Introduction
Myelin is a fatty white substance that surrounds the
axons of neurons. In the central nervous system (CNS),
oligodendrocytes supply the myelin, which provides metabolic
support to the axon and allows for the rapid transmission of action
potentials along the axon (1).
Immune responses, inherited abnormalities or trauma may result in
oligodendrocyte necrosis and dysfunction, and resultant
demyelination in cases of multiple sclerosis and spinal cord
injury. The current treatment strategy for demyelination is based
on remyelination, a process that may restore metabolic support to
the axon to limit axonal degeneration and restore the nodes that
are required to facilitate conduction and, therefore, function
(2).
Although the CNS has little capacity for
regeneration, a number of studies have demonstrated that myelin may
be regenerated by oligodendrocytes (3–5). In
the CNS, endogenous oligodendrocyte precursor cells (OPCs)
contribute toward the replacement of oligodendrocytes required for
remyelination following demyelination (6). OPCs are able to proliferate and
differentiate into mature oligodendrocytes to repair injured myelin
following demyelination. However, the extent and quality of
endogenous remyelination is suboptimal (6). Therefore, numerous studies have aimed
to promote the maturation, proliferation and differentiation of
OPCs in order to improve remyelination (7–10).
In addition, efforts have also been made to enhance oligodendrocyte
replacement through cell transplantation by the authors of the
present study and others (11–14).
Although positive results have been achieved in
preclinical studies using rodent animal models, a clinical trial
using human CNS stem cells that have the ability to differentiate
into oligodendrocytes was performed in children with demyelination,
and the result, which exhibited a modest degree of remyelination
near the injection site, was far from being satisfactory (15). The different degrees of myelination
between rodent-based preclinical studies and human-based clinical
trials may be ascribed to the limited migration of the transplanted
cells in the human brain (2).
Notably, Boyd et al (16)
reported that insufficient OPC migration into demyelinated lesions
may be a critical cause of poor remyelination in multiple
sclerosis. Therefore, developing effective approaches to regulate
the migration of OPCs is urgently required and important for the
treatment of demyelination.
C-X-C motif chemokine ligand 12 (CXCL12; formerly
known as stromal cell-derived factor 1) is a well-identified
chemokine that serves an important role in mediating the migration
ability of multiple normal and tumor cells (17,18).
Previous studies have demonstrated that CXCL12 regulates the
proliferation and differentiation of OPCs (19,20).
Notably, CXCL12 promotes the migration of OPCs and improves
remyelination in vivo (20–23).
However, the underlying mechanism of the CXCL12-induced migration
of OPCs remains unclear. Considering that CXCL12 induces the
invasion of tumors via C-X-C motif chemokine receptor 4 (CXCR4; a
receptor of CXCL12) and the dual specificity mitogen-activated
protein kinase kinase 1 (MEK) and phosphoinositide 3-kinase (PI3K)
pathways (24–26), the present study assessed the
importance of the MEK and PI3K pathways in CXCL12/CXCR4-regulated
migration of OPCs.
Materials and methods
Isolation and culture of rat OPCs
The isolation and culture of OPCs was performed as
previously described (27,28). The cortical tissues of 8–12
neonatal Sprague-Dawley rats (postnatal day 2; 30) were purchased
from the experimental animal centre of Third Military Medical
University (1:1, male: female; weight 7–10 g). The rats were housed
at 25°C, 50% humidity, 12-h light/dark cycle and ad libitum
access to food and water. Cortical tissues of 8–12 rats per repeat
were resected to prepare a cell suspension with a 74 µM filter.
Following centrifugation at 200 × g for 10 min at 4°C, the cell
pellet was resuspended and cultured in a poly-lysine-coated culture
flask for 10 days at 37°C in 5% CO2. To purify OPCs, the
flask was placed onto a rotary shaker at 180 rpm for 1 h.
Subsequently, the supernatant in the flask was replaced with fresh
OPC proliferation medium to remove the dislodged cells (~90%
microglia). Following regular culture for 2 h, the flask was placed
onto a rotary shaker at 180 rpm for 18 h. The following day, the
supernatant was collected to pass through a cell strainer.
Following centrifugation at 200 × g for 10 min at 4°C, the pelleted
cells were resuspended and cultured in a new flask for 1 h. The
flask was gently agitated to remove loosely adherent cells. The
supernatant was collected, centrifuged at 200 × g for 10 min and
cultured in a new poly-lysine-coated culture flask.
The OPC proliferation medium contained Dulbecco's
modified Eagle's medium (DMEM) supplemented with 0.5% fetal bovine
serum (FBS), 10 ng/ml basic fibroblast growth factor, 10 ng/ml
platelet-derived growth factor (PDGF)-AA, 10 µg/ml insulin, 30 nM
sodium selenite, 0.5 µg/ml transferrin, 30 nM thyroiodine, 4 mM
L-glutamine, 5 mM sodium pyruvate, 50 U/ml penicillin and 50 µg/ml
streptomycin (all Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Differentiation of OPCs
To induce OPC differentiation, OPCs were cultured in
differentiation medium for 3 days. The differentiation medium for
OPCs consisted of DMEM, 0.5% FBS, 10 µg/ml insulin, 30 nM sodium
selenite, 0.5 µg/ml transferrin, 30 nM thyroiodine, 4 mM
L-glutamine, 5 mM sodium pyruvate, 50 U/ml penicillin and 50 µg/ml
streptomycin (all Invitrogen; Thermo Fisher Scientific, Inc.).
Immunostaining
Cells on cover slips were fixed with 4%
paraformaldehyde for 20 min at 4°C, permeabilized using 0.1% Triton
X-100 for 15 min and blocked for 60 min at 23°C with 5% goat serum
(Wuhan Boster Biological Technology, Ltd., Wuhan, China). Primary
antibodies against neural/glial antigen 2 (NG2; cat. no.
