair chondrocyte growth and matrix repair and to indirectly
promote polymeric precipitation inhibitor supersaturation in bovine
ageing and osteoarthritis cartilage (10). Furthermore, CILP overexpression in
transgenic mice has been reported to lead to a significantly
decreased signal intensity of IVDs on magnetic resonance imaging,
representing an early sign of degeneration (11,12).
These studies have suggested that CILP may not only function as a
marker for but also have involvement in the pathogenesis of
IDD.
The development of IDD in humans is a complex
process that involves cell autophagy, cellular senescence, invasion
of inflammatory mediators, up-regulation of catabolic enzymes, such
as matrix metalloproteinases (MMPs) and a disintegrin and
metalloproteinase with a thrombospondin motifs (ADAMTs), and
downregulation of matrix proteins, such as aggrecan and collagen II
(1,2). Amongst the many events that occur in
IDD, the disruption of homeostasis of the extracellular matrix
(ECM) is regarded as one of the most important (2). Homeostasis of the ECM is important to
maintain the normal structure and function of IVDs by allowing for
interactions between resident cells and the extracellular
environment (13). Once IDD is
initiated, this homeostasis gradually breaks as an imbalance
develops between the rates of production and breakdown of matrix
components at early stages, leading to a series of events including
alterations in the pH and osmotic pressure, thereby creating a
harsh extracellular environment for resident cells in NP tissues.
Furthermore, the ECM under healthy conditions is able to retain
enough hydration to withstand and absorb the mechanical loading
experienced in daily life (14).
However, in IDD, synthesis of ECM components, including aggrecan
and collagen II, which constitute the majority of the ECM, is
decreased, leading to reduced hydration. Dehydration of the NP
weakens its mechanical support of IVDs and exposes NP cells to
excessive mechanical loading, which then stimulates increased
catabolic responses in NP cells (14,15).
Hence, abnormal mechanical loading is one of the major factors
promoting loss of the ECM in IDD, and it is an important mechanism
through which mechanical loading contributes to IDD (14–16).
Although CILP has been suggested to interfere with
the expression of matrix components, as mediated by several growth
factors, in rabbit and mouse NP cells (10,17),
there is no direct evidence that CILP has a negative effect on
homeostasis of the ECM in human IVDs. Furthermore, examination of
alterations in CILP expression in human NP cells in response to
different mechanical stimuli is required to obtain further insights
into the regulation of CILP to support its potential role in IDD.
To address these issues, the present study collected human NP cells
from patients undergoing lumbar spinal surgery for degenerative
disc disease (DDD) and detected the responses in NP cells,
including the expression of CILP, aggrecan and collagen II, to
mechanical stresses, including cyclic tensile strain (CTS) and
cyclic compressive stress. In addition, the levels of aggrecan and
collagen II, the main components of the ECM and traditional markers
for IDD, were detected following treatment of NP cells with RNAi
and rhCILP).
Materials and methods
Antibodies and reagents
Rabbit monoclonal anti-human CILP (cat. no.
ab192881), collagen II (cat. no. ab188570) and polyclonal
anti-human aggrecan (cat. no. ab36861) antibodies were purchased
from Abcam (Cambridge, MA, USA). rhCILP was obtained from R&D
Systems (Minneapolis, MN, USA). Specific small interfering (si)RNA
duplexes targeting human CILP and reduced-serum transfection medium
were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA), and the siRNA transfection reagent was purchased from
Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Ethics statement
Human IVDs were obtained with the approval of the
Ethical Committee of Xinqiao Hospital (Chongqing, China) and with
informed consent, in accordance with the Helsinki Declaration, from
individuals undergoing surgery for IDD.
Tissue collection and isolation and
culturing of NP cells
Human IVD tissues were collected from 20 patients
(Male: Female, 1:1) ranging in age from 30–60 years who were
undergoing lumbar spinal surgery for DDD at the Department of
Orthopaedics between January 2016 and May 2016, The Second
Affiliated Hospital (Chongqing, China). The NP tissues were
carefully minced and digested to obtain NP cells, as previously
reported (18). Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen;
Thermo Fisher Scientific, Inc.) supplemented with penicillin (50
U/ml), streptomycin (50 U/ml), and 10% fetal bovine serum (FBS;
Invitrogen; Thermo Fisher Scientific, Inc.). Subconfluent NP cells
(passage number <3) were trypsinized (Gibco; Thermo Fisher
Scientific, Inc.) and resuspended in complete medium to prepare for
subsequent experiments.
Application of CTS
An equal amount of each cell suspension was
transferred onto a silicone membrane of a BioFlex 6-well tension
plate (Flexcell International, Dunn Labortechnik, Asbach, Germany)
at a density of 5×105 cells/well and were allowed to
adhere for 48 h. The medium was replenished at 12 h before
application of a CTS. A CTS of 10% at a 1.0 Hz frequency was
delivered to the bases of the silicone membranes within the Bioflex
culture plates using an FX-5000C™ Flexcell system, and
NP cells adhered to the membranes via computer-controlled negative
pressure for different durations (0, 6, 12, 24 and 48 h). Cells
without tensile loading (control group) were cultured in the same
culture plates as the cells in the stimulation groups. Mechanically
stimulated and unstimulated NP cells were incubated at 37°C with 5%
CO2 during stimulation.
