Introduction
Fatty liver diseases, whose prevalence is
continuously rising worldwide, especially in developed countries,
are characterized by excessive hepatic fat accumulation (1). Various causes, including a
high-fat/calorie diet, alcoholism and other metabolic disorders,
can lead to fatty liver diseases. According to different causes,
fatty liver can be divided into alcoholic fatty liver disease and
non-alcoholic fatty liver disease (NAFLD) (2). NAFLD, covering a spectrum of chronic
liver diseases, including simple fatty liver, fatty hepatitis,
fibrosis, cirrhosis, and hepatic carcinoma, affects approximately
20–40% of the general population (3). NAFLD is considered to be the hepatic
manifestation of metabolic dysfunction and is strongly related to
obesity, insulin resistance (IR), and dyslipidemia (4,5).
Studies have shown that the accumulation of fat in
hepatocytes mainly comprises triglyceride (TG) deposition (6). Diet, de novo synthesis and adipose
tissue are three major sources of the acquisition of hepatic TG.
Likewise, total cholesterol (TC) is related to the severity of
NAFLD (7). In addition, it has
been reported that increased concentrations of alanine
aminotransferase (AST) and alanine aminotransferase (ALT) in serum
commonly lead to NAFLD (8). Sterol
response element-binding protein (SREBP)-1c, which is primarily
present in the liver, is negatively regulated by AMP-activated
protein kinase (AMPK) (9) and
usually regulates lipogenesis as a transcription factor binding to
target lipogenic genes, such as fatty acid synthase (FAS) (10,11).
Furthermore, SPEBP-1c has been suggested to stimulate the synthesis
of TG with the synergistic effect of ChREBP (12).
Toll-like receptor 4 (TLR4), the most characteristic
member of the TLR family, commonly functions as a
pattern-recognition receptor. Studies have closely related TLR4 to
the pathogenesis of NAFLD, and it has further been shown that NAFLD
may be prevented by allelic variants of TLR4 in humans (13). Additionally, it is well-known that
the nuclear factor (NF)-κB pathway, a master proinflammatory
switch, is of great importance to the entire process from simple
steatosis to steatohepatitis (14). The binding of various homo- or
heterodimers, such as p65 formed by NF-κB proteins to a consensus
sequence of NF-κB, usually regulates the transcription of many
factors functioning in NAFLD (15). Furthermore, the NF-κB pathway is
reported to be activated by a cascade of downstream signals due to
the activation of TLR4, which further activates the production of
tumor necrosis factor (TNF)-α, interleukin (IL)-6 and other
proinflammatory molecules (16).
Studies have shown that TNF-α acts as a key role in NAFLD
progression both in humans and animals (17), and anti-TNF-α drugs improve
HFD-induced alterations of the liver (18). For IL-6, due to its ability to
reduce oxidative stress and avoid mitochondrial dysfunction, it was
initially thought to be a hepatoprotective factor in hepatic
steatosis (19,20), which was confirmed by further
studies in other liver disease models (21–23).
Later, it was reported that IL-6 increased in human nonalcoholic
steatohepatitis, and deficiency of IL-6 inhibited the progression
of NAFLD (24,25).
Metformin, first introduced in the 1950s in the
clinic, is an effective drug for NAFLD treatment, although its
efficacy is limited and it has considerable side effects (26). In addition, berberine (BBR)
isolated from Rhizoma Coptidis has been reported to have
anti-diabetic and anti-hyperlipidemic effects (27–29).
Furthermore, numerous studies have demonstrated that BBR can
attenuate hepatic steatosis and hyperlipidemia by inhibiting the
synthesis and accumulation of lipid in hepatocytes, which prevents
the development of NAFLD (30,31).
Therefore, both drugs have effects on the treatment of NAFLD
(32–34). However, the specific mechanism
remains unclear.
C-C motif ligand 19 (CCL19), a CC-type chemokine
also known as macrophage inflammatory protein-3β (MIP-3β), is
produced by macrophages, dendritic cells and other different cells,
and it usually binds to CC chemokine receptor 7 (CCR7). CCL19 has
been implicated in chronic inflammation, lymphoid neogenesis, and
dendritic extension (35,36). Studies have indicated that lack of
CCL19-CCR7 signaling has a protective effect on diet-induced
obesity and insulin resistance (37). However, further research on the
effect of CCL19 on fatty liver disease is still lacking.
In this paper, high expression levels of CCL19,
TLR4, NF-κB-p56, IL-6, and TNF-α were detected in NAFLD patients
and increased with the severity of NAFLD. This suggested that CCL19
may be implicated in NAFLD progression. In a rat model of NAFLD,
dyslipidemia and the liver histopathology of steatosis were
improved by metformin and BBR. Furthermore, gene expression
analysis uncovered that high expression of CCL19 and related
factors were also significantly decreased by metformin and BBR,
whereas p-AMPK levels were increased. Therefore, we speculate that
metformin and BBR improves NAFLD may through AMPK activation, and
the inhibition of CCL19 is related to NAFLD treatment.
Materials and methods
Samples of peripheral blood and liver
tissues of patients with fatty liver diseases
Based on specific criteria, the severity of NAFLD
was classified as absent, mild NAFLD (Liver echogen slightly
increased, good visualization of intrahepatic vessels and
diaphragms), moderate NAFLD (Increased liver echogenicity,
disappearance of intrahepatic vascular echo lines) and severe NAFLD
(Significant increase in liver echogenicity, poor penetration of
the right hepatic lobe, and blurred vision of the diaphragm)
(38,39). After obtaining written and informed
consent, peripheral blood and liver tissues were collected from 30
healthy volunteers, 30 mild NAFLD patients and 30 severe NAFLD
patients. All of the patients enrolled in this study were treated
in Putuo District People's Hospital of Shanghai City (Shanghai,
China). After processing the samples, ELISA and western blot
analyses were carried out to examine relative gene or factor
expression. All experiments in this study were approved by the
Ethics Committee of Putuo District People's Hospital of Shanghai
City.
Experimental group
For follow-up animal experiments, a rat model of
fatty liver was induced by feeding a high-fat diet (HFD). Sixty
Sprague-Dawley (SD) rats from the Experimental Animal Center
(Shanghai, China) were housed in a room with a constant temperature
of 22–25°C, a 12/12 h light/dark cycle and a relative humidity of
60±10%. The sixty rats were fed randomly with normal chow diet
(n=12) or HFD (n=48) for 4 weeks to induce NAFLD. Then, the 48
HFD-fed rats were further randomized and continually fed with HFD
(HFD group), with HFD and treatment of 150 mg/kg/d metformin (HFD +
metformin), with HFD and treatment of 250 mg/kg/d BBR (HFD + BBR),
or with HFD and treatment of 150 mg/kg/d metformin plus 250 mg/kg/d
BBR (HFD + metformin + BBR) for 4 and 8 weeks. Additionally, rats
fed a normal chow diet constantly without exposure to HFD were
regarded as controls. At the end of 4 and 8 weeks of treatment, the
serum of these rats was collected for biochemical and specific
ELISA testing, while the liver tissues obtained at 8 weeks were
utilized for hematoxylin and eosin (H&E) staining, RT-PCR as
well as western blot analyses. All of the experimental procedures
were approved by the Experimental Animal Research Committee of
Putuo District People's Hospital of Shanghai (Shanghai, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was applied to isolate the
total RNA from the liver tissues. After quantification, the
integrity of RNA was confirmed by electrophoresis using a 1%
agarose gel. Then, using a reverse transcriptase kit (Fermentas;
Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA), cDNA was
generated from isolated RNA through reverse transcription.
