Introduction
Leukemia is an aggressive hematologic malignancy,
with a poor prognosis. The poor clinical outcomes that are observed
in patients with leukemia are associated with the high proportion
of patients who develop resistance to Adriamycin (ADR) therapy
(1,2). The development of ADR resistance can
be attributed to a number of different factors including, increased
drug efflux, changes in the expression of chemoresistance
associated genes and the enhancement of DNA damage repair inside
cells, as well as the inhibition of apoptosis (3,4).
Among these, the reversal of chemoresistance by attenuating DNA
damage repair and inducing apoptosis have recently attracted
widespread attention amongst researchers in this area of
medicine.
Poly (adenosine diphosphate-ribose) polymerase
(PARP) 1 inhibitors are used as a monotherapy to induce cell death
in tumors with BRCA mutations (5),
or those with a BRCA-like phenotype (also known as BRCAness)
(6). PARP inhibitors have a
cytotoxic effect via several mechanisms, including inhibition of
PARP1/2, auto-PARylation, blocking PARP1/2 release from substrate
DNA, or hypersensitivity to trapped PARP1-DNA complexes (7). Olaparib (Lynparza; AstraZeneca,
Cambridge, UK) is an oral, first-in-class PARP inhibitor, which is
approved by the US Food and Drug Administration (FDA) for the
treatment of patients with advanced germline BRCA-mutated ovarian
cancer (8). Olaparib has antitumor
activity in sporadic cases of metastatic- and castration-resistant
prostate cancer with DNA-repair defects, as determined by a phase
II trial of Olaparib in patients with advanced metastatic resistant
prostate cancer (9). The FDA has
stated that it plans to approve Olaparib for use in
castration-resistant prostate cancers, beyond germline BRCA
mutations (10).
Previously published studies by the authors have
revealed that the Fanconi anemia (FA)/BRCA signaling pathway is
involved in the acquired ADR resistance of leukemia cells (11), and that extensive crosstalk exists
within the DNA repair pathway (12). PARP1 inhibitors can strengthen DNA
damage when combined with traditional chemotherapy drugs. Recent
studies have suggested that PARP inhibitors are able to reverse
drug resistance by targeting the FA/BRCA signaling pathway
(13), and that disruption of FA-
or other HR-associated genes also sensitizes cells to PARP
inhibitors (14,15). This provides strong rationale for
the development of PARP inhibitors as a cancer therapy. PARP1
inhibitors also appear to exhibit a degree of protection against
ADR-induced cardiotoxicity (16).
By combining PARP1 inhibitors with traditional chemotherapy, it may
be possible to lower the dosage and reduce the toxic side effects
commonly associated with many chemotherapeutic agents. Therefore,
the aim of the present study was to explore whether PARP1
inhibitors enhance sensitivity to ADR in ADR-resistant leukemia
cells. Furthermore, the synergistic effects of ADR on apoptosis
were also investigated.
Materials and methods
Cell culture patient samples and
drugs
Bone marrow samples of three patients with
chemoresistant leukemia from The Third Xiangya Hospital of Central
South University (Hunan, China) were collected once written
informed consent was obtained (Table
I). Primary refractory or resistant disease was defined as not
achieving complete remission (CR; i.e., a remaining blast count of
≥5% following 1 to 2 cycles of intense induction therapy) (17) The characteristics of patients with
leukemia are presented in Table I.
The present study was approved by the Central South University, and
the approved protocol was in accordance with the Declaration of
Helsinki. Mononuclear cells were isolated using a lymphocyte
separation medium (Mediatech, Manassas, VA, USA) (18).
 | Table I.Characteristics of patients with
leukemia. |
Table I.
Characteristics of patients with
leukemia.
| Patient | Age (years) | Sex | FAB type | Blast count
(%) | IC50
(µmol) |
|---|
| 1 | 21 | Female | AML-M1 | 10 | 21.85 |
| 2 | 36 | Male | AML-M2b | 8 | 38.19 |
| 3 | 52 | Male | AML-M5 | 15 | 14.17 |
The K562/ADR and K562 human leukemic cell lines were
a generous gift from the Cell Center of Xiangya School of Medicine
(Changsha, Hunan, China). K562 cell lines were grown in RPMI-1640
medium supplemented with 10% heat-inactivated fetal bovine serum
(both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
K562/ADR cells were grown in culture medium containing 1 µM ADR
(Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China) to maintain
drug resistance. Cells were grown at 37°C in an atmosphere of 95%
O2 and 5% CO2.