Application of cyclic compressive
stress
Cell suspensions at a concentration of
2×106/ml were prepared in tubes and were then mixed with
carbonate buffer solution (pH 5.5), maleimide-polymer (vinyl
alcohol), water and PEG-Link according to the manufacturer's
protocol (3D Life Dextran-PEG Hydrogel kit G90-1, Cellendes,
Germany). The reaction was mixed via rapid vortexing for no more
than 5 sec. Each cell suspension (1 ml) was obtained using a
1,000-µl pipette tip (Corning Incorporated, Corning, NY, USA) and a
pipette (Eppendorf, Hamburg, Germany) prior to gel formation. The
front end of the pipette tip was used to obtain a cross-section of
5 mm in diameter. The gel in the pipette tip was then extruded and
sliced into cylinders with a height of 3 mm that were suitable for
placement in the inner foam rings of Bioflex 6-well compression
plates (Flexcell International, Dunn Labortechnik, Asbach,
Germany). These cylinders were placed into the Bioflex plates
according to the manufacturer's protocol. DMEM-F12 containing 10%
serum was added to each plate to immerse the cylinders in medium. A
compressive loading of 1–2.5 MPa at a 1.0 Hz frequency was
delivered to the bases of the silicone membranes within the Bioflex
plates using an FX-5000C™ Flexercell system and then to
NP cells residing in the gel. The stimulation groups were subjected
to stimuli for different periods of time (6, 12, 24 and 48 h), and
cells without a tensile load (control group) were cultured in the
same culture plates as those in the other groups. Mechanically
stimulated and unstimulated NP cells were incubated at 37°C and 5%
CO2 during stimulation.
Extraction of cells from the gel
following compressive loading
The gels in the plates were transferred to sterile
centrifuge tubes, and 100 µl glucanase each tube, provided in a 3D
Life Dextran-PEG Hydrogel kit G90-1 (Cellendes GmbH, Reutlingen,
Germany), was added to the tubes. Once the gels had dissolved, the
suspended cells were harvested via centrifugation (500 × g, 6 min,
room temperature).
Transfection with siRNA
Subconfluent NP cells (passage number <4) were
equally divided into the siRNA group, the negative control group
and the blank control group, and then were transferred to cell
culture plates at a density of 5×105 cells/well. siRNA
transfection was conducted using Lipofectamine iMax (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The final siRNA (cat. no. sc-60384; Santa Cruz
Biotechnology, Inc.) concentration was 100 nM. Cells in the control
group were cultured in the same culture medium without siRNA.
Negative control siRNA (cat. no. sc-37007; Santa Cruz
Biotechnology, Inc.) was used to evaluate the off-target effects of
RNAi and to verify the accuracy of gene-specific siRNA-dependent
RNAi. Knockdown efficiency was detected at 48 h after transfection
by reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blotting.
Treatment with rhCILP at various
concentrations
Subconfluent NP cells (passage number <4) were
equally divided into the treatment groups and the control group and
then were transferred to cell culture plates at a density of
5×105 cells/well. The cells were allowed to adhere for
24 h prior to treatment with rhCILP (10, 100 and 1,000 ng/ml). An
untreated group served as the control group. The cells were
incubated at 37°C and 5% CO2 for 72 h after
treatment.
Extraction of total RNA and
RT-qPCR
Total RNA was extracted using an RNeasy Mini Kit
(Qiagen, Valencia, CA), and cDNA was reverse transcribed from 1 µg
total RNA using an Omniscript Reverse Transcription kit (Qiagen,
Inc., Valencia, CA, USA) according to the manufacturer's protocol.
RT-qPCR was performed with a ViiA7 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and a
QuantiNova™ SYBR Green PCR kit (Qiagen, Inc.). Primers
with the following sequences were used: Human GAPDH: F:
5′CCAGCAAGAGCACAAGAGGAAGAG3′, R: 5′GGTCTACATGGCAACTGTGAGGAG3′,
human CILP: F: 5′AGCGGTGTACGGAAACTCG3′, R: 5′ACGGCACTCCCCTTCTTGT3′,
human aggrecan: F: 5′TGAGGAGGGCTGGAACAAGTACC3′, R:
5′GGAGGTGGTAATTGCAGGGAACA3′ and human collagen II: F:
5′TTTCCCAGGTCAAGATGGTC3′, R: 5′TCACCTGGTTTTCCACCTTC3′.
qPCR reaction was performed in triplicate. The 20 ml
reaction volume was applied. The reaction parameters were 95°C for
30 sec followed by 40 cycles of 95°C for 5 sec for template
denaturation and 60°C for 34 sec for annealing and extension. The
results are presented as a Cq value, which is the cycle number at
which the amplified product was first detected. The mean Cq value
was obtained from three repeated experiments. The expression levels
of the target genes were normalized to that of the endogenous
control (GAPDH). The relative target mRNA expression levels were
calculated using the 2−ΔΔCq method (19).
Western blot analysis
Total protein was extracted using
radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) with Halt™
Protease/Phosphatase Inhibitor (Thermo Fisher Scientific, Inc.) at
4°C, with frequent agitation for 30 min. Cell lysates were cleared
of insoluble debris via centrifugation at 12,000 × g for 30 min at
4°C. The amount of total protein was determined by bicinchoninic
acid protein assay (Pierce; Thermo Fisher Scientific, Inc.). Equal
amounts of protein (20 µg) were separated by 10% SDS-PAGE and
transferred onto polyvinylidene difluoride membranes (Immobilon;
EMD Millipore, Billerica, MA, USA), which were blocked with 5% milk
for 1 h at 37°C. The filters were incubated overnight at 4°C with
primary rabbit antibodies diluted 1:1,000 and then with horseradish
peroxidase-conjugated secondary antibodies (cat. no. ZB-2301;
Origene Technologies, Inc., Beijing, China) diluted 1:2,500 for 1 h
at room temperature. Bands were detected using an enhanced
chemiluminescence system (EMD Millipore) and scanned using an
ImageQuant LAS4000 imaging system (GE Healthcare, Chicago, IL,
USA). The optical density (OD) of the bands was measured using
ImageJ software (National Institutes of Health, Bethesda, MD,
USA).