14-6504-80; 1:200; Thermo Fisher Scientific, Inc.), PDGF receptor-α
(PDGFR-α; cat. no. sc-31178; 1:200; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), oligodendrocyte marker O4 (O4; cat. no. MAB1326;
1:400; R&D Systems, Inc., Minneapolis, MN, USA) and myelin
basic protein (MBP; cat. no. sc-13526; 1:200; Santa Cruz
Biotechnology, Inc.) were diluted in 5% goat serum and incubated
with the samples overnight at 4°C. The following day, the cells
were incubated with secondary antibodies including goat anti-mouse
IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor
Plus 488 (cat. no. A32723; Thermo Fisher Scientific, Inc.; 1:500),
goat anti-mouse IgM (Heavy chain) cross-adsorbed secondary
antibody, Alexa Fluor 488 (cat. no. A-21042; Thermo Fisher
Scientific, Inc.; 1:500) and donkey anti-goat IgG (H+L)
cross-adsorbed secondary antibody, Alexa Fluor 488 (cat. no.
A-11055; Thermo Fisher Scientific, Inc.; 1:400) for 1 h at 37°C.
The nuclei were stained with DAPI (Invitrogen; Thermo Fisher
Scientific, Inc.) for 10 min at 23°C. Fluorescence images were
acquired using a fluorescence microscope (Olympus Corporation,
Tokyo, Japan; magnification, ×200).
Migration assay
The migration of OPCs was assessed using a Boyden
chamber, which contained 48-well inserts with an 8-µm pore-size
filter (Neuro Probe, Inc., Gaithersburg, MD, USA). A total of
5×104 OPCs were seeded in the top of the insert in
proliferation medium, while different concentrations of CXCL12 (0,
5, 10 and 20 ng/ml; PeproTech, Inc., Rocky Hill, NJ, USA) diluted
in proliferation medium were placed in the well below as a
chemoattractant. After 48 h, the top surface of the filter was
cleared with a cotton swab. Following fixing in methanol for 20 min
at 23°C, the back of the filter was stained at 23°C for 15 min with
0.1% crystal violet to observe the migrated OPCs. For quantitative
analysis, five random images of 480×360 µm were captured of the
filter. Each group had three replicate filters. To counteract
observer bias, the migrated OPCs (positive for crystal violet) were
counted by an individual who was blinded to the grouping of the
samples.
To test the effect of CXCR4 on the CXCL12-induced
migration of OPCs, CXCR4 short hairpin RNAs (shRNAs) were used to
downregulate the expression of CXCR4, while an shRNA control
(shRNAcon) served as the control (methods described below). The
untreated, shRNAcon-treated and CXCR4 shRNA-treated OPCs were used
to perform the 20 ng/ml CXCL12-induced migration assay, as
described above.
To test the effect of the MEK/extracellular
signal-regulated kinase (ERK) and PI3K/RAC-α
serine/threonine-protein kinase (AKT) pathways on the
CXCL12-induced migration of OPCs, OPCs were divided into four
groups: OPCs; OPCs treated with U1026 (inhibitor of the MEK/ERK
pathway; 10 µM; Invitrogen; Thermo Fisher Scientific, Inc.); OPCs
treated with LY294002 (inhibitor of the PI3K/AKT pathway; 10 µM;
Invitrogen; Thermo Fisher Scientific, Inc.); and OPCs treated with
U1026 (10 µM) and LY294002 (10 µM). Subsequently, the four groups
of OPCs were used to perform the 20 ng/ml CXCL12-induced migration
assay, as described above.
Western blotting
Cells were lysed in radioimmunoprecipitation assay
protein lysis buffer (Beyotime Institute of Biotechnology, Haimen,
China) and centrifuged at 12,000 × g for 5 min at 4°C. The
supernatant was collected to determine the protein concentration
using a bicinchoninic acid protein assay kit (CWBIO, Beijing,
China). Total proteins (20 µg/group) were separated using 12%
SDS-PAGE and transferred onto polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA). Following blocking with 5%
skimmed milk in TBS with Tween-20 (TBST) for 1 h at 23°C, the
membranes were incubated with antibodies against CXCR4 (1:1,000;
cat. no. PA1237; Wuhan Boster Biological Technology, Co., Ltd.,
Wuhan, China), AKT1 (1:2,000; cat. no. ab235958; Abcam, Cambridge,
UK), phosphorylated (p)-AKT1 (cat. no. ab81283; 1:2,000; Abcam),
ERK1/2 (1:2,000; cat. no. sc-93; Santa Cruz Biotechnology, Inc.),
p-ERK1/2 (1:2,000; cat. no. sc-16982-R; Santa Cruz Biotechnology,
Inc.) or GAPDH (1:5,000; cat. no. 10494-1-AP; ProteinTech Group,
Inc., Chicago, IL, USA) at 4°C overnight. The following day, the
membranes were washed with TBST three times and further incubated
with a horseradish peroxidase (HRP)-conjugated secondary antibody
(1:1,000; cat. no. A0208; Beyotime Institute of Biotechnology) at
37°C for 2 h. Finally, the protein bands were detected using an
enhanced chemiluminescence reagent (Pierce; Thermo Fisher
Scientific, Inc.). Densitometry was performed using Quantity one
v4.6.7 software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Knockdown of CXCR4
A lentivirus-based CXCR4 shRNA vector was
constructed as previously described (29). A total of three types of CXCR4
small interfering (si)RNA were designed and chemically synthesized
by Shanghai SunBio Biotechnology Co., Ltd. (Shanghai, China). The
sequences were as follows: CXCR4 siRNA 1,
5′-GGAUAACUACUCCGAAGAAdTdT-3′; CXCR4 siRNA 2,
5′-CCAACAAGGAACCCTGCTTdTdT-3′; and CXCR4 siRNA 3,
5′-CCCTCAAGACTACGGTCATdTdT-3′. Following transfection into OPCs
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), CXCR4 siRNA 2 exhibited the best efficiency at
downregulating the expression of CXCR4. As a result, complementary
DNA oligonucleotides of CXCR4 siRNA 2 were subcloned into a
lentiviral vector to construct CXCR4 shRNA. A vector containing a
scrambled sequence served as a CXCR4 shRNA control. Finally, OPCs
were transfected with CXCR4 shRNA or shRNAcon for 72 h and
subjected to western blot analysis or a migration assay. Based on
the sequence of 5′-GCAAGAUCACACACCUCAUdTdT-3′, siRNA of atypical
chemokine receptor 3 (CXCR7) was chemically synthesized and
subcloned into shRNA. The knockdown of CXCR7 was performed as
described above.