Drug treatment and survival assay
The stock solution of Olaparib (Selleck Chemicals,
Houston, TX, USA) was prepared by dissolving 10 mg of the drug in
50 µl of dimethyl sulfoxide (Amresco, LLC, Solon, OH, USA). Then it
was diluted with RPMI to a concentration of 5 µmol. The aliquots
were stored at −80°C. For each experiment a new aliquot was thawed
and used.
Cell viability was measured using Cell Counting
kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) according to the manufacturer's instructions. Cells were
seeded at 1×105 cells/ml in duplicate for each time
point (24 h, 48 h and 72 h) in a 96-well culture plate. K562 and
K562/ADR cells were exposed to 0, 2, 4, 8, 16 and 32 µmol/l ADR at
37°C for 24, 48 and 72 h respectively. K562/ADR cells at
1×105 cells/ml in a 96-well culture plate were exposed
to 0, 1.25, 2.5, 5 and 10 µmol/l Olaparib at 37°C for 72 h. Then 10
µl CCK-8 solution was added to each well and plates were incubated
at 37°C for 4 h. The absorbance at 570 nm was then measured using a
microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
All experiments were performed in triplicate, and three independent
experiments were conducted.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
A Total RNA Extractor (Omega Bio-Tek, Inc.,
Norcross, GA, USA) and RT-PCR kit (Toyobo Life Science, Osaka,
Japan) were used for cDNA synthesis (25°C for 10 min and 42°C for
50 min, followed by 70°C for 15 min). SYBR-Green reagents (Toyobo
Life Science, Osaka, Japan) were used to determine PARP1 and γ-H2A
histone family member X (H2AX) expression. The thermocycling
conditions consisted of the following: Initial denaturation for 2
min 30 sec at 95°C, followed by 40 cycles of denaturation for 30
sec at 95°C, annealing for 30 sec at 58°C and extension for 30 sec
at 72°C, then a final extension at 72°C for 10 min. The gene
expression level was normalized using the endogenous control gene
glyceraldehyde 3-phosphate dehydrogenase (GAPDH). RT-qPCR reactions
were performed using a Master Cycler Ep Realplex (Eppendorf,
Hamburg, Germany). Relative expression was evaluated using the
2−ΔΔCq method (17).
All PCR assays were performed three times. The primers for the
individual genes were as follows: PARP1 forward,
5′-TACCATCCAGGCTGCTTTGTCA-3′ and reverse,
5′-CTTCGCCACTTCATCCACTCCA-3′; γ-H2AX forward,
5′-GGCCTCCAGTTCCCAGTG-3′ and reverse, 5′-TCAGCGGTGAGGTACTCCAG-3′;
and GAPDH forward, 5′-GACTCATGACCACAGTCCATGC-3′ and reverse,
5′-AGAGGCAGGGATGATGTTCTG-3′.
Western blot analysis
Cells (5×105/ml) were exposed to 2 µM
ADR, 5 µM Olaparib, or a combination of the two drugs at 37°C for 3
days. Subsequently, they were collected by centrifugation (30,000 ×
g for 5 min at 4°C), washed with cold PBS, and finally lysed with a
cell lysis buffer (Cell Signaling Technology, Inc., Danvers, MA,
USA). The protein concentrations were determined using a BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.). Equal amounts
of protein lysates were then mixed with SDS-PAGE protein loading
buffer, boiled for 5 min, and then run on 10% SDS-PAGE. Proteins
were then transferred onto polyvinylidene fluoride membranes, which
were blocked with 5% skimmed milk powder in Tris-buffered saline
with 0.1% Tween 20 (Beijing Solarbio Science & Technology Co.,
Ltd.) for 2 h at 4°C. Membranes were incubated with the following
primary antibodies at 4°C overnight: PARP1 (1:1,000; cat. no.
YT595; Immunoway Biotechnology Company, Plano, TX, USA); γ-H2AX
(1:1,000; cat. no. pSer139; Novus Biologicals, LLC, Littleton, CO,
USA); caspase 3 (1:1,000; cat. no. ab32351), cleaved-PARP (1:1,000;
cat. no. ab4830; both Abcam, Cambridge, UK); cyclinB1 (1:1,000;
cat. no. 4138); cleaved-caspase 3 (1:2,000; cat. no. 1177; both
CST, Biological Reagents Co., Ltd., Shanghai, China); and β-actin
(1:3,000; cat. no. 20536-1-AP; ProteinTech Group, Inc., Chicago,
IL, USA). The next morning, the membranes were incubated with
horseradish peroxidase (HRP)-conjugated goat anti-mouse or rabbit
secondary antibody (cat. no. sc-2031; 1:10,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) for 2 h at room temperature.