Subsequently, using a SYBR Green PCR kit (Thermo Fisher Scientific,
Inc.) and cDNA as the template, a 25 µl volume of RT-PCR reaction
was assessed by a ABI Prism 7300 real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
After the procedure, using GAPDH as an internal control and using
the 2−ΔΔCq method (40), the mRNA expression of CCL19,
SREBP-1c, FAS, TLR4 and NF-κB-p65 was determined. The sequences of
primers used were as follows: GAPDH forward (F):
5′-GGAGTCTACTGGCGTCTTCAC-3′ and reverse (R):
5′-ATGAGCCCTTCCACGATGC-3′; CCL19 F: 5′-CCTTCCGCTACCTTCTTATC-3′ and
R: 5′-CTCTTCTGGTCCTTGGTTTC-3′; SREBP-1c F:
5′CGCTACCGTTCCTCTATCAATG3′ and R: 5′-TCTGGTTGCTGTGCTGTAAG-3′; FAS
F: 5′-TGGTTCATTCCGTGACTG-3′ and R: 5′-TGTCCTTCGGTTCTTTCC-3′; TLR4
F: 5′-ATGCTAAGGTTGGCACTCTC-3′ and R: 5′-CAGGCAGGAAAGGAACAATG-3′;
NF-κB-p65 F: 5′-AGACCTGGAGCAAGCCATTAG-3′ and R:
5′-CGGACCGCATTCAAGTCATAG-3′. The RT-PCR procedure was as follows:
95°C for 10 min; followed by 95°C for 15 sec, 60°C for 45 sec for
40 cycles; 95°C for 15 sec and 60°C for 1 min for one cycle; and
95°C for 15 sec and 60°C for 15 sec for one cycle (41).
Western blot analysis
The collected liver tissues were cut into tiny
pieces, lysed by RIPA buffer (Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China) containing protease and
phosphatase inhibitors, and completely homogenized with a
homogenizer. Using a prechilled centrifuge, the proteins in the
supernatant of the solution were acquired after centrifugation for
15 min at 12,000 × g. Later, following quantification with a BCA
kit (Thermo Fisher Scientific, Inc.), the proteins were separated
by electrophoresis using 10 and 15% gels, followed by a semi-dry
transfer onto polyvinylidene fluoride (PVDF) membranes (EMD
Millipore, Billerica, MA, USA). Next, the membranes were blocked
with 5% skim milk (BD Biosciences, Franklin Lakes, NJ, USA) for 1 h
at room temperature (or overnight at 4°C). After blocking, several
specific primary antibodies against the target genes CCL19 (1:500,
cat. no. bs-2454R; BIOSS, Beijing, China), TLR4 (1:500, cat. no.
Sc-293072; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
NF-κB-p65 (1:1,000, cat. no. 8242; Cell Signaling Technology, Inc.,
Danvers, MA, USA), SREBP-1c (1:1,000, cat. no. Ab28481; Abcam,
Cambridge, MA, USA), FAS (1:1,000, cat. no. Ab82419; Abcam), AMPK
(1:800, Ab110036; Abcam), p-AMPK (1:1,000, cat. no. AF3423;
Affinity Biosciences, Zhenjiang, China), and GAPDH (1:2,000, cat.
no. 5174; Cell Signaling Technology, Inc.) were incubated with the
membranes at 4°C overnight with gentle shaking (or 2 h at room
temperature). Subsequently, after 5–6 washes, the membranes were
incubated with corresponding secondary antibodies (1:1,000
dilution; Beyotime, Shanghai, China) according to the sources of
primary antibodies for 1 h at 37°C. Finally, after 5 min of
incubation of the chemiluminescence detection reagent (cat. no.
WBKLS0100; EMD Millipore) in the dark, the bands of target proteins
were visualized through an ECL imaging system (Tanon, Shanghai,
China).
Enzyme-linked immunosorbent assay
(ELISA)
The peripheral blood samples from patients and
treated rats were harvested. Following centrifugation at
2,000–3,000 × g for approximately 20 min, the supernatants of the
samples were obtained. Using a double antibody sandwich ELISA kit,
the concentrations of CCL19, IL-6 and TNF-α in the supernatant of
peripheral blood were detected.
H&E staining
As previously described, the liver tissues collected
from treated rats were embedded in paraffin and sliced into 4–7 µm
sections by a paraffin slicer. The liver sections were then stained
with double dyes of hematoxylin and eosin (BASO) (42). After staining, each section was
assessed and photographed by a 10×200 light microscope. Following
the criteria, the severity of fatty liver was scored: Grade 0, no
steatosis; grade 1, less than 10% of hepatic parenchyma is fatty
hepatocytes; grade 2, occupying 10–30%; grade 3, occupying 30–60%;
grade 4, more than 60% is fatty hepatocytes (43).
Biochemical analysis
After 4 and 8 weeks of drug treatment, the serum of
these rats was collected for biochemical analysis. A specific AST
kit (C010-1) was used to analyze the activity of AST in serum
following the recommended protocol. Likewise, the activity of ALT,
TG and TC was also respectively detected by specific ALT (C009-1),
TG (A110-2) and TC (A111-2) testing kit which were provided by
Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
Statistical analysis
GraphPad Prism 7.0 software (GraphPad Software,
Inc., La Jolla, CA, USA) was used to carry out statistical
analyses. One way analysis of variance followed by Tukey's post hoc
test was used to examine multiple comparisons. With at least three
independent experiments, all results were presented as the mean ±
standard deviation, and P<0.05 was considered to indicate a
statistically significant difference.
Results
High levels of CCL19, TLR4, NF-κB-p65,
IL-6, and TNF-α in NAFLD patients
Studies have correlated signal pathways
(TLR4/NF-κB-p65) and proinflammatory cytokines (IL-6/TNF-α) to
NAFLD progression (13,14,17,24).
Thus, peripheral blood samples were collected from 30 healthy
controls, 30 mild NAFLD patients, and 30 severe NAFLD patients.
After preprocessing, the concentrations of CCL19, IL-6 and TNF-α in
these samples were measured by specific ELISA. As shown in Fig. 1A-C, the levels of CCL19, IL-6 and
TNF-α were much higher in the blood of NAFLD patients than in that
of healthy controls. Furthermore, the levels of CCL19, IL-6 and
TNF-α progressively increased with the severity of fatty liver.
Through western blot analysis, we found that the protein levels of
CCL19, TLR4 and NF-κB-p56 in liver tissues collected from NAFLD
patients were also markedly increased with the severity of fatty
liver (Fig. 1D). These data
indicate that CCL19, TLR4, NF-κB-p65, IL-6, and TNF-α may function
in fatty liver.
