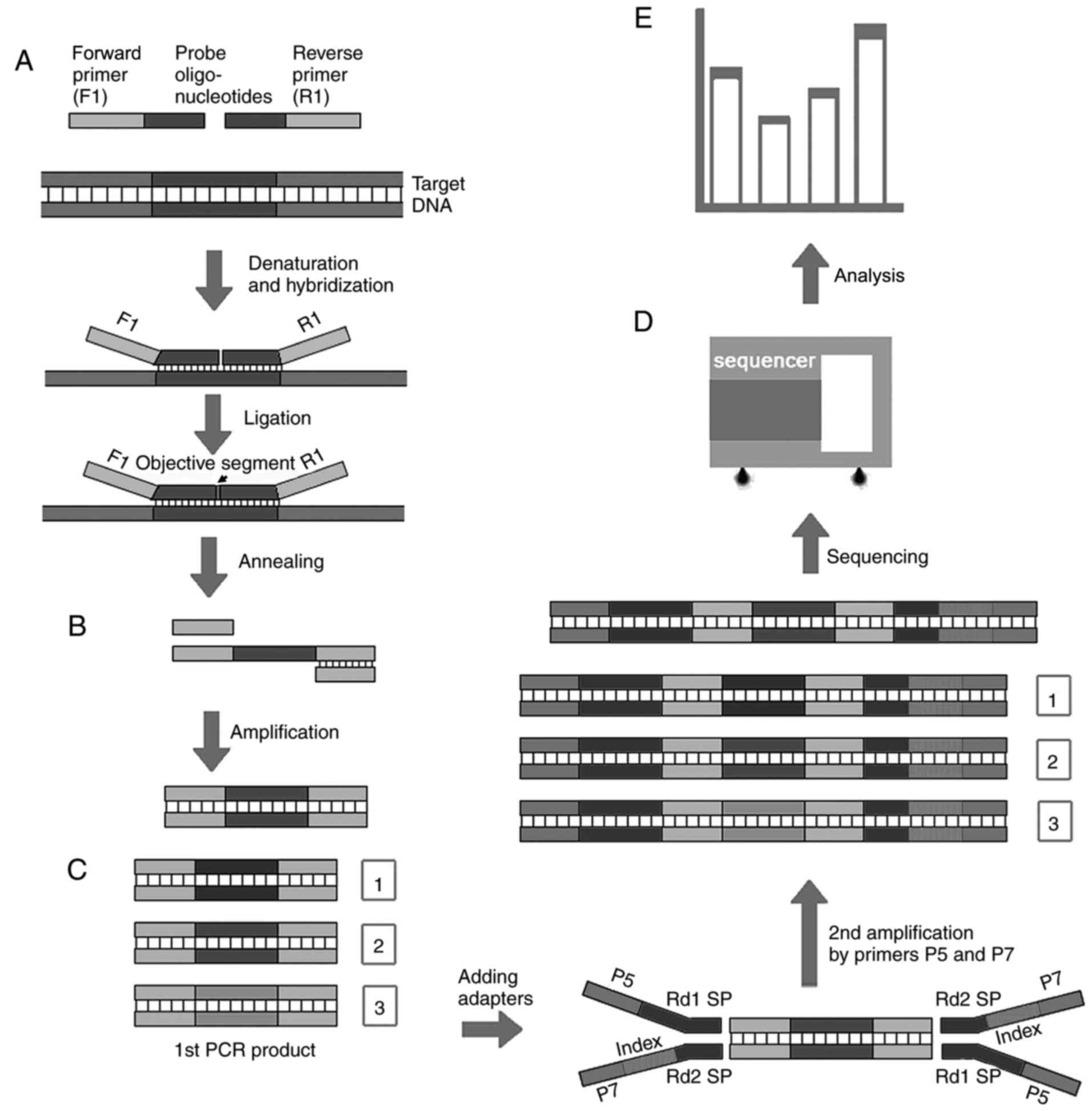
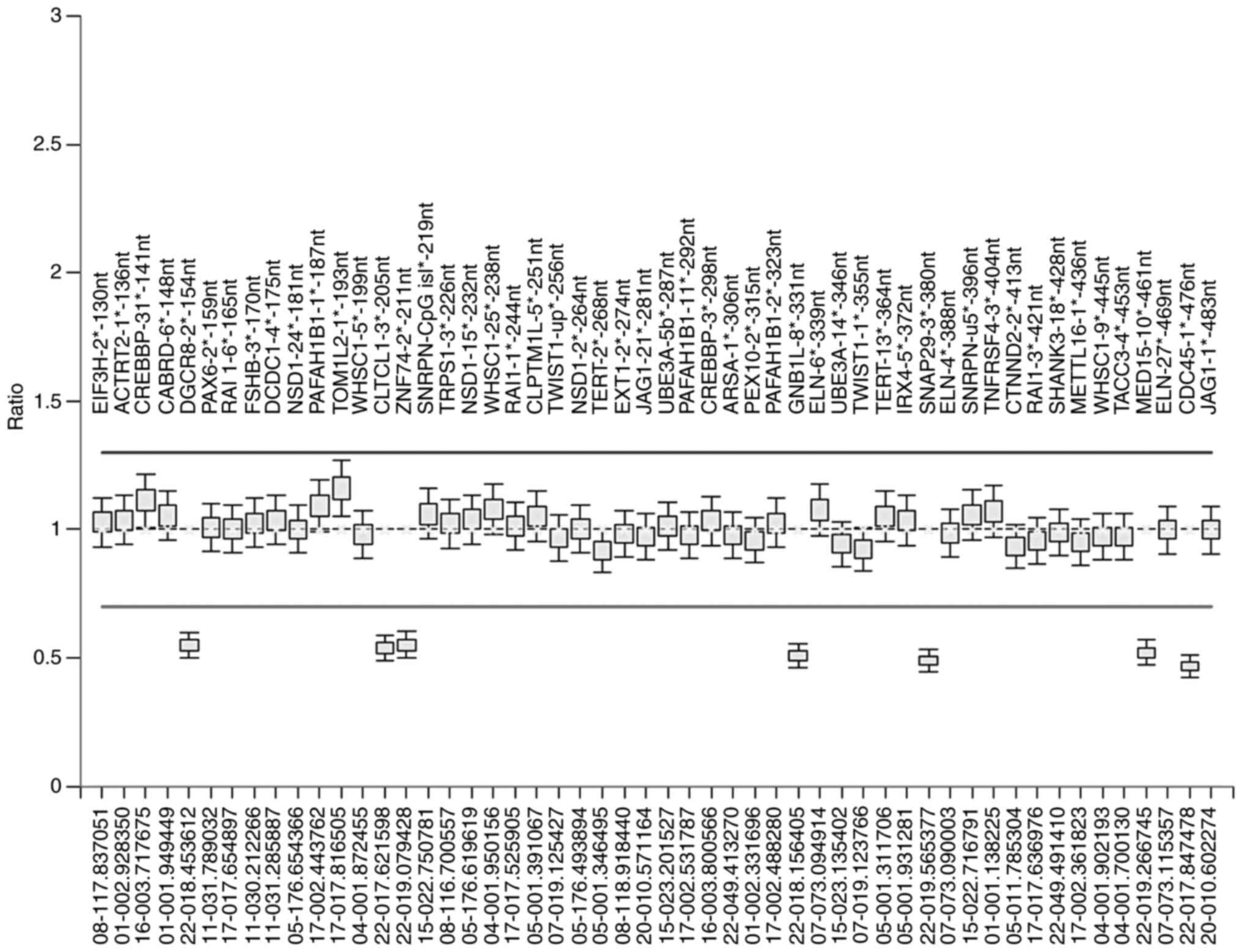
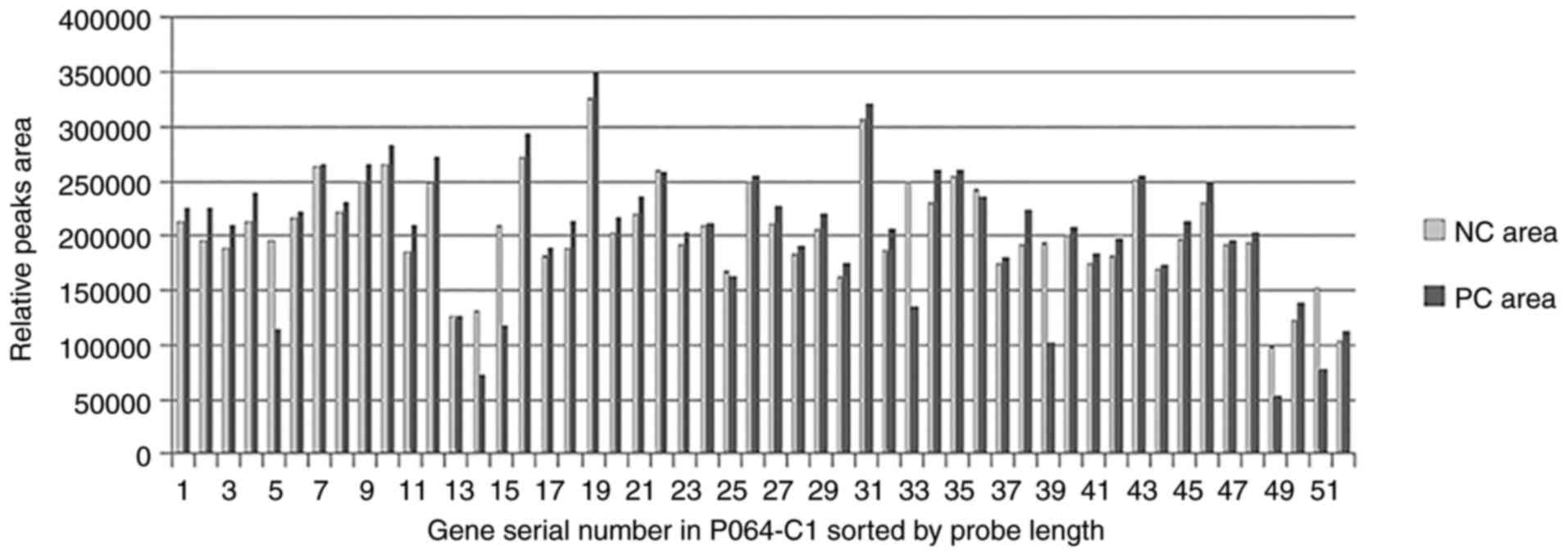
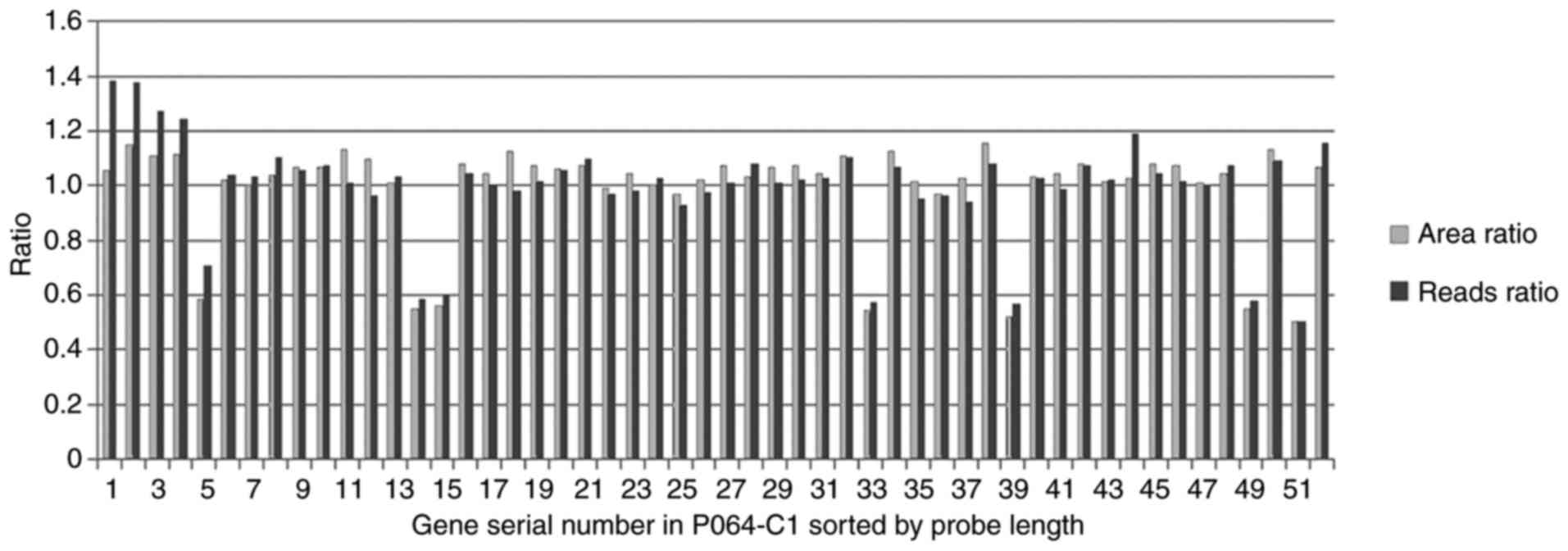
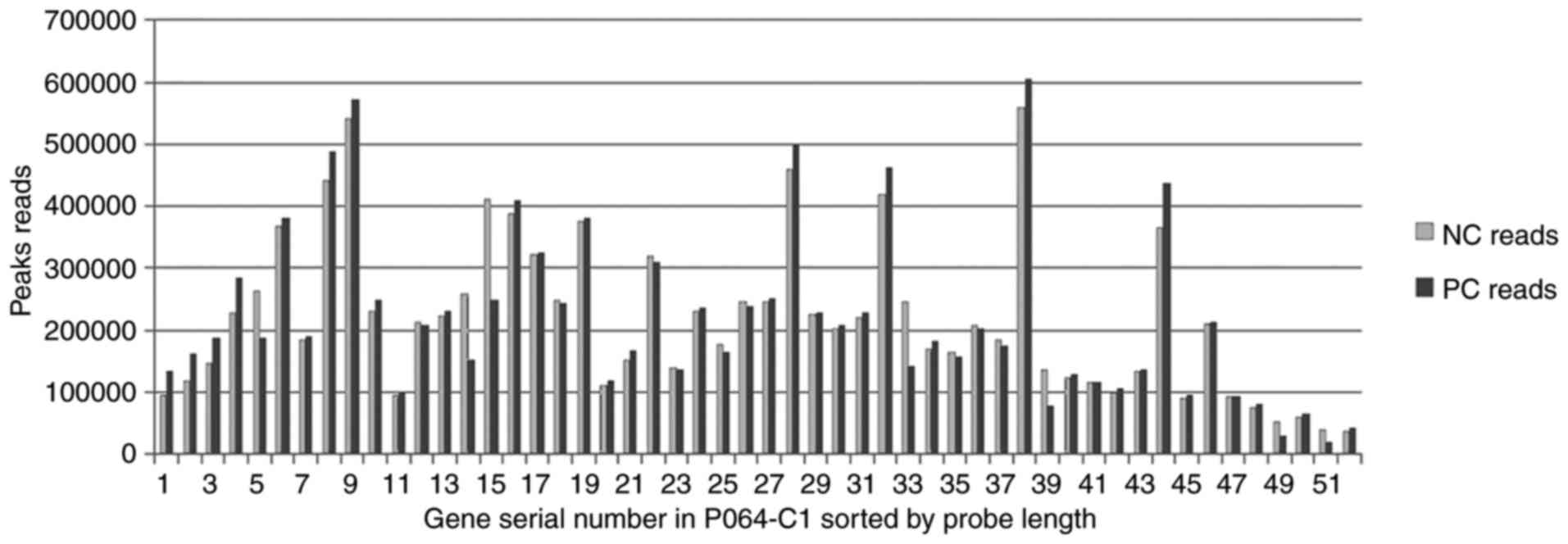
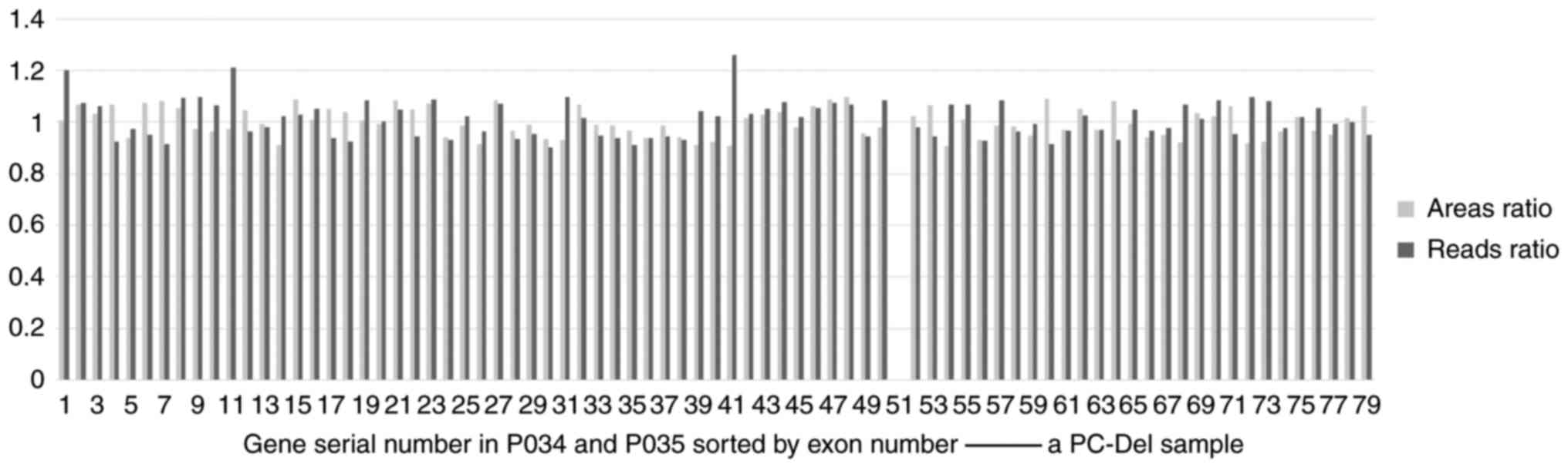
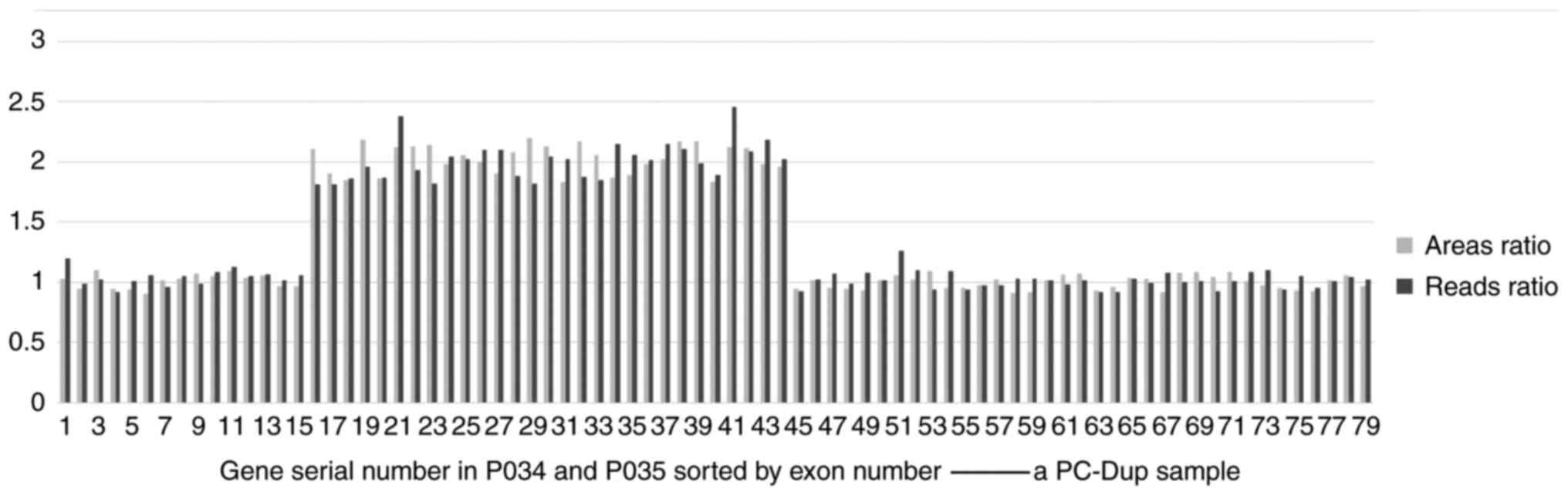
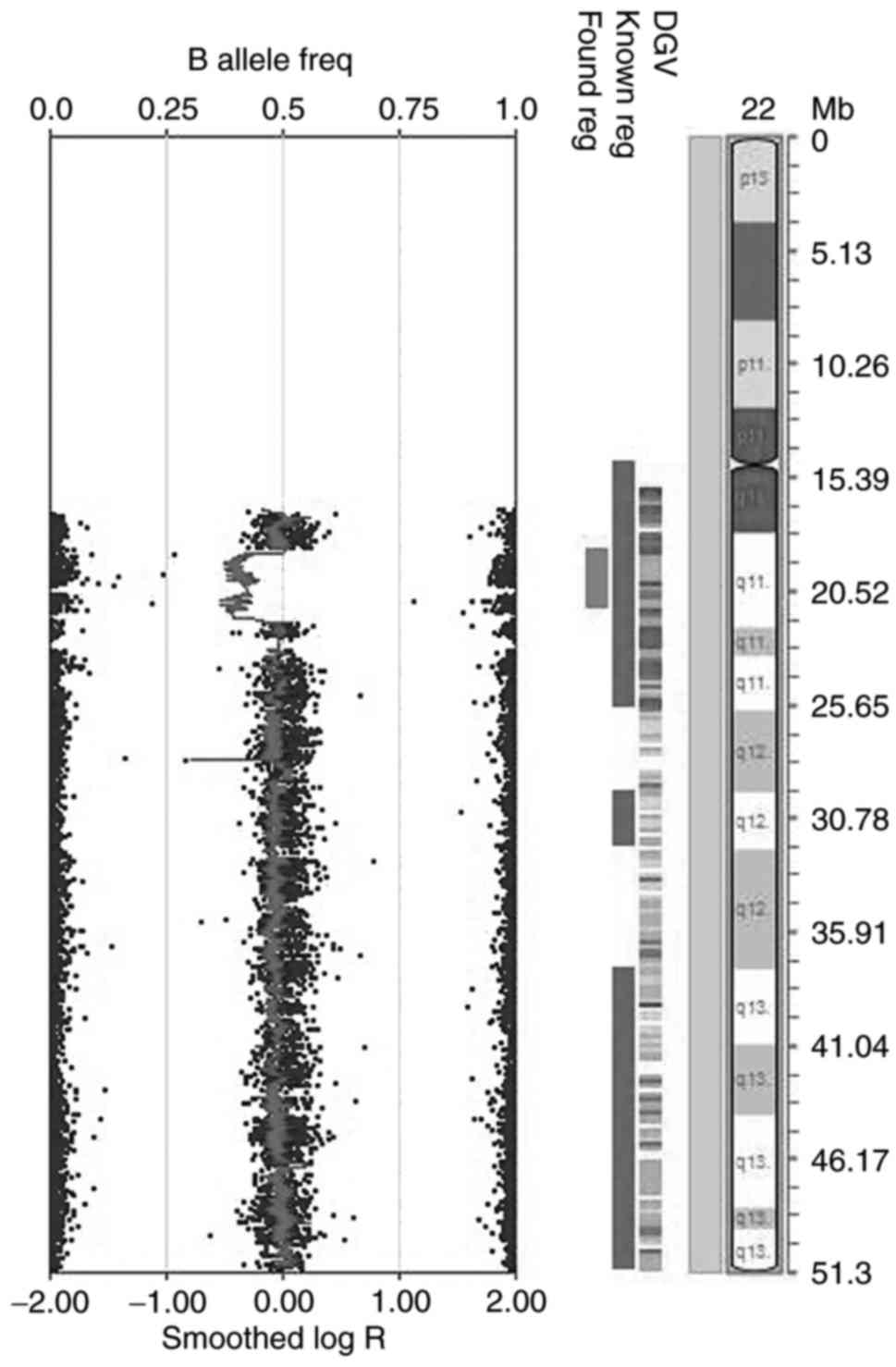
|
1
|
Shaikh TH: Copy number variation
disorders. Curr Genet Med Rep. 5:183–190. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hastings PJ, Lupski JR, Rosenberg SM and