Statistical analysis
Experimental results are presented as the mean ±
standard deviation and were analyzed using SPSS 19.0 statistical
software (IBM Corp., Armonk, NY, USA) with one-way analysis of
variance (ANOVA). Following ANOVA, the least significant difference
post hoc test was used. Experiments were repeated three times.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Isolation, culturing and
identification of rat OPCs
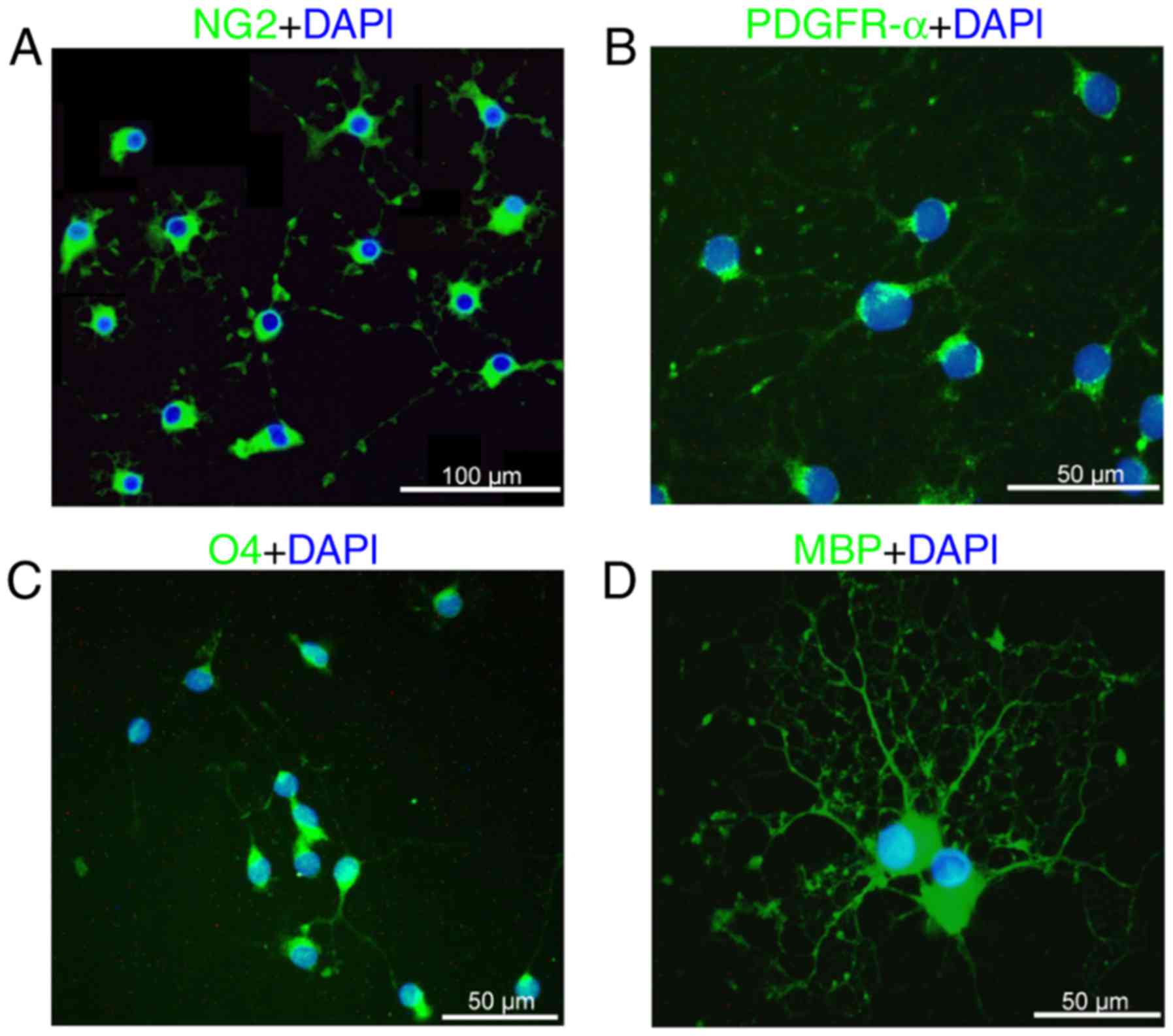
The present study isolated primary rat OPCs, as
previously described (27,28). The isolated cells harbored the
typical morphology of OPCs, exhibiting a round body with bipolar
and tripolar processes. The immunostaining assay confirmed that the
isolated OPCs expressed NG2 and PDGFR-α, OPC-specific markers
(Fig. 1A and B), although they
were negative for O4 and MBP, as markers of mature oligodendrocytes
(data not shown). Following culturing in differentiation medium for
3 days, the OPCs were differentiated into mature oligodendrocytes,
which highly expressed O4 and MBP (Fig. 1C and D). These findings confirmed
that the isolated cells acquired the OPC phenotype.