Statistical analysis
GraphPad Prism 6 (GraphPad Software, Inc., La Jolla,
CA, USA) and SPSS version 22.0 statistical software (IBM Corp.,
Armonk, NY, USA) were used to analyse and display the data of this
study. Each condition was performed in triplicate, and the mean was
determined. All of the results are presented as the mean ± standard
deviation. Statistical comparisons were performed using a one-way
analysis of variance, followed by Bonferroni's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
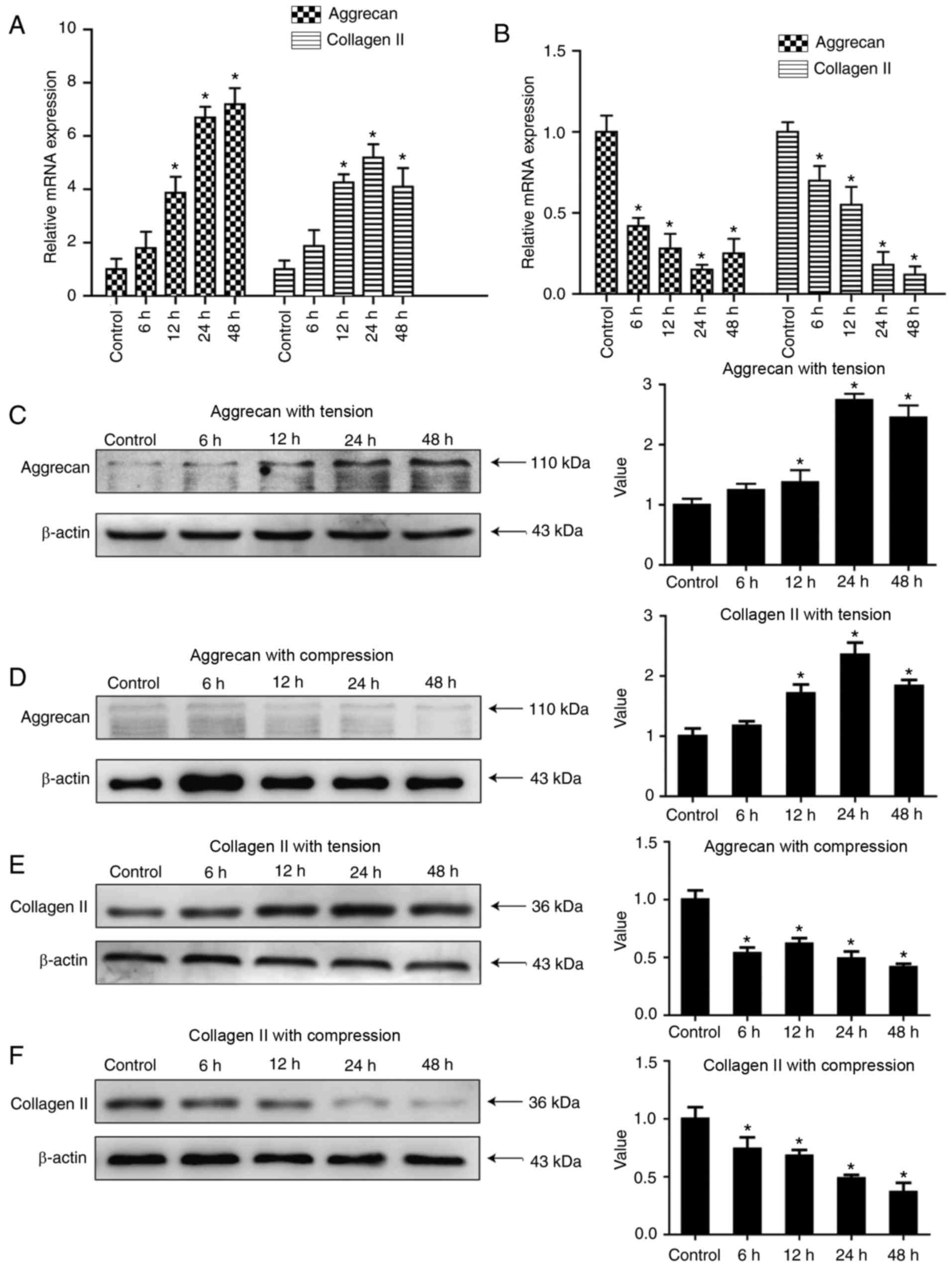
Aggrecan and collagen II expression is
significantly increased under tensile loading and decreased under
compressive loading
Aggrecan and collagen II are the main components of
the ECM, as well as traditional degenerative markers for IDD, and
their expression levels decrease with ageing and degeneration. As
presented in Fig. 1A, which
demonstrated that there was a significant increase in mRNA level of
aggrecans and collagen II subjected to tensile loading, with time
(only up to 24 h in collagen II) was observed (P<0.05). The peak
expression was observed in the 24 h group for collagen II, and in
the 48-h group for aggrecan. With regard to the effect of
compressive loading on matrix gene expression in human NP cells,
the mRNA expression levels of aggrecan and collagen II were
significantly decreased in all groups, and the lowest expression
was observed in the 48-h group for collagen II, and in the 24-h
group for aggrecan (P<0.05; Fig.