Ira G: Mechanisms of change in gene copy number. Nat Rev Genet.
10:551–564. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang F, Gu W, Hurles ME and Lupski JR:
Copy number variation in human health, disease, and evolution. Annu
Rev Genomics Hum Genet. 10:451–481. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Girirajan S, Campbell CD and Eichler EE:
Human copy number variation and complex genetic disease. Annu Rev
Genet. 45:203–226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iourov IY, Vorsanova SG and Yurov YB:
Molecular cytogenetics and cytogenomics of brain diseases. Curr
Genomics. 9:452–465. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang X, Du R, Li S, Zhang F, Jin L and
Wang H: Evaluation of copy number variation detection for a SNP
array platform. BMC Bioinformatics. 15:502014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stuppia L, Antonucci I, Palka G and Gatta
V: Use of the MLPA assay in the molecular diagnosis of gene copy
number alterations in human genetic diseases. Int J Mol Sci.
13:3245–3276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsuchiya KD, Shaffer LG, Aradhya S,
Gastier-Foster JM, Patel A, Rudd MK, Biggerstaff JS, Sanger WG,
Schwartz S, Tepperberg JH, et al: Variability in interpreting and
reporting copy number changes detected by array-based technology in
clinical laboratories. Genet Med. 11:866–873. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Manning M and Hudgins L: Professional
Practice and Guidelines Committee: Array-based technology and
recommendations for utilization in medical genetics practice for
detection of chromosomal abnormalities. Genet Med. 12:742–745.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schouten JP, McElgunn CJ, Waaijer R,
Zwijnenburg D, Diepvens F and Pals G: Relative quantification of 40
nucleic acid sequences by multiplex ligation-dependent probe
amplification. Nucleic Acids Res. 30:e572002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Banerjee S, Oldridge D, Poptsova M,
Hussain WM, Chakravarty D and Demichelis F: A computational
framework discovers new copy number variants with functional
importance. PLoS One. 6:e175392011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eijk-Van Os PG and Schouten JP: Multiplex
ligation-dependent probe amplification (MLPA®) for the
detection of copy number variation in genomic sequences. Methods
Mol Biol. 688:97–126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deveson IW, Chen WY, Wong T, Hardwick SA,
Andersen SB, Nielsen LK, Mattick JS and Mercer TR: Representing
genetic variation with synthetic DNA standards. Nat Methods.