CXCL12 induces the migration of
OPCs
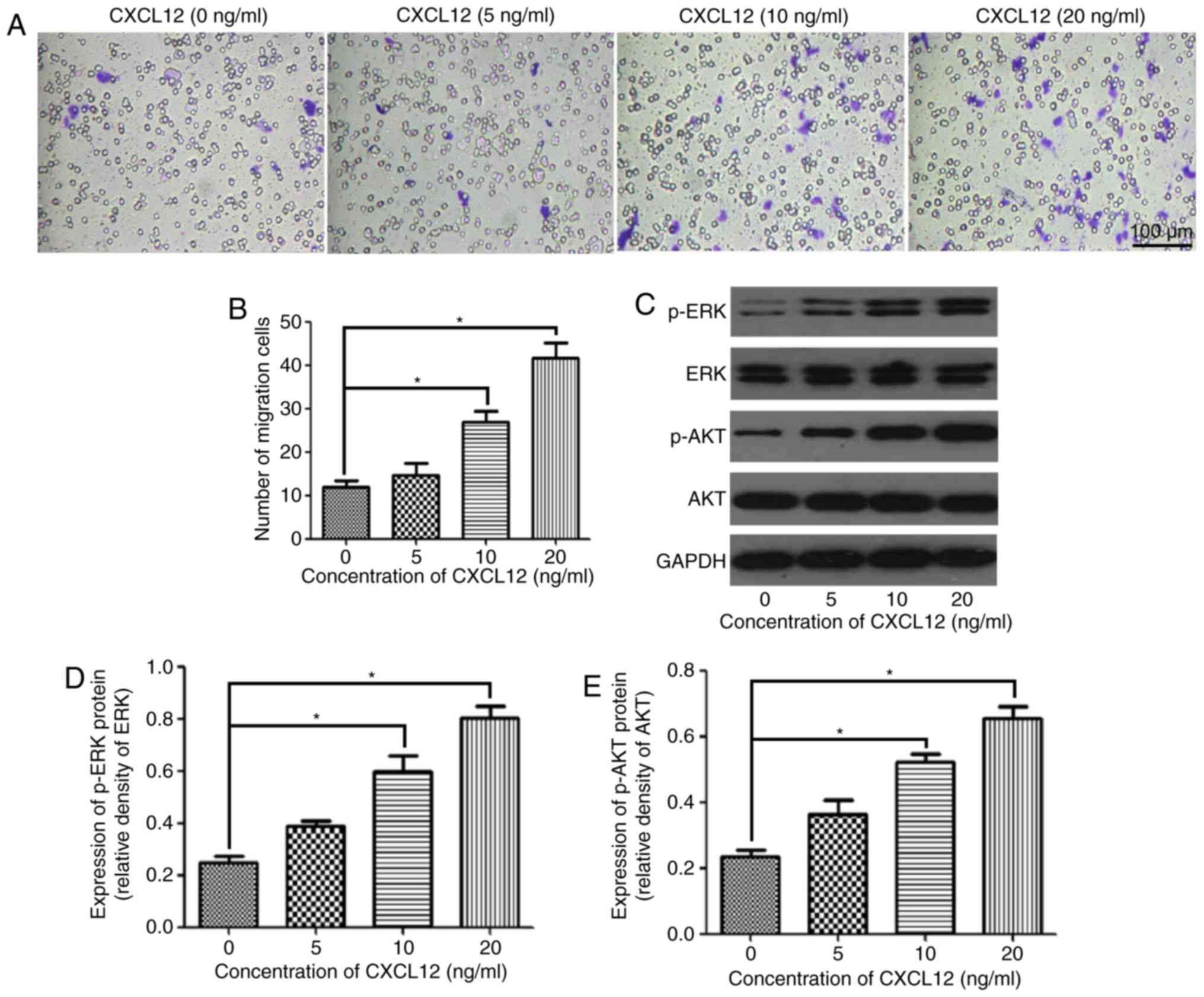
The present study assessed the effect of CXCL12 on
the migration of OPCs using the Boyden chamber assay. The migration
of OPCs was gradually enhanced with the increasing concentration of
CXCL12 (Fig. 2A). According to the
statistical analysis, the migration of OPCs was significantly
increased by the chemoattractant effects of 10 and 20 ng/ml CXCL12
compared with the control group (0 ng/ml CXCL12) (Fig. 2B). To further investigate the
downstream mechanism of the CXCL12-induced migration of OPCs, the
present study examined the expression of p-ERK and p-AKT, and
demonstrated that p-ERK and p-AKT were significantly upregulated
following treatment with 10 and 20 ng/ml CXCL12 (Fig. 2C-E). However, treatment with CXCL12
did not markedly affect the expression of ERK and AKT in OPCs
(Fig. 2C-E). These findings
suggested that the MEK/ERK and PI3K/AKT pathways are likely to be
the downstream mechanism through which CXCL12 induces the migration
of OPCs.
Knockdown of CXCR4 inhibits the
migration of OPCs
Additionally, the effect of knockdown of CXCR4 on
the CXCL12-induced migration of OPCs was assessed. A total of three
types of CXCR4 siRNA were designed to knock down the expression of
CXCR4, and CXCR4 siRNA 2 exhibited the best efficiency at
downregulating the expression of CXCR4 (Fig. 3A and B). CXCR4 shRNA was
constructed based on CXCR 4 siRNA 2. Under the chemotactic effects
of 20 ng/ml CXCL12, the migration of CXCR4 shRNA-transfected OPCs
was significantly decreased compared with the untreated and
shRNAcon-treated OPCs (Fig. 3C and
D). However, knockdown of CXCR7 (another receptor of CXCL12)
did not affect CXCL12-induced migration (data not shown). In
addition, CXCR4 shRNA significantly downregulated the expression of
CXCR4 following treatment with 20 ng/ml CXCL12 (Fig. 3E and F). Notably, the expression of
p-ERK and p-AKT was also downregulated by treatment with CXCR4
shRNA, which supported the hypothesis that the MEK/ERK and PI3K/AKT
pathways may be downstream of CXCR4 in OPCs (Fig. 3G and H). These data demonstrated
that CXCL12 induced the migration of OPCs via CXCR4.