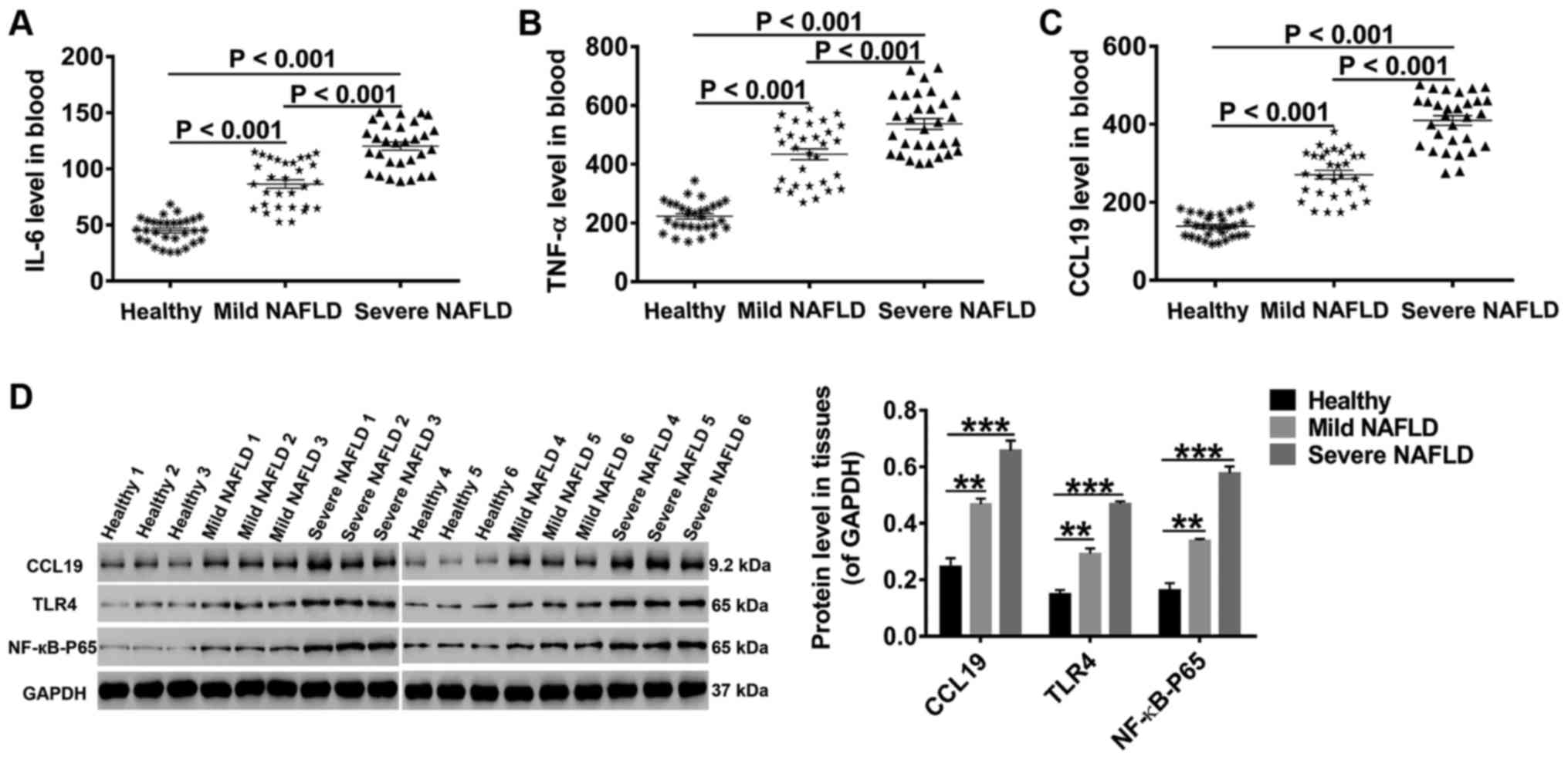 | Figure 1.High levels of CCL19, TLR4,
NF-κB-p65, IL-6 and TNF-α in NAFLD patients. Following processing,
the concentrations of (A) IL-6, (B) TNF-α and (C) CCL19 in the
blood were detected by ELISA. (D) The protein levels of CCL19, TLR4
and NF-κB-p65 in liver tissues were quantified by western blot
analysis. All data are presented as the mean ± standard deviation.
**P<0.01 and ***P<0.001 as indicated. CCL19, C-C motif ligand
19; TLR4, Toll-like receptor 4; NF-κB, nuclear factor-κB; IL,
interleukin; TNF-α, tumor necrosis factor-α; NAFLD, non-alcoholic
fatty liver disease. |
The levels of plasma AST, ALT, TC and
TG in HFD rats and drug-treated HFD rats
To verify the rat model of NAFLD and the effect of
metformin or BBR on NAFLD, the levels of AST, ALT, TC and TG in the
sera of HFD and drug-treated HFD rats were measured at 4 and 8
weeks by specific biochemical testing. As shown in Fig. 2, HFD rats displayed dyslipidemia
with elevated levels of plasma AST, ALT, TC and TG, which was
similar to the human dyslipidemia of NAFLD. In addition, metformin
or BBR led to an inhibiting effect on HFD-induced dyslipidemia, and
the joint effect of the two drugs was better. These results suggest
that dyslipidemia of NAFLD patients can be simulated by HFD feeding
and improved by metformin and BBR treatment.
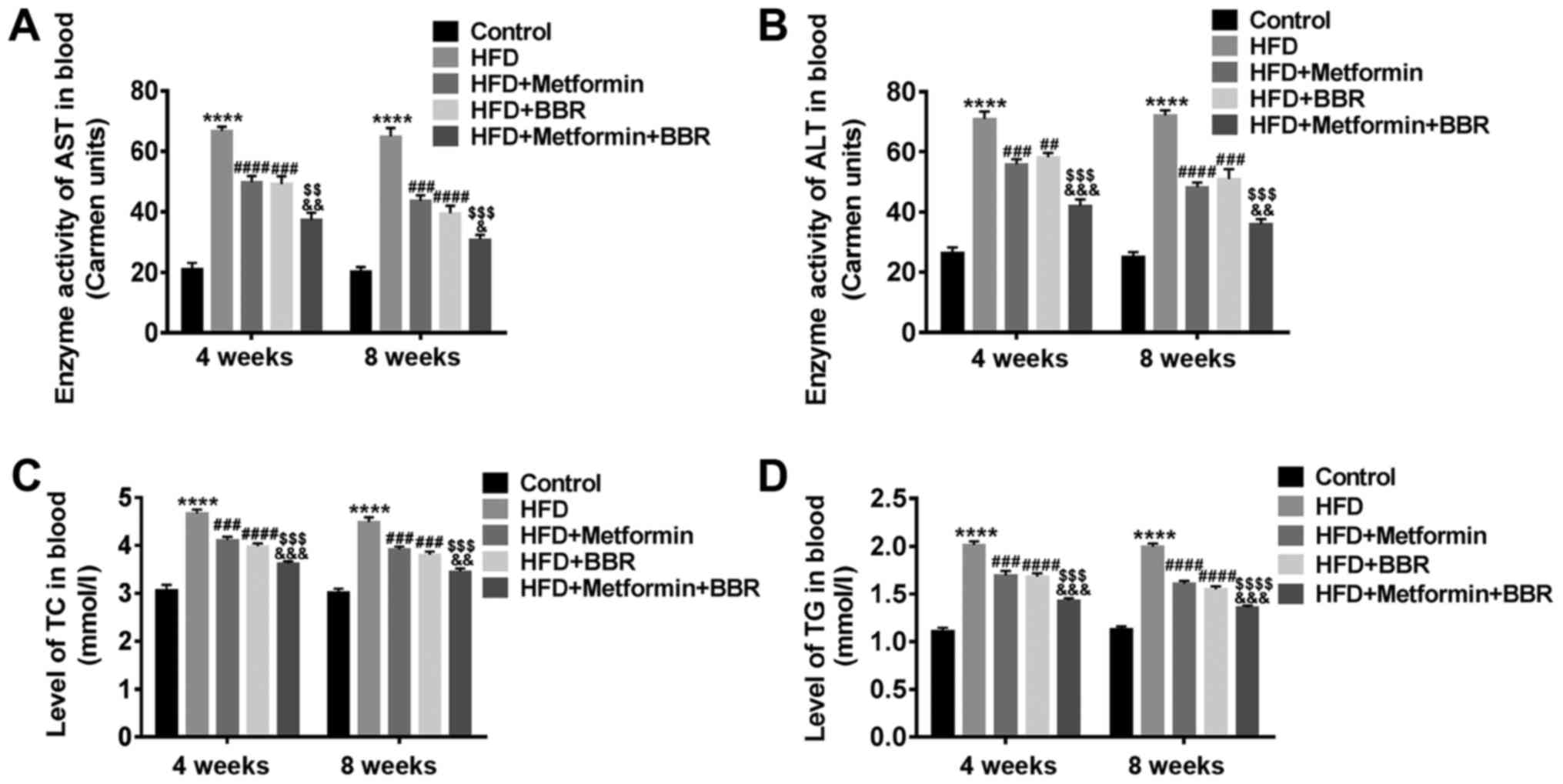 | Figure 2.Levels of plasma AST, ALT, TC and TG
in HFD rats and drug-treated HFD rats. The serum of Control, HFD,
HFD + metformin, HFD + BBR and HFD + metformin + BBR rats was
obtained at 4 and 8 weeks. Then, the levels of plasma (A) AST, (B)
ALT, (C) TC and (D) TG in these rats were analyzed by specific
biochemical testing. The data are presented as the mean ± standard
deviation. ****P<0.0001 vs. Control; ###P<0.001