13:784–791. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Xu Y, Liu D, Geng J, Chen S,
Jiang Z, Fu Q and Sun K: A modified multiplex ligation-dependent
probe amplification method for the detection of 22q11.2 copy number
variations in patients with congenital heart disease. BMC Genomics.
16:3642015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gross SJ, Ryan A and Benn P: Noninvasive
prenatal testing for 22q11.2 deletion syndrome: Deeper sequencing
increases the positive predictive value. Am J Obstet Gynecol.
213:254–255. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chung JH, Cai J, Suskin BG, Zhang Z,
Coleman K and Morrow BE: Whole-genome sequencing and integrative
genomic analysis approach on two 22q11.2 deletion syndrome family
trios for genotype to phenotype correlations. Hum Mutat.
36:797–807. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Nettleton D and Ying K: Copy
number variation detection using next generation sequencing read
counts. BMC Bioinformatics. 15:1092014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bunyan DJ, Skinner AC, Ashton EJ,
Sillibourne J, Brown T, Collins AL, Cross NC, Harvey JF and
Robinson DO: Simultaneous MLPA-based multiplex point mutation and
deletion analysis of the dystrophin gene. Mol Biotechnol.
35:135–140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Naoufal R, Legendre M, Couet D,
Gilbert-Dussardier B, Kitzis A, Bilan F and Harbuz R: Association
of structural and numerical anomalies of chromosome 22 in a patient
with syndromic intellectual disability. Eur J Med Genet.
59:483–487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiong B, Tan K, Tan YQ, Gong F, Zhang SP,
Lu CF, Luo KL, Lu GX and Lin G: Using SNP array to identify
aneuploidy and segmental imbalance in translocation carriers. Genom
Data. 2:92–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Belfield EJ, Brown C, Gan X, Jiang C,
Baban D, Mithani A, Mott R, Ragoussis J and Harberd NP:
Microarray-based optimization to detect genomic deletion mutations.
Genom Data. 2:53–54. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gilbert DC, McIntyre A, Summersgill B,
Missiaglia E, Goddard NC, Chandler I, Huddart RA and Shipley J:
Minimum regions of genomic imbalance in stage I testicular
embryonal carcinoma and association of 22q loss with relapse. Genes
Chromosomes Cancer. 50:186–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chan LF, Campbell DC, Novoselova TV, Clark
AJ and Metherell LA: Whole-exome sequencing in the differential
diagnosis of primary adrenal insufficiency in children. Front
Endocrinol (Lausanne). 6:1132015.PubMed/NCBI
|
|
24
|
Nimkarn S, Gangishetti PK, Yau M and New
MI: 21-hydroxylase-deficient congenital adrenal
hyperplasiaGeneReviews®. Adam MP, Ardinger HH, Pagon RA,
Wallace SE, Bean LJH, Stephens K and Amemiya A: Seattle (WA):
1993
|
|
25
|
Wong A, Martin Lese C, Heretis K, Ruffalo
T, Wilber K, King W and Ledbetter DH: Detection and calibration of
microdeletions and microduplications by array-based comparative
genomic hybridization and its applicability to clinical genetic
testing. Genet Med. 7:264–271. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aten E, White SJ, Kalf ME, Vossen RH,
Thygesen HH, Ruivenkamp CA, Kriek M, Breuning MH and den Dunnen JT:
Methods to detect CNVs in the human genome. Cytogenet Genome Res.
123:313–321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen Y and Wu BL: Designing a simple
multiplex ligation-dependent probe amplification (MLPA) assay for
rapid detection of copy number variants in the genome. J Genet
Genomics. 36:257–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bremer A, Giacobini M, Nordenskjöld M,
Brøndum-Nielsen K, Mansouri M, Dahl N, Anderlid B and Schoumans J:
Screening for copy number alterations in loci associated with
autism spectrum disorders by two-color multiplex ligation-dependent
probe amplification. Am J Med Genet B Neuropsychiatr Genet.