1B). WB results demonstrated aggrecan expression under tensile
loading was significantly increased (P<0.05; Fig. 1C) and decreased significantly under
compression loading (P<0.05; Fig.
1D), Similarly, the protein level of collagen II under tensile
loading was significantly increased (P<0.05; Fig. 1E) while decreased significantly
under compression loading (P<0.05; Fig. 1F).
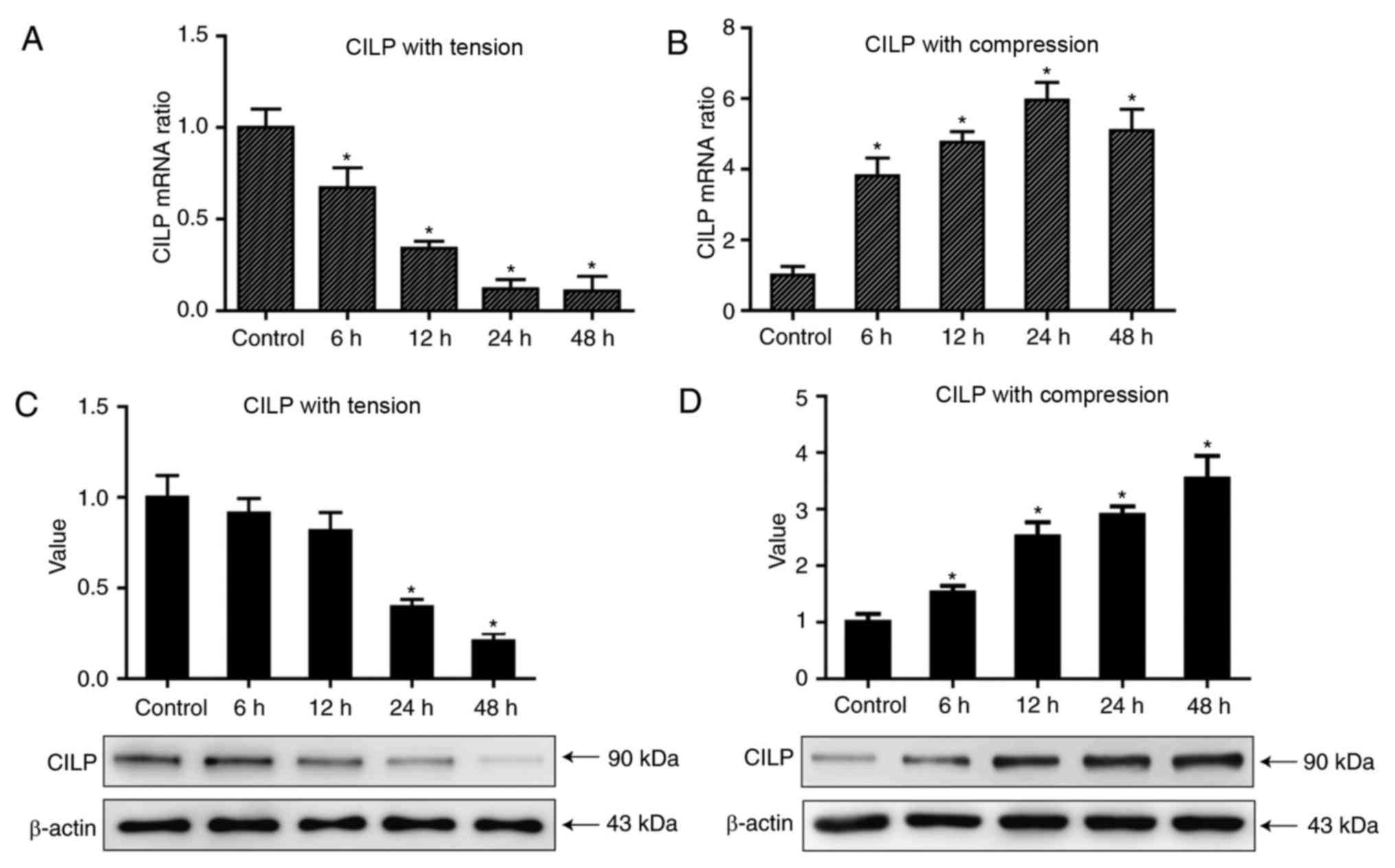
CILP expression is decreased under
tensile loading and increased under compressive loading in contrast
with that of aggrecan and collagen II
The regulations of mechanical loading on the
synthesis of ECM by NP cells was revealed previously; therefore,
the present study aimed to investigate whether mechanical loading
can regulate CILP expression. The expression of CILP demonstrated
an opposite trend as that of the matrix genes in tensile loading;
they decreased in a time-dependent manner following compressive
loading (Fig. 2A; P<0.05), and
increased in a time-dependent manner following tensile loading
(Fig. 2B; P<0.05). The protein
expression levels demonstrated the same trend (Fig. 2C and D, respectively; P<0.05).
Considering the ability of healthy NP tissues to buffer and
transform mechanical loading to other mechanical loading including
tension, it is reasonable to speculate the relatively low level of
CILP in healthy IVDs compared with degenerative IVDs is partly a
result of tensile loading endured in daily life.