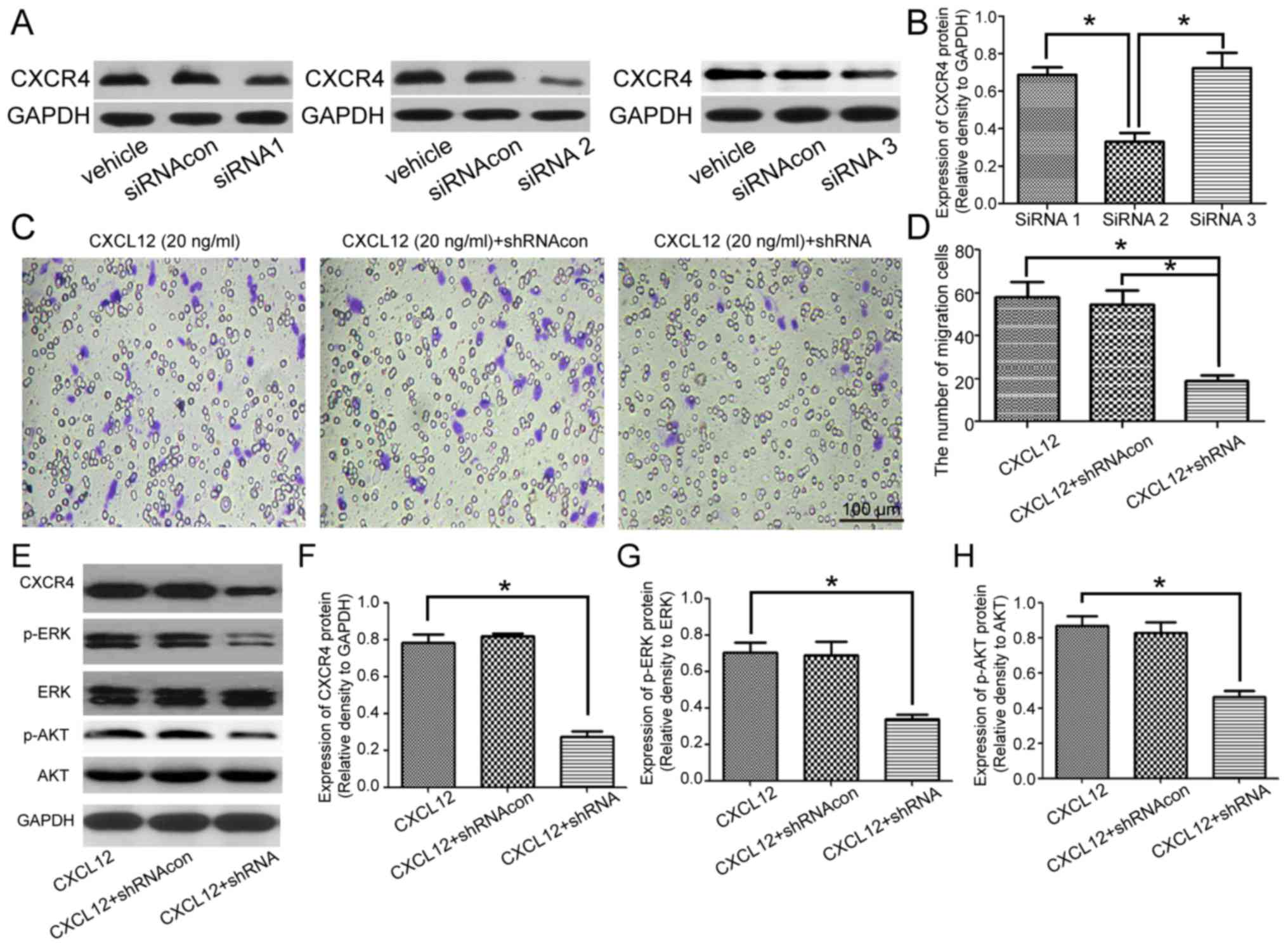 | Figure 3.Knockdown of CXCR4 inhibits the
CXCL12-induced migration of OPCs. (A) Western blot analysis
demonstrated the expression of CXCR4 in the OPCs treated with
vehicle (PBS), siRNAcon and each siRNA (siRNA1, siRNA2 and siRNA3).
(B) Relative expression of CXCR4 in the OPCs treated with siRNA1,
siRNA2 and siRNA3. (C) Transwell migration assay of OPCs treated
with CXCL12 (20 ng/ml) and CXCR4 shRNA or shRNAcon. (D) Analysis of
the migration data for each group. (E) Western blot analysis
demonstrated the expression of CXCR4, p-ERK, ERK, p-AKT and AKT in
the OPCs treated with CXCL12 (20 ng/ml) and CXCR4 shRNA or
shRNAcon. GAPDH served as a loading control. (F) Group data for the
relative expression of CXCR4. (G) Group data for the relative
expression of p-ERK. (H) Group data for the relative expression of
p-AKT. Data are presented as the mean ± standard deviation. n=3.
*P<0.05. CXCL12, C-X-C motif chemokine ligand 12; CXCR4, C-X-C
motif chemokine receptor 4; siRNA, small interfering RNA; con,
control; shRNA, short hairpin RNA; OPCs, oligodendrocyte precursor
cells; p, phosphorylated; ERK, extracellular signal-regulated
kinase; AKT, RAC-α serine/threonine-protein kinase. |
CXCL12-induced migration of OPCs is
regulated by the MEK/ERK and PI3K/AKT pathways
To further confirm that the MEK/ERK and PI3K/AKT
pathways were involved in the CXCL12-induced migration of OPCs,
U1026 and LY294002 were used as specific kinase inhibitors of the
MEK/ERK and PI3K/AKT pathways to respectively block each signaling
pathway. As expected, treatment with U1026 or LY294002 was able to
significantly inhibit the CXCL12-induced migration of OPCs
(Fig. 4A and B). Furthermore,
western blot analysis demonstrated that U1026 specifically
downregulated the expression of p-ERK in OPCs following treatment
with 20 ng/ml CXCL12 (Fig. 4C and
D). Likewise, LY294002 specifically downregulated the
expression of p-AKT in OPCs (Fig. 4C
and E). Taken together, these results suggested that the
CXCL12-induced migration of OPCs was regulated by the MEK/ERK and
PI3K/AKT pathways.