and ####P<0.0001 vs. HFD; $$P<0.01,
$$$P<0.001 and $$$$P<0.0001 vs. HFD +
metformin; &P<0.05,
&&P<0.01 and
&&&P<0.001 vs. HFD + BBR. AST, aspartate
aminotransferase; ALT, alanine aminotransferase; TC, total
cholesterol; TG, triglyceride; HFD, high-fat diet; BBR,
berberine. |
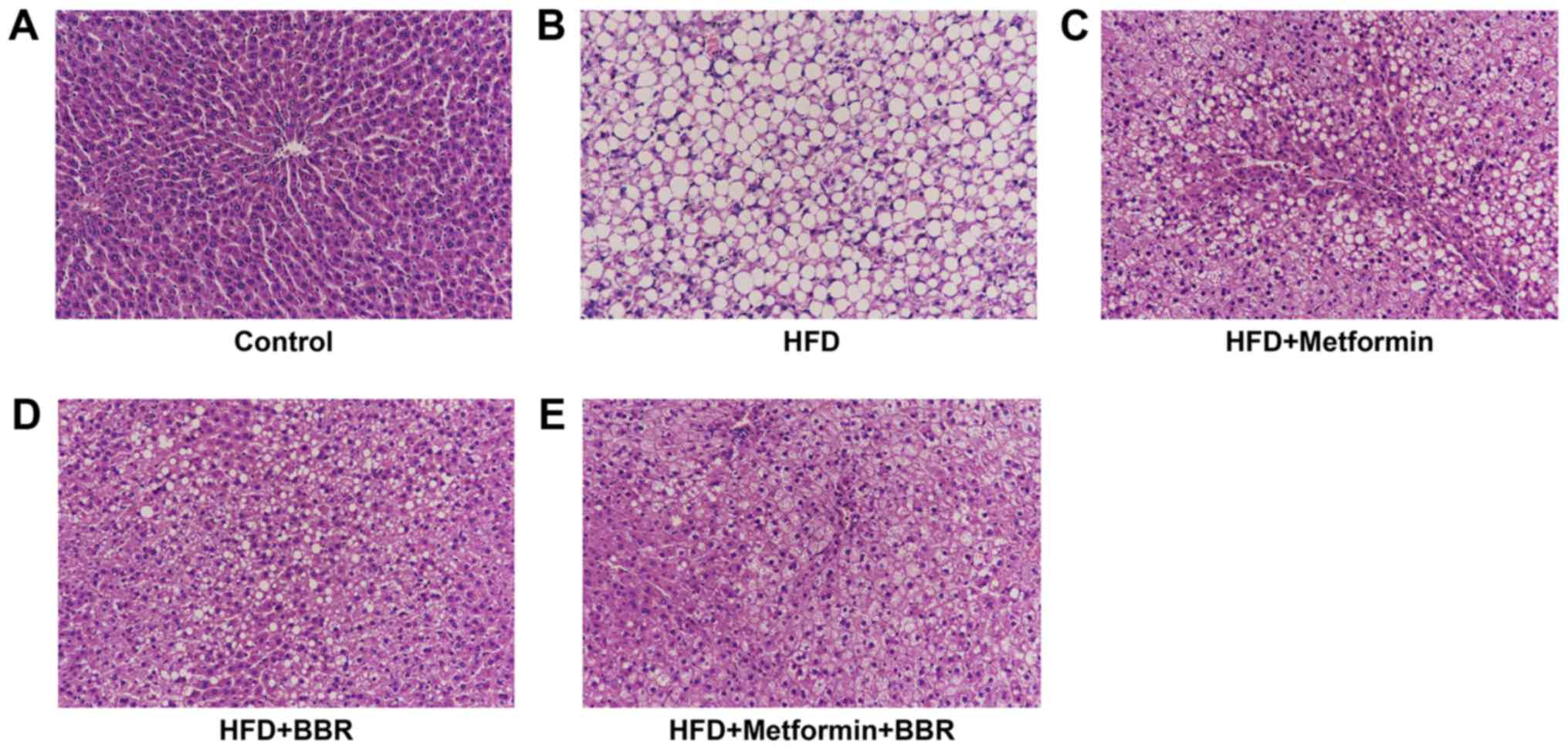
Liver histopathology of HFD rats and
drug-treated HFD rats
To further study HFD-induced NAFLD as well as the
effects of the two drugs on NAFLD, the liver histopathology of the
treated rats was analyzed by H&E staining at 8 weeks. As shown
in Fig. 3, we found significantly
higher microvesicular and macrovesicular steatosis in liver tissues
after HFD feeding than in the control, which further evidenced that
HFD feeding can simulate steatosis to human NAFLD (Fig. 3A and B). In Fig. 3C-E, HFD-induced steatosis was
remarkably eliminated by metformin or BBR treatment, and the joint
effect of the two drugs was better. This further demonstrates that
metformin and BBR contribute to NAFLD treatment.
The concentrations of CCL19, IL-6 and
TNF-α in serum of HFD rats and drug-treated HFD rats
Experimental animals fed with HFD are considered
good models of NAFLD. Here, we further explored the effect of CCL19
in these models. After 4 and 8 weeks of treatment, the
concentrations of CCL19, IL-6 and TNF-α in the sera of these
treated rats were measured by specific ELISA. As shown in Fig. 4, elevated levels of CCL19, IL-6 and
TNF-α, similar to those observed in NAFLD patients, were also
present in HFD rats. Metformin and BBR markedly inhibited the
HFD-induced levels of CCL19, IL-6 and TNF-α. These results
demonstrate that the release of CCL19 and the proinflammatory
cytokines IL-6 and TNF-α may enhance the progression of NAFLD and
is inhibited by metformin and BBR, which may be closely related to
the treatment of NAFLD.