The present study further investigated the effect of
compressive loading on CILP expression. A compressive loading was
applied at 1.0–2.5 MPa at 1 Hz, which is higher than that endured
by NP cells in healthy IVDs in daily life. Consequently,
compressive loading led to decreased aggrecan and collagen II
expression, as well as markedly upregulated CILP expression, with
peak expression detected in the 48-h group (P<0.05; Figs. 1 and 2). The results above demonstrated that
CILP expression is regulated by mechanical stimuli, and the
regulation is dependent on the mechanical types. Furthermore, these
results suggested it is the harsh mechanical loading in IVD that
contributes to the abnormal high expression of CILP in degenerative
IVDs.
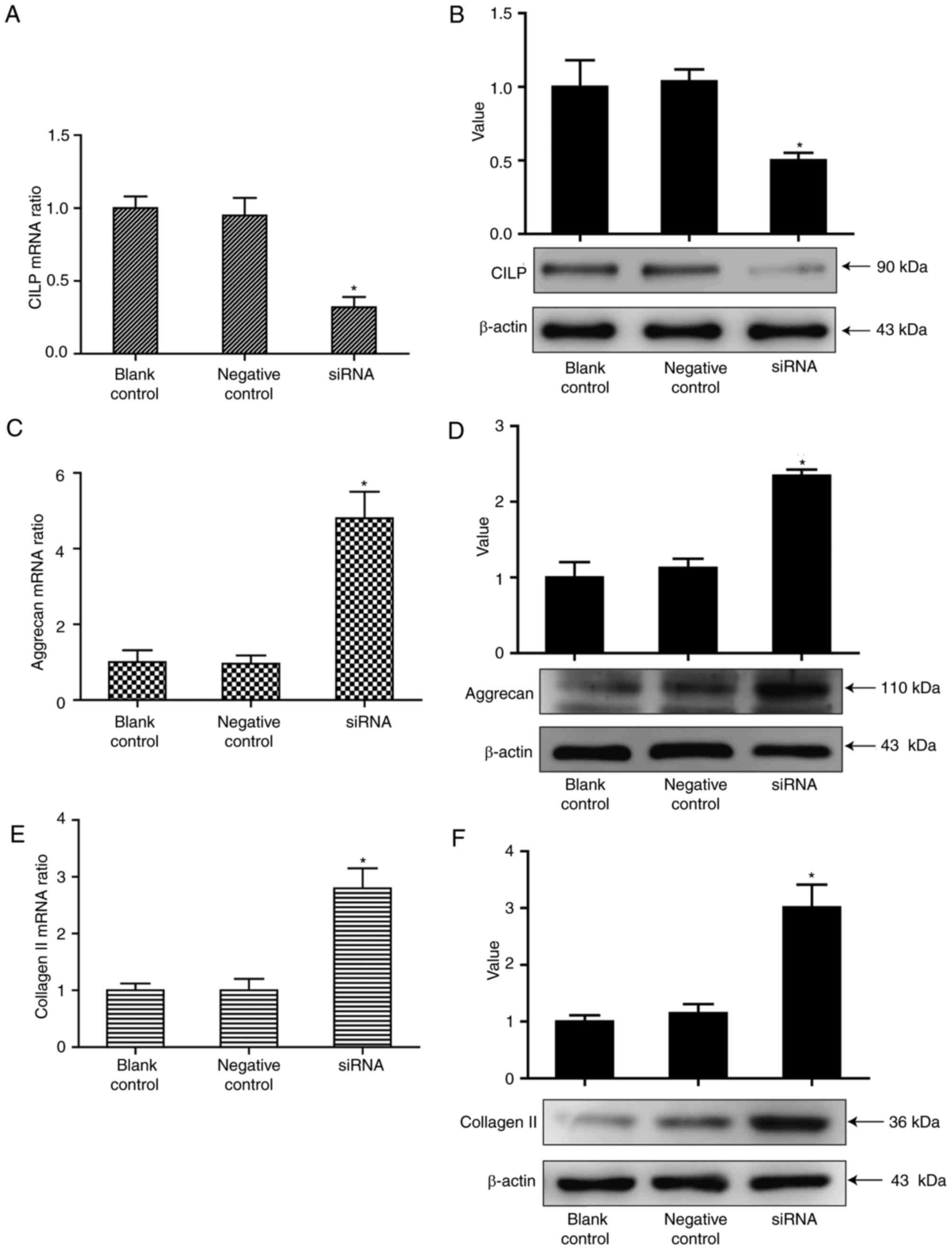
Aggrecan and collagen II levels are
increased following the silencing of CILP expression by RNAi
It is important to investigate whether excessive
CILP expression by NP cells contributes to IDD. As previous studies
have revealed the antagonism of CILP to TGF-β in rabbit NP cells
(5), The present study
hypothesized that CILP may function as a negative factor in ECM
synthesis in human NP cells. To prove the assumption, the present
study used RNAi to inhibit CILP expression. The NP cells were
transfected with 100 nm siRNA targeting the CILP gene. The results
in Fig. 3 revealed that while CILP
expression was effectively inhibited by siRNA (P<0.05), the
expression of aggrecan and collagen II were markedly increased
(P<0.05). Therefore, abnormal CILP expression may exert a
negative effect on the synthesis of matrix genes in IDD.