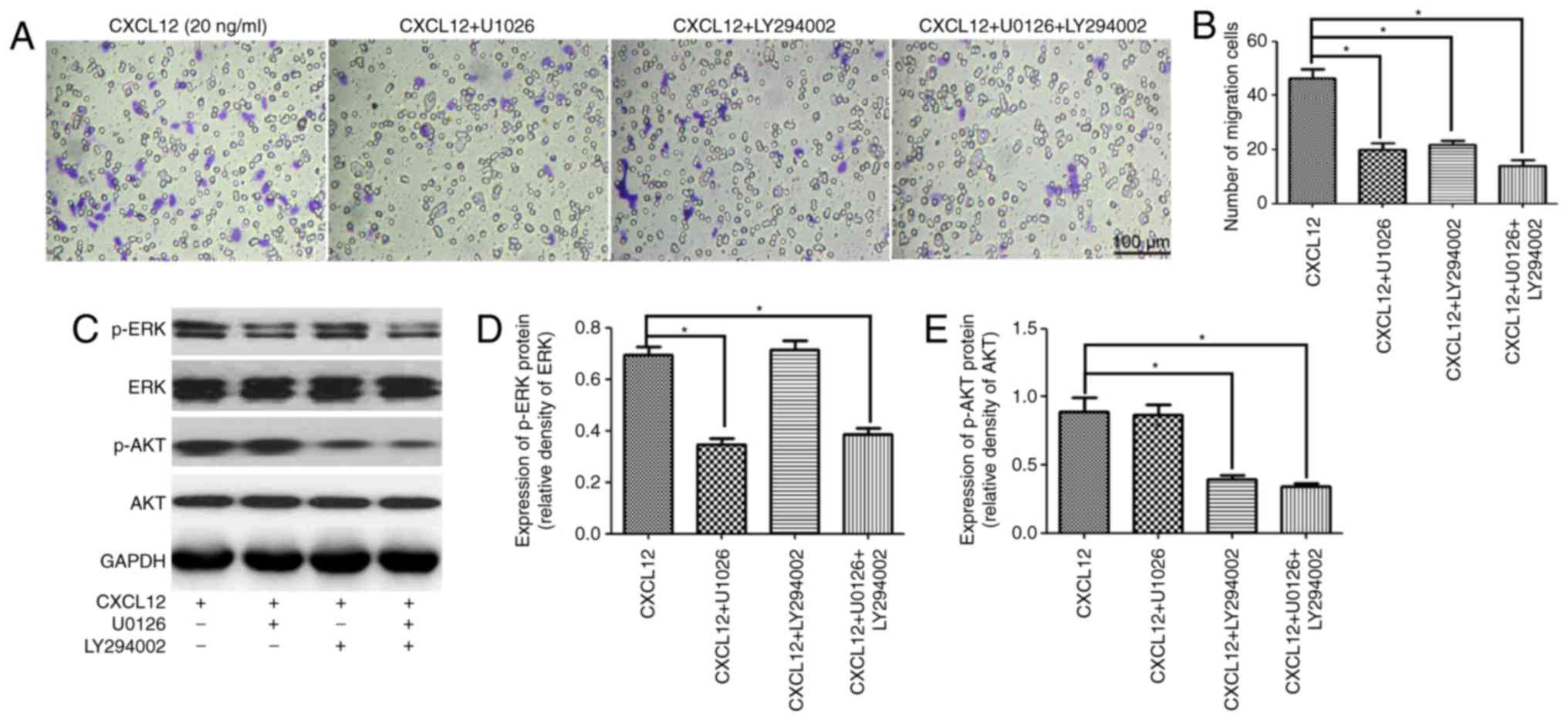 | Figure 4.MEK/ERK and PI3K/AKT pathways are
involved in the CXCL12-induced migration of OPCs. (A) Transwell
migration assay of OPCs treated with CXCL12 (20 ng/ml) and an
inhibitor of the MEK/ERK pathway (U1026), or an inhibitor of the
PI3K/AKT pathway (LY294002). (B) Group migration data. (C) Western
blot analysis demonstrated the expression of p-ERK, ERK, p-AKT and
AKT in the OPCs treated with CXCL12 (20 ng/ml) and U1026 or
LY294002. GAPDH served as a loading control. (D) Group data for the
relative expression of p-ERK. (E) Group data for the relative
expression of p-AKT. Data are presented as the mean ± standard
deviation. n=3. *P<0.05. CXCL12, C-X-C motif chemokine ligand
12; OPCs, oligodendrocyte precursor cells; p, phosphorylated; ERK,
extracellular signal-regulated kinase; AKT, RAC-α
serine/threonine-protein kinase; MEK, dual specificity
mitogen-activated protein kinase kinase 1; PI3K, phosphoinositide
3-kinase. |
Discussion
Investigating the migration of OPCs is of great
importance to improve remyelination in the CNS. The results of the
present study demonstrated that CXCL12 induced the migration of
OPCs via the CXCR4-activated MEK/ERK and PI3K/AKT pathways.
According to a well-established protocol, neonatal
OPCs were separated by shaking and differential adhesion (27,28).