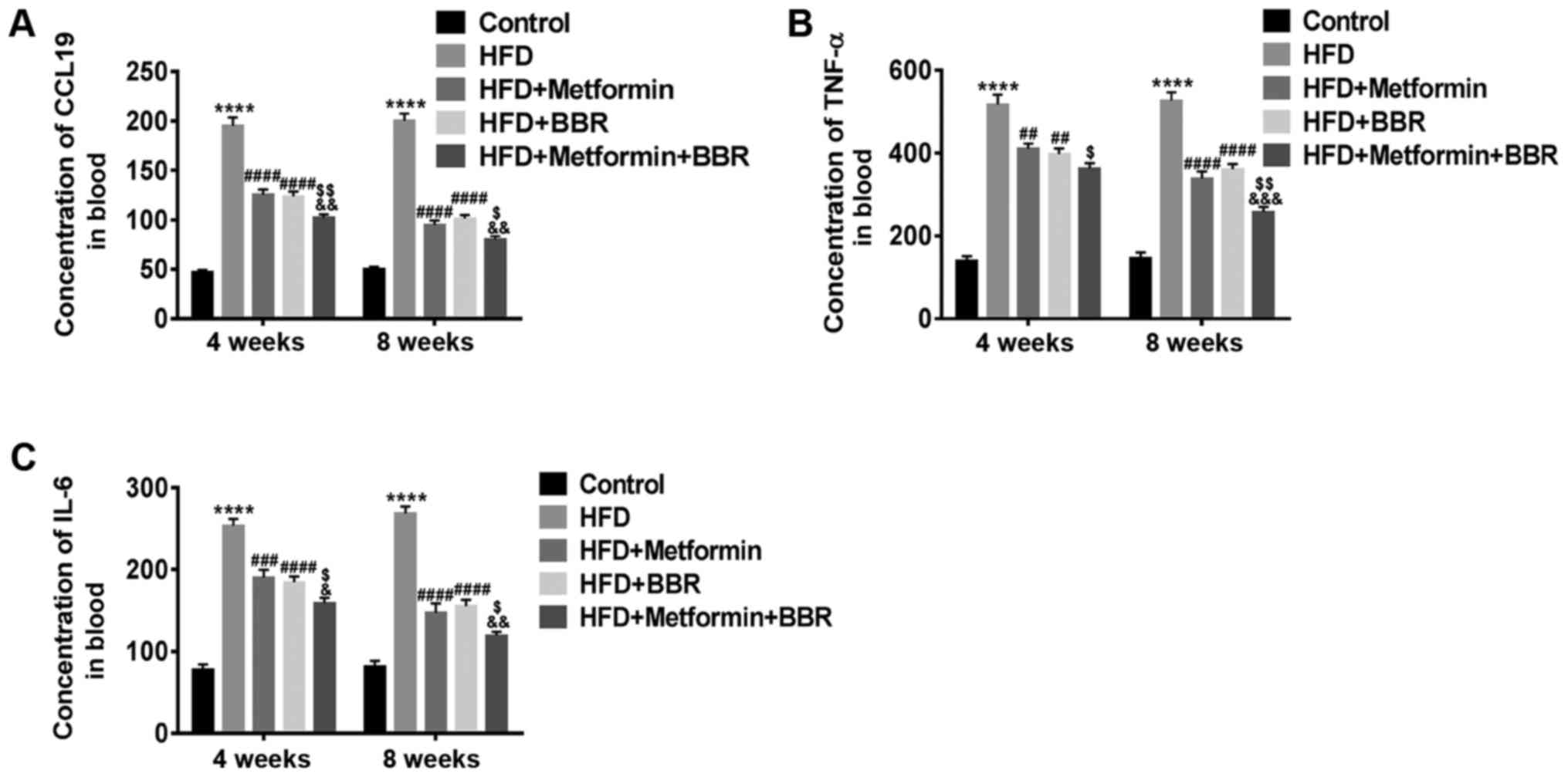 | Figure 4.Concentrations of CCL19, IL-6 and
TNF-α in the sera of HFD- and drug-treated HFD rats. The serum
concentrations of (A) CCL19, (B) TNF-α and (C) IL-6 in Control,
HFD, HFD + metformin, HFD + BBR and HFD + metformin + BBR rats were
measured by ELISA at 4 and 8 weeks. The data are expressed as the
mean ± standard deviation. ****P<0.0001 vs. Control;
##P<0.01, ###P<0.001 and
####P<0.0001 vs. HFD; $P<0.05 and
$$P<0.01 vs. HFD + metformin;
&P<0.05, &&P<0.01 and
&&&P<0.001 vs. HFD + BBR. CCL19, C-C
motif ligand 19; IL, interleukin; TNF-α, tumor necrosis factor-α;
HFD, high-fat diet; BBR, berberine. |
The expression of CCL19, TLR4,
NF-κB-p65, SREBP-1c, FAS, p-AMPK, and AMPK in the liver tissues of
HFD rats and drug-treated HFD rats
For further study, the levels of CCL19, signaling
pathways (TLR4/NF-κB-p65) and lipid metabolism factors (SREBP-1c
and FAS) in the liver tissues of these treated rats were quantified
by RT-PCR and western blot. As shown in Fig. 5, the HFD-induced expression of
CCL19, TLR4, NF-κB-p65, SREBP-1c, and FAS was inhibited by the two
drugs at both the transcriptional and translational levels, and the
joint effect of the two drugs was better. In addition, HFD
suppression of phosphorylated-AMPK (p-AMPK) levels was
significantly increased by treatment with the two drugs, whereas
AMPK expression was unchanged. These results further clarified that
the expression of CCL19, TLR4, NF-κB-p65, SREBP-1c, FAS, and p-AMPK
was related to the NAFLD process. Metformin and BBR improved NAFLD
may through the activation of AMPK and the inhibition of
TLR4/NF-κB-p65 signaling. Meantime, CCL19 expression was inhibited
by metformin and BBR, indicating that inhibition of CCL19 may be
benefit NAFLD treatment.
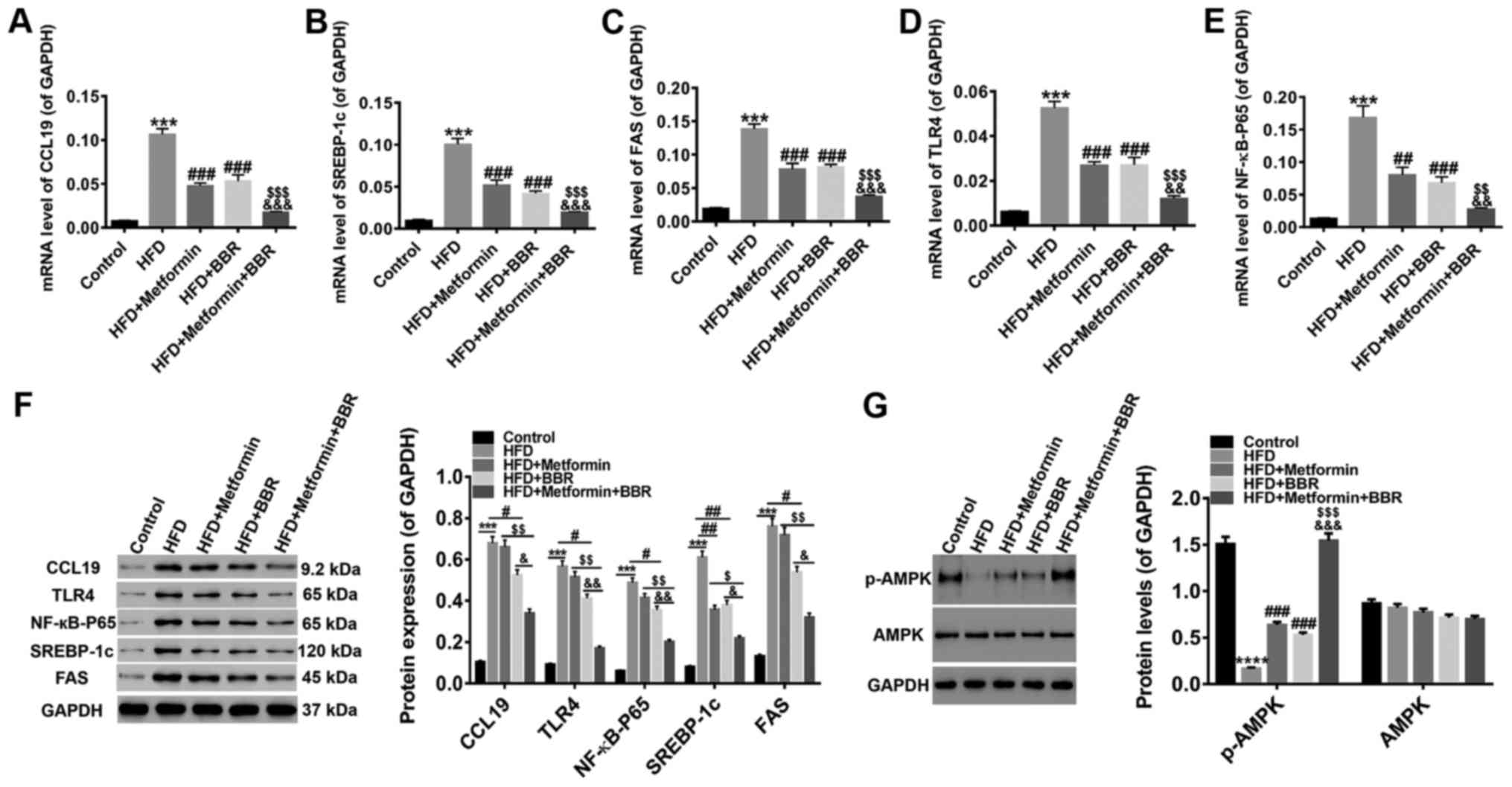 | Figure 5.Expression of CCL19, TLR4, NF-κB-p65,
SREBP-1c, FAS, p-AMPK and AMPK in the liver tissues of HFD- and
drug-treated HFD rats Following 8 weeks of drug treatment, the mRNA
levels of (A) CCL19, (B) SREBP-1c, (C) FAS, (D) TLR4 and (E)
NF-κB-p65 were quantified by reverse transcription-quantitative
polymerase chain reaction, while (F) western blotting was applied
to detect the protein levels. (G) In addition, the levels of p-AMPK
and AMPK proteins were detected. All data are presented as the mean
± standard deviation. ***P<0.001 and ****P<0.0001 vs.