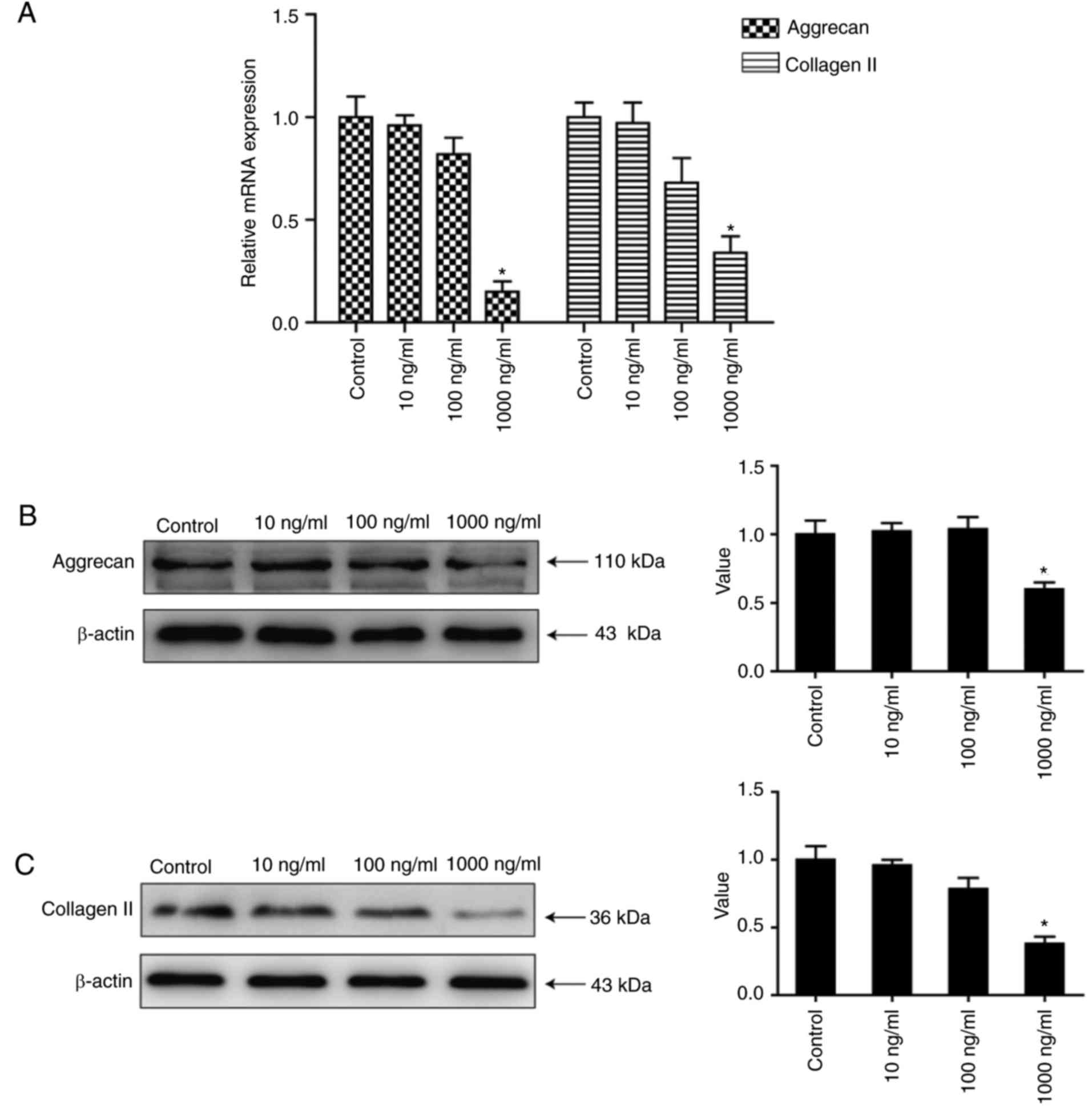
Aggrecan and collagen II levels are
increased following treatment of NP cells with high concentrations
of rhCILP
To further verify the negative regulation of CILP on
the synthesis of ECM, human NP cells were treated with various
concentrations of rhCILP. The results in Fig. 4 demonstrated that expression of the
matrix genes was markedly downregulated in the 1,000 ng/ml groups
(P<0.05), illustrating that a high level of CILP expression
inhibits the expression of matrix genes. No significant differences
were detected in the 10 and 100 ng/ml groups compared with the
control group (P>0.05), implying that the inhibition only has an
effect when CILP expression is upregulated to a high degree. These
results, along with the above findings, indicated that CILP
expression is strongly associated with the homeostasis of ECM in
IVDs, and a high CILP level in IDD may accelerate the degeneration
by disrupting ECM homeostasis.
Discussion
The present study aimed to investigate the
regulation of CILP by mechanical stress, and the association of
CILP expression with the synthesis of aggrecan and collagen II,
which are main components of the ECM that serve important roles in
maintaining the normal structure and function of human IVDs.
Consequently, to the best of our knowledge, the present study is
the first to demonstrate that CILP expression is regulated by
mechanical stress, and that its regulation varies with the type of
mechanical stress. Furthermore, the results indicated that altered
CILP expression has negative effects on the synthesis of aggrecan
and collagen II.
Preliminary examinations of the correlation of CILP
with IDD have been performed in previous studies. At the genetic
level, CILP gene expression has been associated with IDD in
Japanese and Finnish individuals (6–9). In
rabbit NP cells, a functional SNP of the CILP gene has been
reported to increase the direct interaction of CILP with TGF-β and
to interfere with the binding of TGF-β1 to its receptors. This
interference leads to suppression of the phosphorylation of mothers
against decapentaplegic (Smad)2/3, which are key factors in the
TGF-β/Smad signalling pathway (3).