153B:280–285. 2010.PubMed/NCBI
|
|
29
|
Cai G, Edelmann L, Goldsmith JE, Cohen N,
Nakamine A, Reichert JG, Hoffman EJ, Zurawiecki DM, Silverman JM,
Hollander E, et al: Multiplex ligation-dependent probe
amplification for genetic screening in autism spectrum disorders:
Efficient identification of known microduplications and
identification of a novel microduplication in ASMT. BMC Med
Genomics. 1:502008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Slater H, Bruno D, Ren H, La P, Burgess T,
Hills L, Nouri S, Schouten J and Choo KH: Improved testing for
CMT1A and HNPP using multiplex ligation-dependent probe
amplification (MLPA) with rapid DNA preparations: Comparison with
the interphase FISH method. Hum Mutat. 24:164–171. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Herodez Stangler S, Zagradisnik B, Skerget
Erjavec A, Zagorac A and Vokac Kokalj N: Molecular diagnosis of
PMP22 gene duplications and deletions: Comparison of different
methods. J Int Med Res. 37:1626–1631. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Q and Keleş S: CNV-guided multi-read
allocation for ChIP-seq. Bioinformatics. 30:2860–2867. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duan J, Zhang JG, Deng HW and Wang YP:
Detection of common copy number variation with application to
population clustering from next generation sequencing data. Conf
Proc IEEE Eng Med Biol Soc. 2012:1246–1249. 2012.PubMed/NCBI
|
|
34
|
de Ligt J, Boone PM, Pfundt R, Vissers LE,
de Leeuw N, Shaw C, Brunner HG, Lupski JR, Veltman JA and Hehir-Kwa
JY: Platform comparison of detecting copy number variants with
microarrays and whole-exome sequencing. Genom Data. 2:144–146.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
de Ligt J, Boone PM, Pfundt R, Vissers LE,
Richmond T, Geoghegan J, O'Moore K, de Leeuw N, Shaw C, Brunner HG,
et al: Detection of clinically relevant copy number variants with
whole-exome sequencing. Hum Mutat. 34:1439–1448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu
X, Guo H, Jiang Q, Allen AS and Zhu M: An evaluation of copy number
variation detection tools from whole-exome sequencing data. Hum
Mutat. 35:899–907. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Samarakoon PS, Sorte HS, Kristiansen BE,
Skodje T, Sheng Y, Tjønnfjord GE, Stadheim B, Stray-Pedersen A,
Rødningen OK and Lyle R: Identification of copy number variants
from exome sequence data. BMC Genomics. 15:6612014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fromer M, Moran JL, Chambert K, Banks E,
Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O'Donovan MC,
Owen MJ, et al: Discovery and statistical genotyping of copy-number
variation from whole-exome sequencing depth. Am J Hum Genet.
91:597–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu J, Grzeda KR, Stewart C, Grubert F,
Urban AE, Snyder MP and Marth GT: Copy number variation detection
from 1000 Genomes project exon capture sequencing data. BMC
Bioinformatics. 13:3052012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo Y, Sheng Q, Samuels DC, Lehmann B,
Bauer JA, Pietenpol J and Shyr Y: Comparative study of exome copy
number variation estimation tools using array comparative genomic
hybridization as control. Biomed Res Int. 2013:9156362013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sathya B, Dharshini AP and Kumar GR: NGS
meta data analysis for identification of SNP and INDEL patterns in
human airway transcriptome: A preliminary indicator for lung
cancer. Appl Transl Genom. 4:4–9. 2014.PubMed/NCBI
|
|
42
|
Liu B, Madduri RK, Sotomayor B, Chard K,
Lacinski L, Dave UJ, Li J, Liu C and Foster IT: Cloud-based
bioinformatics workflow platform for large-scale next-generation
sequencing analyses. J Biomed Inform. 49:119–133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li H and Durbin R: Fast and accurate short
read alignment with burrows-wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Brandariz-Fontes C, Camacho-Sanchez M,
Vilà C, Vega-Pla JL, Rico C and Leonard JA: Effect of the enzyme
and PCR conditions on the quality of high-throughput DNA sequencing
results. Sci Rep. 5:80562015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen CP, Huang JP, Chen YY, Chern SR, Wu
PS, Su JW, Chen YT, Chen WL and Wang W: Chromosome 22q11.2 deletion
syndrome: Prenatal diagnosis, array comparative genomic
hybridization characterization using uncultured amniocytes and
literature review. Gene. 527:405–409. 2013. View Article : Google Scholar : PubMed/NCBI
|