Control; #P<0.05, ##P<0.01 and
###P<0.001 vs. HFD; $P<0.05,
$$P<0.01 and $$$P<0.001 vs. HFD +
metformin; &P<0.05,
&&P<0.01 and
&&&P<0.001 vs. HFD + BBR. CCL19, C-C
motif ligand 19; TLR4, Toll-like receptor 4; NF-κB, nuclear
factor-κB; TNF-α, tumor necrosis factor-α; SREBP-1c, sterol
response element-binding protein-1c; FAS, fatty acid synthase; p-,
phosphorylated; AMPK, adenosine monophosphate-activated protein
kinase; HFD, high-fat diet; BBR, berberine. |
Discussion
Fatty liver disease remains the most common chronic
liver disease and is a serious threat to human health. Previous
research has demonstrated that various factors, including cytokines
and chemokines, are involved in fatty liver diseases. For instance,
high expression of CCL2 (also known as MCP-1) was noted in the
steatotic livers of NAFLD patients (44) and could mediate the inflammatory
response of NAFLD (45). In
addition, CCL5 (also known as RANTES) was also expressed highly in
NAFLD patients (46), and it has
been reported that CCL20 (also known as MIP-3α) was enriched in the
necrotic regions of liver portions from hepatitis patients
(47).
In our study, the levels of CCL19, TLR4/NF-κB-p65
and IL-6/TNF-α were markedly increased in NAFLD patients,
indicating that these molecules may be implicated in NAFLD
progression. Furthermore, high release of CCL19 may offer a novel
diagnostic blood indicator of NAFLD. In a rat model of NAFLD,
metformin or BBR could ameliorate NAFLD by reducing the
dyslipidemia and steatosis, and the effect of the two drugs
combined was much better. Additionally, high levels of CCL19,
TLR4/NF-κB-p65, SREBP-1c, FAS and IL-6/TNF-α were also remarkably
decreased, whereas HFD-suppressed p-AMPK levels were increased by
drug treatment. Based on these data, we inferred that metformin and
BBR treatment improved NAFLD may through the activation of AMPK and
the inactivation of TLR4/NF-κB-p65 signaling. Meantime, metformin
and BBR could inhibit CCL19, which related CCL19 inhibition to the
treatment of NAFLD. As shown in Fig.
6, the experiments indicated that the NF-κB-p65 signaling
pathway was activated by upstream TLR4 via signal transduction,
which further promoted the secretion of proinflammatory cytokines
IL-6 and TNF-α (48). By
regulating the chronic inflammatory response, IL-6 and TNF-α have
been demonstrated to be essential for fatty liver (49,50).
In addition, SREBP-1c was reported to upregulate the expression of
several prolipogenic genes, such as FAS, and be responsible for the
modulation of TG synthesis in the liver. AMPK has been suggested to
regulate cellular metabolism, such as lipid metabolism (51); by downregulating SREBP-1c, AMPK
activation inhibits FAS expression, enhancing TG deposition and
further causing hepatic lipogenesis and hepatic steatosis (52). Thus, TLR4/NF-κB-p65/IL-6/TNF-α and
p-AMPK/SREBP-1c/FAS play important roles in NAFLD progression,
which is consistent with the results of our study. Furthermore, the
CCL19 promoter has been reported to have two putative NF-κB binding
sites, and CCL19 expression is regulated by several transcription
factors, including NF-κB. Activity of the CCL19 promoter was
induced by ectopic expression of TLR3/4 (15). Therefore, combining our results
that CCL19 expression was inhibited by metformin and BBR, it is
likely that CCL19 functions in NAFLD, which probably involve
TLR4/NF-κB-p65 pathway, but further studies are needed. Thus,
targeting CCL19 is potential to provide a novel effective therapy
for NAFLD.
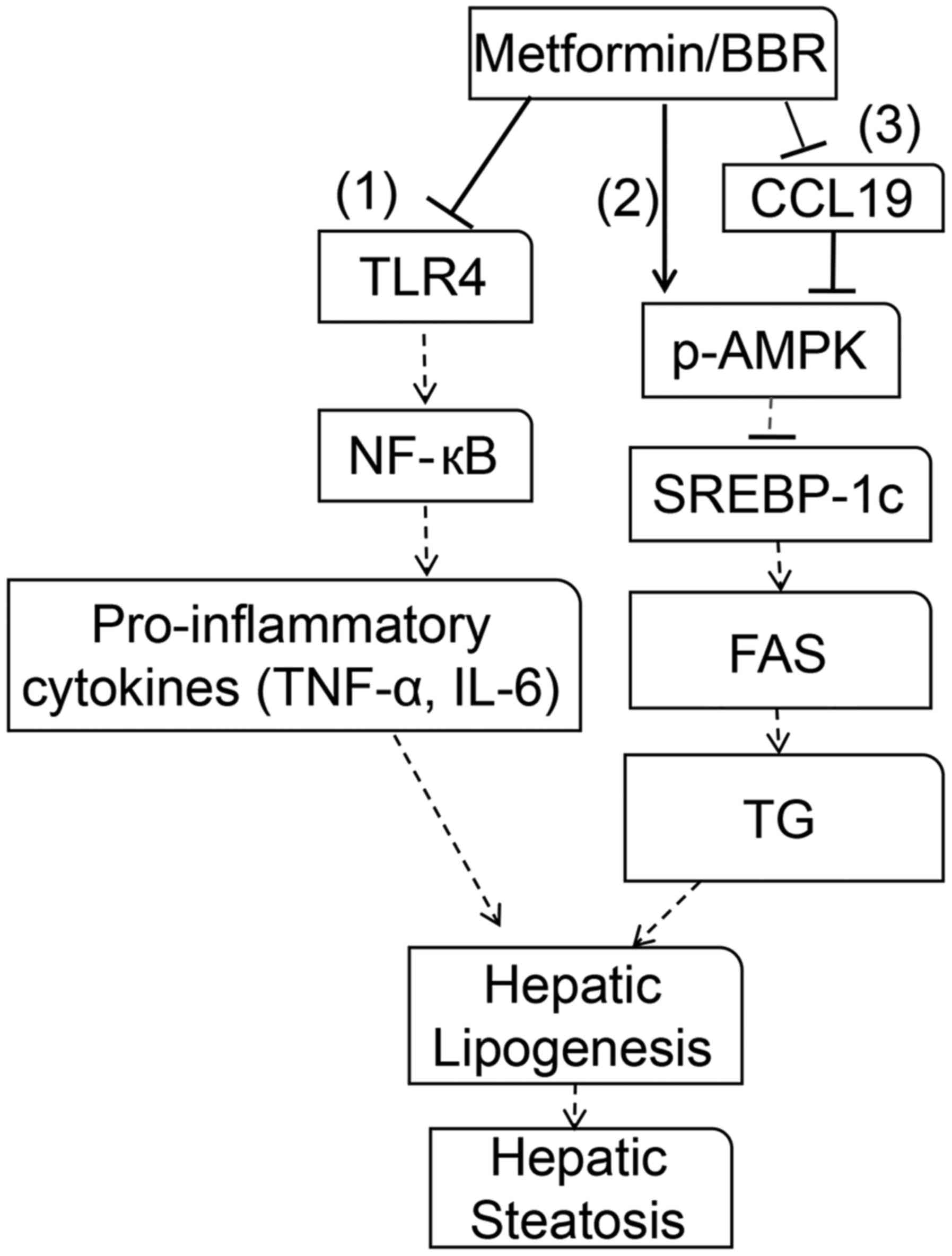 | Figure 6.Molecular mechanisms of metformin or
BBR in reverting dysfunction in NAFLD. Metformin or BBR induced a
series of alterations, including proinflammatory cytokine
production and lipogenesis in the liver. In protecting NAFLD,
metformin or BBR is directed toward several pathways. (1) Metformin or BBR inhibits TLR4, which
further inhibits NF-κB-p65 pathway activation, thereby reducing
IL-6/TNF-α production. (2)
Metformin or BBR phosphorylates AMPK, and the activation of AMPK
inhibits SREBP-1c expression, which further inhibits lipogenesis by
inhibiting FAS expression. (3)
Metformin or BBR phosphorylates AMPK by regulating CCL19. Hepatic
lipogenesis and hepatic steatosis are suppressed via these
pathways. BBR, berberine; NAFLD, non-alcoholic fatty liver disease;
TLR4, Toll-like receptor 4; NF-κB, nuclear factor-κB; IL-6,
interleukin 6; TNF-α, tumor necrosis factor-α; p-, phosphorylated;
AMPK, adenosine monophosphate-activated protein kinase; SREBP-1c,
sterol response element-binding protein-1c; FAS, fatty acid
synthase; TG, triglyceride; CCL19, C-C motif ligand 19. |
In conclusion, we found that metformin and BBR could
improve NAFLD by p-AMPK activation and TLR4/NF-κB-p65 inactivation.