Similarly, increased CILP expression exerts an inhibitory effect on
chondrocyte IGF-1 responsiveness and impairs IGF-1-mediated
chondrocyte growth and matrix repair in bovine chondrocytes
(10). Thus, it seems that CILP
may affect matrix synthesis via its interactions with several
growth factors. However, there is no direct evidence of a role of
CILP in matrix synthesis in human NP cells. To the best of our
knowledge, these results are the first to demonstrate that in human
NP cells, excessive CILP expression negatively regulates matrix
synthesis. The results further imply that abnormal CILP expression
may be one of the reasons for IDD, as suggested by previous
studies. However, it remains unknown whether this negative control
is a direct effect of interactions with NP cells or an indirect
effect of interactions with several growth factors; further study
is necessary to clarify this issue.
IVDs are load-bearing structures that undergo
various deformations in response to the spine load in daily life,
and transfer the external mechanical load to resident cells in NP
tissues (15–17,20).
These cells are susceptible to changes in the mechanical
environment, and exposure to excessive stress promotes increased
catabolic responses in NP cells, including upregulation of ADAMTs
and MMPs and downregulation of aggrecan and collagen II (15–17,20–22).
Therefore, homeostasis of the internal environment around NP cells
relies on the dynamic balance between the external mechanical
stimuli and the ability to buffer and absorb stress (13,15,16,21,22).
Once degeneration is initiated in IVDs, the aggrecan and collagen
II levels in the ECM decrease during the early stages, resulting in
reduced hydration (1), which
weakens the ability of IVDs to buffer mechanical stress and causes
NP cells to be exposed to more stressful stimuli, especially direct
compressive loading, further promoting the catabolic responses in
the ECM (1,13). In summary, mechanical stress,
especially excessive stress, is a primary factor promoting IDD.
Therefore, it is of significance to examine the regulation of CILP
expression by mechanical stresses. Mechanical stimuli have various
effects on metabolism in NP cells depending on the stimulus type,
and it is impossible to completely simulate the internal mechanical
environment in vitro. The present study selected compression
and tension, which are the main mechanical stimuli exerted on NP
cells, to determine the response of CILP expression in NP cells to
mechanical stress. Previous studies have demonstrated that the
effect of compression on NP cells relies on the magnitude,
frequency and duration. The compression experienced in daily life
ranges from 0.1–1.3 MPa at a frequency 0.1–1.0 Hz (22), and compression of >1 MPa at a
frequency of 1 Hz has been reported to increase catabolism in NP
cells (15), while compression of
0.25 MPa at a frequency of 0.1 Hz results in increased anabolism in
these cells (21). Given that the
NP cells used in the present study were derived from patients with
degenerative IVDs compression of 1.0–2.5 MPa at a frequency of 1 Hz
was selected to simulate the excessive stress faced by NP cells in
these IVDs. With regard to tension, previous studies concerning
tensile strain have demonstrated that 10% elongation evokes
significant changes in cells, and the loading of cells with a 3–10%
strain primarily leads to anabolic responses in chondrocyte cells
(23). To examine the effect of
tensile stimuli on CILP expression, the present study selected 10%
elongation at a frequency of 1 Hz. The results revealed that under
10% tensile loading at 1 Hz, mechanical stimuli that promoted the
expression of matrix genes significantly decreased that of CILP. In
addition, compressive loading of 1.0–2.5 MPa, an abnormal stimulus
for human NP cells, resulted in the marked upregulation of CILP
expression. These results demonstrated that CILP expression is
regulated not only by ageing or growth factors (5,10,17),
but also by mechanical stress, and that the effect of mechanical
regulation is dependent on the mechanical stress type. Given that
the NP cells in degenerative IVDs are exposed to excessive
compressive loading and insufficient tensile loading resulting from
the weakened ability of IVDs to suffer and transform external
mechanical stress. These results suggested that the high level of
CILP in IDD is more likely to be a result of abnormal mechanical
stress endured in daily life. Combined with the previous results
concerning the effect of CILP on the synthesis of ECM, these
experiments provided a novel explanation for the differential
expression of CILP in IVDs with differing degrees of IDD: It is the
abnormal mechanical loading leading to the excessive expression of
CILP in degenerative IVDs, thereby promoting IDD by exerting an
inhibitory effect on matrix synthesis.
However, there is a limitation to the present study
study; the role of CILP in mechanical stress-mediated regulation of
the ECM was not examined, of which the occurrence is implied by the
findings of our study but is not directly demonstrated, which need
further experiments to display. In conclusion, these results have
suggested that CILP may be a potential therapeutic target for
preventing loss of the ECM in IDD.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81071497).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
JH was involved in the conception and design of the
study, collection and/or assembly of data, data analysis and
interpretation, and manuscript writing. CF was involved in the
collection and/or assembly of data, and data analysis and
interpretation. KL and JS were involved in the collection and/or
assembly of data, and provision of study materials and patients. TC
was involved in the conception and design of the study, and the
revision of the manuscript. YZ was involved in the conception and
design of the study, and provided final approval of the manuscript.
YP provided final approval of the manuscript, was involved in the
conception and design of the study, and provided financial and
administrative support.