The obtained OPCs were positive for NG2 and PDGFR-α and were able
to differentiate into mature oligodendrocytes, which were positive
for O4 and MBP. In addition, it was observed that 10 and 20 ng/ml
CXCL12 significantly promoted the migration of OPCs in
vitro, which is consistent with the results of a previous study
(22). By contrast, another study
demonstrated that CXCL12 inhibits the migration of OPCs and
augments the differentiation of OPCs into mature oligodendrocytes
(30). It has been reported that
the expression of CXCR4 decreases gradually during the
differentiation of OPCs (22).
CXCR4, as a receptor of CXCL12, serves an important role in the
migration of OPCs. As a result, when CXCL12 induced the
differentiation of OPCs, CXCL12 was insufficient in promoting the
migration of OPCs due to the downregulation of CXCR4. However,
CXCL12 promoted the migration of OPCs in the present study as the
OPCs were prevented from differentiating in the proliferation
medium. In addition, it was additionally demonstrated that
knockdown of CXCR4 inhibited the CXCL12-induced migration of OPCs
in vitro. Dziembowska et al (22) also demonstrated that
CXCR4−/− mice exhibit defective migration of OPCs in
vivo. This suggests that CXCL12 may induce the migration of
OPCs through CXCR4.
Furthermore, the present study demonstrated that the
MEK/ERK and PI3K/AKT pathways were downstream of CXCL12/CXCR4.
Treatment with CXCL12 was able to activate the MEK/ERK and PI3K/AKT
pathways, while knockdown of CXCR4 inhibited the MEK/ERK and
PI3K/AKT pathways. Using high-throughput quantitative
phosphoproteomic analysis, Yi et al (31) demonstrated that the MEK/ERK pathway
is downstream of CXCL12/CXCR4 in breast cancer stem cells. Notably,
specific inhibitors of the MEK/ERK and PI3K/AKT pathways
significantly reduced the migration of OPCs, which supported the
hypothesis that the MEK/ERK and PI3K/AKT pathways were involved in
the migration of OPCs. This finding was in agreement with previous
results demonstrating that the CXCL12/CXCR4-activated MEK/ERK and
PI3K/AKT pathways regulated the migration of cancer cells (24–26,32,33).
Considering the fact that CXCL12/CXCR4 increases the
phosphorylation of a number of cell migration- and
invasion-associated proteins in breast cancer stem cells (31), there may be other pathways involved
in the CXCL12/CXCR4-induced migration of OPCs.
Taken together, the results of the present study
confirmed that CXCL12 induces the migration of OPCs through the
CXCR4-activated MEK/ERK and PI3K/AKT pathways. This study provides
an experimental basis for the improved understanding of the
CXCL12-induced migration of OPCs, which is of translational
importance in improving remyelination.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81471262).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XR and TJ conceived and designed the experiments.
YT, XD and BT performed the experiments. YT and HY analyzed the
data. XR and TJ contributed reagents and materials. HY and TJ wrote
the paper.
Ethics approval and consent to
participate
All procedures were performed according to protocols
approved by the Institutional Review Board of Third Military
Medical University and conformed to the National Institutes of
Health (Bethesda, MD, USA) guide for the care and use of laboratory
animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CXCL12
|
C-X-C motif chemokine ligand 12
|
|
CXCR4
|
C-X-C motif chemokine receptor 4
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
OPCs
|
oligodendrocyte precursor cells
|
|
CNS
|
central nervous system
|
|
PDGFR-α
|
platelet-derived growth factor
receptor-α
|
|
MBP
|
myelin basic protein
|
|
AKT
|
RAC-α serine/threonine-protein
kinase
|
References
|
1
|
Nave K: Myelination and the trophic
support of long axons. Nat Rev Neurosci. 11:275–283. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Franklin RJM ffrench-Constant C, .
Regenerating CNS myelin-from mechanisms to experimental medicines.
Nat Rev Neurosci. 18:753–769. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Young KM, Psachoulia K, Tripathi RB, Dunn
SJ, Cossell L, Attwell D, Tohyama K and Richardson WD:
Oligodendrocyte dynamics in the healthy adult CNS: Evidence for
myelin remodeling. Neuron. 77:873–885. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De La Fuente AG, Lange S, Silva ME,
Gonzalez GA, Tempfer H, van Wijngaarden P, Zhao C, Di Canio L,
Trost A, Bieler L, et al: Pericytes stimulate oligodendrocyte
progenitor cell differentiation during CNS remyelination. Cell Rep.
20:1755–1764. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo YE, Suo N, Cui X, Yuan Q and Xie X:
Vitamin C promotes oligodendrocytes generation and remyelination.
Glia. 66:1302–1316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alizadeh A, Dyck SM and Karimi-Abdolrezaee
S: Myelin damage and repair in pathologic CNS: Challenges and
prospects. Front Mol Neurosci. 8:352015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tokunaga H, Seiwa C, Yoshioka N, Mizoguchi
K, Yamamoto M, Asou H and Aiso S: An extract of chinpi, the dried
peel of the citrus fruit unshiu, enhances axonal remyelination via
promoting the proliferation of oligodendrocyte progenitor cells.