Meantime, CCL19 levels were inhibited by metformin and BBR,
indicating that CCL19 inhibition may contribute to NAFLD treatment.
Although the mechanisms are still unclear, these data reveal that
CCL19 may act as a new target for therapy of fatty liver
diseases.
Acknowledgements
Not applicable.
Funding
The present study was funded by Shanghai Grassroots
Senior Experts in Traditional Chinese Medicine Heritage Research
Studio Construction Projects (grant no. JCZYGZS-020) and Putuo
District People's Hospital Fund for Distinguished Young Scholars
(grant no. PRYYC2016-A001).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JZ and JH conceived and designed the study. JZ, YW,
XW, PT, YY, SG and DH performed the experiments. JZ and JH wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments in the present study were approved
by the Ethics Committee of Putuo District People's Hospital of
Shanghai City (Shanghai, China); written informed consent was
obtained from all patients. All of the experimental procedures
involving animals were approved by the Experimental Animal Research
Committee of Putuo District People's Hospital of Shanghai.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Angulo P: Nonalcoholic fatty liver
disease. N Engl J Med. 346:1221–1231. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ding RB, Bao J and Deng CX: Emerging roles
of SIRT1 in fatty liver diseases. Int J Biol Sci. 13:852–867. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vernon G, Baranova A and Younossi ZM:
Systematic review: The epidemiology and natural history of
non-alcoholic fatty liver disease and non-alcoholic steatohepatitis
in adults. Aliment Pharmacol Ther. 34:274–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alisi A, Cianfarani S, Manco M, Agostoni C
and Nobili V: Non-alcoholic fatty liver disease and metabolic
syndrome in adolescents: Pathogenetic role of genetic background
and intrauterine environment. Ann Med. 44:29–40. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perticone M, Cimellaro A, Maio R, Caroleo
B, Sciacqua A, Sesti G and Perticone F: Additive effect of
non-alcoholic fatty liver disease on metabolic syndrome-related
endothelial dysfunction in hypertensive patients. Int J Mol Sci.
17:4562016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cohen JC, Horton JD and Hobbs HH: Human
fatty liver disease: Old questions and new insights. Science.
332:1519–1523. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu KT, Kuo PL, Su SB, Chen YY, Yeh ML,
Huang CI, Yang JF, Lin CI, Hsieh MH, Hsieh MY, et al: Nonalcoholic
fatty liver disease severity is associated with the ratios of total
cholesterol and triglycerides to high-density lipoprotein
cholesterol. J Clin Lipidol. 10:420–425. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kotronen A, Peltonen M, Hakkarainen A,
Sevastianova K, Bergholm R, Johansson LM, Lundbom N, Rissanen A,
Ridderstråle M, Groop L, et al: Prediction of non-alcoholic fatty
liver disease and liver fat using metabolic and genetic factors.
Gastroenterology. 137:865–872. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou G, Myers R, Li Y, Chen Y, Shen X,
Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al: Role of
AMP-activated protein kinase in mechanism of metformin action. J
Clin Invest. 108:1167–1174. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Walker AK and Näär AM: SREBPs: Regulators
of cholesterol/lipids as therapeutic targets in metabolic
disorders, cancers and viral diseases. Clin Lipidol. 7:27–36. 2012.
View Article : Google Scholar
|
|
11
|
Horton JD: Sterol regulatory
element-binding proteins: Transcriptional activators of lipid
synthesis. Biochem Soc Trans. 30:1091–1095. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bricambert J, Miranda J, Benhamed F,
Girard J, Postic C and Dentin R: Salt-inducible kinase 2 links
transcriptional coactivator p300 phosphorylation to the prevention
of ChREBP-dependent hepatic steatosis in mice. J Clin Invest.
120:4316–4331. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kiziltas S, Ata P, Colak Y, Mesçi B,
Senates E, Enc F, Ulasoglu C, Tuncer I and Oguz A: TLR4 gene
polymorphism in patients with nonalcoholic fatty liver disease in
comparison to healthy controls. Metab Syndr Relat Disord.
12:165–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao CY, Yan L, Wang YD, Wang W, Zhou JY
and Zhen Z: Role of resistin in inflammation of hepatocytes in
nonalcoholic steatohepatitis. Zhonghua Gan Zang Bing Za Zhi.
17:683–687. 2009.(In Chinese). PubMed/NCBI
|
|
15
|
Pietilä TE, Veckman V, Lehtonen A, Lin R,
Hiscott J and Julkunen I: Multiple NF-kappaB and IFN regulatory
factor family transcription factors regulate CCL19 gene expression
in human monocyte-derived dendritic cells. J Immunol. 178:253–261.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zuany-Amorim C, Hastewell J and Walker C:
Toll-like receptors as potential therapeutic targets for multiple
diseases. Nat Rev Drug Discov. 1:797–807. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hotamisligil GS, Shargill NS and
Spiegelman BM: Adipose expression of tumor necrosis factor-alpha:
Direct role in obesity-linked insulin resistance. Science.