Ethics approval and consent to
participate
Human IVDs were obtained with the approval of the
Ethical Committee of Xinqiao Hospital (Chongqing, China) and with
informed consent, in accordance with the Helsinki Declaration, from
individuals undergoing surgery for IDD.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Colombini A, Lombardi G, Corsi MM and
Banfi G: Pathophysiology of the human intervertebral disc. Int J
Biochem Cell Biol. 40:837–842. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Daly C, Ghosh P, Jenkin G, Oehme D and
Goldschlager T: A review of animal models of intervertebral disc
degeneration: Pathophysiology, regeneration, and translation to the
clinic. Biomed Res Int. 2016:1–14. 2016. View Article : Google Scholar
|
|
3
|
Seki S, Kawaguchi Y, Chiba K, Mikami Y,
Kizawa H, Oya T, Mio F, Mori M, Miyamoto Y, Masuda I, et al: A
functional SNP in CILP, encoding cartilage intermediate layer
protein, is associated with susceptibility to lumbar disc disease.
Nat Genet. 37:607–612. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yee A, Lam MP, Tam V, Chan WC, Chu IK,
Cheah KS, Cheung KM and Chan D: Fibrotic-like changes in degenerate
human intervertebral discs revealed by quantitative proteomic
analysis. Osteoarthritis Cartilage. 24:503–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Z, Kim JH, Higashino K, Kim SS, Wang
S, Seki S, Hutton WC and Yoon ST: Cartilage intermediate layer
protein (CILP) regulation in intervertebral discs. The effect of
age, degeneration, and bone morphogenetic protein-2. Spine (Phila
Pa 1976). 37:E203–E208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Min SK, Nakazato K, Yamamoto Y, Gushiken
K, Fujimoto H, Fujishiro H, Kobayakawa Y and Hiranuma K: Cartilage
intermediate layer protein gene is associated with lumbar disc
degeneration in male, but not female, collegiate athletes. Am J
Sports Med. 38:2552–2557. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Min SK, Nakazato K, Ishigami H and
Hiranuma K: Cartilage intermediate layer protein and asporin
polymorphisms are independent risk factors of lumbar disc
degeneration in male collegiate athletes. Cartilage. 5:37–42. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang W, Hao J, Zheng S, Xiao X, Wen Y, He
A, Guo X and Zhang F: Association between cartilage intermediate
layer protein and degeneration of intervertebral disc: A
Meta-analysis. Spine (Phila Pa 1976). 41:E1244–E1248. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mayer JE, Iatridis JC, Chan D, Qureshi SA,
Gottesman O and Hecht AC: Genetic polymorphisms associated with
intervertebral disc degeneration. Spine J. 13:299–317. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnson K, Farley D, Hu SI and Terkeltaub
R: One of two chondrocyte-expressed isoforms of cartilage
intermediate-layer protein functions as an insulin-like growth
factor 1 antagonist. Arthritis Rheum. 48:1302–1314. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yao Z, Nakamura H, Masuko-Hongo K,
Suzuki-Kurokawa M, Nishioka K and Kato T: Characterisation of
cartilage intermediate layer protein (CILP)-induced arthropathy in
mice. Ann Rheum Dis. 63:252–258. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seki S, Tsumaki N, Motomura H, Nogami M,
Kawaguchi Y, Hori T, Suzuki K, Yahara Y, Higashimoto M, Oya T, et
al: Cartilage intermediate layer protein promotes lumbar disc
degeneration. Biophys Res Commun. 446:876–881. 2014. View Article : Google Scholar
|
|
13
|
Huang M, Wang HQ, Zhang Q, Yan XD, Hao M
and Luo ZJ: Alterations of ADAMTSs and TIMP-3 in human nucleus
pulposus cells subjected to compressive load: Implications in the
pathogenesis of human intervertebral disc degeneration. J Orthop
Res. 30:267–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Emanuel KS, Vergroesen PP, Peeters M,
Holewijn RM, Kingma I and Smit TH: Poroelastic behaviour of the
degenerating human intervertebral disc: A ten-day study in a loaded
disc culture system. Eur Cell Mater. 29:330–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
MacLean JJ, Lee CR, Alini M and Iatridis
JC: Anabolic and catabolic mRNA levels of the intervertebral disc
vary with the magnitude and frequency of in vivo dynamic
compression. J Orthop Res. 22:1193–1200. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao X, Zhu Q and Gu W: Prediction of
glycosaminoglycan synthesis in intervertebral disc under mechanical
loading. J Biomech. 49:2655–2661. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mori M, Nakajima M, Mikami Y, Seki S,
Takigawa M, Kubo T and Ikegawa S: Transcriptional regulation of the
cartilage intermediate layer protein (CILP) gene. Biochem Biophys
Res Commun. 341:121–127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Le Maitre CL, Frain J, Millward-Sadler J,
Fotheringham AP, Freemont AJ and Hoyland JA: Altered integrin
mechanotransduction in human nucleus pulposus cells derived from
degenerated discs. Arthritis Rheum. 60:460–469. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iorio JA, Jakoi AM and Singla A:
Biomechanics of degenerative spinal disorders. Asian Spine J.
10:377–384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Neidlinger-Wilke C, Würtz K, Urban JP,
Börm W, Arand M, Ignatius A, Wilke HJ and Claes LE: Regulation of
gene expression in intervertebral disc cells by low and high
hydrostatic pressure. Eur Spine J. 15 Suppl 3:S372–S378. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li P, Gan Y, Wang H, Zhang C, Wang L, Xu
Y, Song L, Li S, Li S, Ou Y and Zhou Q: Dynamic compression effects
on immature nucleus pulposus: A study using a novel intelligent and
mechanically active bioreactor. Int J Med Sci. 13:225–234. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bleuel J, Zaucke F, Brüggemann GP and
Niehoff A: Effects of cyclic tensile strain on chondrocyte
metabolism: A systematic review. PLoS One. 10:e01198162015.
View Article : Google Scholar : PubMed/NCBI
|