Evid Based Complement Alternat Med. 2016:86926982016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ossola B, Zhao C, Compston A, Pluchino S,
Franklin RJM and Spillantini MG: Neuronal expression of
pathological tau accelerates oligodendrocyte progenitor cell
differentiation. Glia. 64:457–471. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang S, Tang C, Sun S, Cao W, Qi W, Xu J,
Huang J, Lu W, Liu Q, Gong B, et al: Protective effect of
electroacupuncture on neural myelin sheaths is mediated via
promotion of oligodendrocyte proliferation and inhibition of
oligodendrocyte death after compressed spinal cord injury. Mol
Neurobiol. 52:1870–1881. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hackett AR, Lee DH, Dawood A, Rodriguez M,
Funk L, Tsoulfas P and Lee JK: STAT3 and SOCS3 regulate NG2 cell
proliferation and differentiation after contusive spinal cord
injury. Neurobiol Dis. 89:10–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen LX, Ma SM, Zhang P, Fan ZC, Xiong M,
Cheng GQ, Yang Y, Qiu ZL, Zhou WH and Li J: Neuroprotective effects
of oligodendrocyte progenitor cell transplantation in premature rat
brain following hypoxic-ischemic injury. PLoS One. 10:e01159972015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang S, Bates J, Li X, Schanz S,
Chandler-Militello D, Levine C, Maherali N, Studer L, Hochedlinger
K, Windrem M and Goldman SA: Human iPSC-derived oligodendrocyte
progenitor cells can myelinate and rescue a mouse model of
congenital hypomyelination. Cell Stem Cell. 12:252–264. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Xiong LL, Wang YC, He X, Jiang L,
Fu SJ, Han XF, Liu J and Wang TH: Oligodendrocyte precursor cell
transplantation promotes functional recovery following contusive
spinal cord injury in rats and is associated with altered microRNA
expression. Mol Med Rep. 17:771–782. 2018.PubMed/NCBI
|
|
14
|
Wu B, Sun L, Li P, Tian M, Luo Y and Ren
X: Transplantation of oligodendrocyte precursor cells improves
myelination and promotes functional recovery after spinal cord
injury. Injury. 43:794–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gupta N, Henry RG, Strober J, Kang SM, Lim
DA, Bucci M, Caverzasi E, Gaetano L, Mandelli ML, Ryan T, et al:
Neural stem cell engraftment and myelination in the human brain.
Sci Transl Med. 4:155ra1372012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boyd A, Zhang H and Williams A:
Insufficient OPC migration into demyelinated lesions is a cause of
poor remyelination in MS and mouse models. Acta Neuropathol.
125:841–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Janssens R, Struyf S and Proost P: The
unique structural and functional features of CXCL12. Cell Mol
Immunol. Oct 30–2017.(Epub ahead of print). PubMed/NCBI
|
|
18
|
Meng W, Xue S and Chen Y: The role of
CXCL12 in tumor microenvironment. Gene. 641:105–110. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kadi L, Selvaraju R, de Lys P, Proudfoot
AE, Wells TN and Boschert U: Differential effects of chemokines on
oligodendrocyte precursor proliferation and myelin formation in
vitro. J Neuroimmunol. 174:133–146. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Patel JR, McCandless EE, Dorsey D and
Klein RS: CXCR4 promotes differentiation of oligodendrocyte
progenitors and remyelination. Proc Natl Acad Sci USA.
107:11062–11067. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zilkha-Falb R, Kaushansky N, Kawakami N
and Ben-Nun A: Post-CNS-inflammation expression of CXCL12 promotes
the endogenous myelin/neuronal repair capacity following
spontaneous recovery from multiple sclerosis-like disease. J
Neuroinflammation. 13:72016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dziembowska M, Tham T, Lau P, Vitry S,
Lazarini F and Dubois-Dalcq M: A role for CXCR4 signaling in
survival and migration of neural and oligodendrocyte precursors.
Glia. 50:258–269. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carbajal KS, Miranda JL, Tsukamoto MR and
Lane TE: CXCR4 signaling regulates remyelination by endogenous
oligodendrocyte progenitor cells in a viral model of demyelination.
Glia. 59:1813–1821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sobolik T, Su YJ, Wells S, Ayers GD, Cook
RS and Richmond A: CXCR4 drives the metastatic phenotype in breast
cancer through induction of CXCR2 and activation of MEK and PI3K
pathways. Mol Biol Cell. 25:566–582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kukreja P, Abdel-Mageed AB, Mondal D, Liu
K and Agrawal KC: Up-regulation of CXCR4 expression in PC-3 Cells
by stromal-derived factor-1α (CXCL12) increases endothelial
adhesion and transendothelial migration: Role of MEK/ERK signaling
pathway-dependent NF-kappaB activation. Cancer Res. 65:9891–9898.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang CY, Lee CY, Chen MY, Yang WH, Chen
YH, Chang CH, Hsu HC, Fong YC and Tang CH: Stromal cell-derived
factor-1/CXCR4 enhanced motility of human osteosarcoma cells
involves MEK1/2, ERK and NF-κB-dependent pathways. J Cell Physiol.
221:204–212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Armstrong R: Isolation and
characterization of immature oligodendrocyte lineage cells.
Armstrong RC. 16:282–292. 1998.
|
|
28
|
Itoh K: Culture of oligodendrocyte
precursor cells (NG2+/O1-) and oligodendrocytes (NG2(−)/O1(+)) from
embryonic rat cerebrum. Brain Res Brain Res Protoc. 10:23–30. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu X, Chen D, Zhang Y, Wu X, Huang Z, Zhou
H, Zhang Y and Zhang Z: Overexpression of CXCR4 in mesenchymal stem
cells promotes migration, neuroprotection and angiogenesis in a rat
model of stroke. J Neurol Sci. 316:141–149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maysami S, Nguyen D, Zobel F, Pitz C,
Heine S, Höpfner M and Stangel M: Modulation of rat oligodendrocyte
precursor cells by the chemokine CXCL12. Neuroreport. 17:1187–1190.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle
T, Etzkorn M, Seo HC, Nagiec M, Luna RE, Reinherz EL, et al:
Quantitative phosphoproteomic analysis reveals system-wide
signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem
cells. Proc Natl Acad Sci USA. 111:E2182–E2190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu T, Wu Y, Helman JI, Wen Y, Wang C and
Li L: CXCR4 promotes oral squamous cell carcinoma migration and
invasion through inducing expression of MMP-9 and MMP-13 via the
ERK signaling pathway. Mol Cancer Res. 9:161–172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun X, Wei L, Chen Q and Terek R:
CXCR4/SDF1 mediate hypoxia induced chondrosarcoma cell invasion
through ERK signaling and increased MMP1 expression. Mol Cancer.
9:172010. View Article : Google Scholar : PubMed/NCBI
|