259:87–91. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pinto Lde F, Compri CM, Fornari JV,
Bartchewsky W, Cintra DE, Trevisan M, Carvalho Pde O, Ribeiro ML,
Velloso LA, Saad MJ, et al: The immunosuppressant drug,
thalidomide, improves hepatic alterations induced by a high-fat
diet in mice. Liver Int. 30:603–610. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
El-Assal O, Feng H, Kim WH, Radaeva S and
Gao B: IL-6-deficient mice are susceptible to ethanol-induced
hepatic steatosis: IL-6 protects against ethanol-induced oxidative
stress and mitochondrial permeability transition in the liver. Cell
Mol Immunol. 1:205–211. 2004.PubMed/NCBI
|
|
20
|
Teoh N, Field J and Farrell G:
Interleukin-6 is a key mediator of the hepatoprotective and
pro-proliferative effects of ischaemic preconditioning in mice. J
Hepatol. 45:20–27. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Peters M, Blinn G, Jostock T, Schirmacher
P, Meyer zum Büschenfelde KH, Galle PR and Rose-John S: Combined
interleukin 6 and soluble interleukin 6 receptor accelerates murine
liver regeneration. Gastroenterology. 119:1663–1671. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blindenbacher A, Wang X, Langer I, Savino
R, Terracciano L and Heim MH: Interleukin 6 is important for
survival after partial hepatectomy in mice. Hepatology. 38:674–682.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Klein C, Wüstefeld T, Assmus U, Roskams T,
Rose-John S, Müller M, Manns MP, Ernst M and Trautwein C: The
IL-6-gp130-STAT3 pathway in hepatocytes triggers liver protection
in T cell-mediated liver injury. J Clin Invest. 115:860–869. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mas E, Danjoux M, Garcia V, Carpentier S,
Ségui B and Levade T: IL-6 deficiency attenuates murine
diet-induced non-alcoholic steatohepatitis. PLoS One. 4:e79292009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wieckowska A, Papouchado BG, Li Z, Lopez
R, Zein NN and Feldstein AE: Increased hepatic and circulating
interleukin-6 levels in human nonalcoholic steatohepatitis. Am J
Gastroenterol. 103:1372–1379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lavine JE, Schwimmer JB, Van Natta ML,
Molleston JP, Murray KF, Rosenthal P, Abrams SH, Scheimann AO,
Sanyal AJ, Chalasani N, et al: Effect of vitamin E or metformin for
treatment of nonalcoholic fatty liver disease in children and
adolescents: The TONIC randomized controlled trial. JAMA.
305:1659–1668. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yin J, Xing H and Ye J: Efficacy of
berberine in patients with type 2 diabetes mellitus. Cell Mol
Immunol. 57:712–717. 2008.
|
|
28
|
Kong W, Wei J, Abidi P, Lin M, Inaba S, Li
C, Wang Y, Wang Z, Si S, Pan H, et al: Berberine is a novel
cholesterol-lowering drug working through a unique mechanism
distinct from statins. Nat Med. 10:1344–1351. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong WJ, Zhang H, Song DQ, Xue R, Zhao W,
Wei J, Wang YM, Shan N, Zhou ZX, Yang P, et al: Berberine reduces
insulin resistance through protein kinase C-dependent up-regulation
of insulin receptor expression. Metabolism. 58:109–119. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brusq JM, Ancellin N, Grondin P, Guillard
R, Martin S, Saintillan Y and Issandou M: Inhibition of lipid
synthesis through activation of AMP kinase: An additional mechanism
for the hypolipidemic effects of berberine. J Lipid Res.
47:1281–1288. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang X, Yan H, Fei J, Jiang M, Zhu H, Lu
D and Gao X: Berberine reduces methylation of the MTTP promoter and
alleviates fatty liver induced by a high-fat diet in rats. J Lipid
Res. 51:2504–2515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yuan X, Wang J, Tang X, Li Y, Xia P and
Gao X: Berberine ameliorates nonalcoholic fatty liver disease by a
global modulation of hepatic mRNA and lncRNA expression profiles. J
Transl Med. 13:242015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan HM, Xia MF, Wang Y, Chang XX, Yao XZ,
Rao SX, Zeng MS, Tu YF, Feng R, Jia WP, et al: Efficacy of
berberine in patients with non-alcoholic fatty liver disease. PLoS
One. 10:e01341722015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mazza A, Fruci B, Garinis GA, Giuliano S,
Malaguarnera R and Belfiore A: The role of metformin in the
management of NAFLD. Exp Diabetes Res. 2012:7164042012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luther SA, Bidgol A, Hargreaves DC,
Schmidt A, Xu Y, Paniyadi J, Matloubian M and Cyster JG: Differing
activities of homeostatic chemokines CCL19, CCL21 and CXCL12 in
lymphocyte and dendritic cell recruitment and lymphoid neogenesis.
Journal of Immunology. 169:424–433. 2002. View Article : Google Scholar
|
|
36
|
Yanagawa Y and Onoé K: CCL19 induces rapid
dendritic extension of murine dendritic cells. Blood.
100:1948–1956. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sano T, Iwashita M, Nagayasu S, Yamashita
A, Shinjo T, Hashikata A, Asano T, Kushiyama A, Ishimaru N,
Takahama Y and Nishimura F: Protection from diet-induced obesity
and insulin resistance in mice lacking CCL19-CCR7 signaling.
Obesity (Silver Spring). 23:1460–1471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Borai IH, Shaker Y, Kamal MM, Ezzat WM,
Ashour E, Afify M, Gouda W and Elbrashy MM: Evaluation of
biomarkers in egyptian patients with different grades of
nonalcoholic fatty liver disease. J Clin Transl Hepatol. 5:109–118.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee SH, Yun SJ, Kim DH, Jo HH and Park YS:
Severity of nonalcoholic fatty liver disease on sonography and risk
of coronary heart disease. J Clin Ultrasound. 45:391–399. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hong J, Kang B, Kim A, Hwang S, Ahn J, Lee
S, Kim J, Park JH and Cheon DS: Development of a highly sensitive
real-time one step RT-PCR combined complementary locked primer
technology and conjugated minor groove binder probe. Virol J.
8:3302011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Q, Zhao Y, Zhang DB and Sun LJ:
Effect of Sinai san decoction on the development of non-alcoholic
steatohepatitis in rats. World J Gastroenterol. 11:1392–1395. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Brunt EM, Janney CG, Di Bisceglie AM,
Neuschwander-Tetri BA and Bacon BR: Nonalcoholic steatohepatitis: A
proposal for grading and staging the histological lesions. Am J
Gastroenterol. 94:2467–2474. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Haukeland JW, Damås JK, Konopski Z, Løberg
EM, Haaland T, Goverud I, Torjesen PA, Birkeland K, Bjøro K and
Aukrust P: Systemic inflammation in nonalcoholic fatty liver
disease is characterized by elevated levels of CCL2. J Hepatol.
44:1167–1174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cynis H, Kehlen A, Haegele M, Hoffmann T,
Heiser U, Fujii M, Shibazaki Y, Yoneyama H, Schilling S and Demuth
HU: Inhibition of Glutaminyl Cyclases alleviates CCL2-mediated
inflammation of non-alcoholic fatty liver disease in mice. Int J
Exp Pathol. 94:217–225. 2013.PubMed/NCBI
|
|
46
|
Kirovski G, Gäbele E, Dorn C, Moleda L,
Niessen C, Weiss TS, Wobser H, Schacherer D, Buechler C, Wasmuth HE
and Hellerbrand C: Hepatic steatosis causes induction of the
chemokine RANTES in the absence of significant hepatic
inflammation. Int J Clin Exp Pathol. 3:675–680. 2010.PubMed/NCBI
|
|
47
|
Shimizu Y, Murata H, Kashii Y, Hirano K,
Kunitani H, Higuchi K and Watanabe A: CC-chemokine receptor 6 and
its ligand macrophage inflammatory protein 3alpha might be involved
in the amplification of local necroinflammatory response in the
liver. Hepatology. 34:311–319. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen W, Wang X, Huang LI and Liu BO:
Hepcidin in non-alcoholic fatty liver disease regulated by the
TLR4/NF-κB signaling pathway. Exp Ther Med. 11:73–76. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hassan K, Bhalla V, El Regal ME and
A-kader HH: Nonalcoholic fatty liver disease: A comprehensive
review of a growing epidemic. World J Gastroenterol.
20:12082–12101. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang J, Tan Y, Yao F and Zhang Q:
Polydatin alleviates non-alcoholic fatty liver disease in rats by
inhibiting the expression of TNF-α and SREBP-1c. Mol Med Rep.
6:815–820. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Long YC and Zierath JR: AMP-activated
protein kinase signaling in metabolic regulation. J Clin Invest.
116:1776–1783. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kohjima M, Higuchi N, Kato M, Kotoh K,
Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Enjoji M, et al:
SREBP-1c, regulated by the insulin and AMPK signaling pathways,
plays a role in nonalcoholic fatty liver disease. Int J Mol Med.
21:507–511. 2008.PubMed/NCBI
|