Following washing with PBS (Gibco; Thermo Fisher Scientific, Inc.)
at room temperature for 5 minute, bands were captured with a Gel
imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
relative expression of target proteins was calculated with ImageJ
version 2× (National Institutes of Health, Bethesda, MD, USA) and
normalized to β-actin.
Cell counting assay
For survival assays, stimulated K562/ADR cells were
collected, centrifuged (10,000 × g for 5 min at 4°C), suspended in
a fresh complete medium (time 0 on the graphics), and seeded at
5×105 cells/ml in each well of 6-well culture plates.
K562/ADR cells were divided into four treatment groups: i) a
control group (medium only); ii) 2 µM ADR; iii) 5 µM Olaparib; iv)
and a combination group of the two drugs at 37°C for 3 days. Cells
were counted via a trypan blue dye exclusion assay in
quadruplicate, once the cells were dispersed, 10 µl of cell
suspension was transferred to a micro centrifuge tube,10 µl of
trypan blue (Beijing Solarbio Science & Technology Co., Ltd.;
C0040) was then added and mixed gently with a pipette; 10 µl of
cell/trypan blue suspension was then loaded into a cell counting
chamber and placed under an inverted microscope (Nikon Corporation,
Tokyo, Japan) for counting. The assays were performed 3 days
post-drug exposure. All of the experiments were performed three
times each, and all three experiments were conducted
independently.
Cell apoptosis assay
Cell apoptosis was analyzed using an Annexin
V/propidium iodide (PI) kit (Nanjing KeyGen Biotech, Co., Ltd.)
according to the manufacturer's protocol. Cells were collected and
analyzed on a FACS Canto II (BD Biosciences, San Jose, CA, USA)
flow cytometer. To determine cell cycle distribution,
8×104 K562/ADR cells were seeded in 6 cm dishes for a
total of 3 days following lentiviral infection. Following the 3-day
culture, the cells were washed twice with ice-cold PBS and were
re-suspended in a PBS (ZSGB-BIO; OriGene Technologies, Inc.,
Beijing, China) containing 50 µg/ml RNaseA (Nanjing KeyGen Biotech,
Co., Ltd.) and 50 µg/ml PI (Nanjing KeyGen Biotech, Co., Ltd.).
Cells were incubated at 37°C in the dark for 1 h. The percentage of
cells in each phase of the cell cycle was measured using FACScan
(BD Biosciences) and subsequently the results were analyzed using
ModFit software, version 3.2 (Verity Software House, Inc., Topsham,
ME, USA).
Cell cycle analysis
Following a total of 3 days, the treatment groups
were subjected to cell cycle analysis. Following centrifugation at
300 × g for 5 min at room temperature, cells (1×106)
were fixed with 70% ethanol on ice for 2 h, followed by
centrifugation for 5 min at 300 × g at 4°C. Subsequently, 0.05
mg/ml PI (Nanjing KeyGen Biotech, Co., Ltd.) and 0.1 mg/ml RNAse A
(Nanjing KeyGen Biotech, Co., Ltd.) were added to the samples at
room temperature in the dark for 30 min. Cells were examined using
a BD FACSCalibur flow cytometer (BD Biosciences) and Cell Quest
software version 3.3 (BD Biosciences). The experiments were
repeated three times.
Alkaline comet assay
The alkaline comet assay was used to detect
ADR-induced DNA damage in K562/ADR cells. A total of
1×105 cells were treated with either 2 µM ADR, 5 µM
Olaparib, or a combination of the two drugs for at 37°C for 3 days.
Then, cells were washed in PBS and resuspended in ice-cold PBS. A
total of ~10 µl of the re-suspended cells were mixed with 65 µl of
a low melting point agarose at 4°C. The slides were placed at 4°C
in the dark until gelling occurred, at which point they were
immersed in a pre-chilled lysis buffer at 4°C. Following a 1 h
incubation period, the buffer was aspirated and replaced with
pre-chilled alkaline solution for 90–120 min at 4°C. Following
lysis and DNA double strand unwinding into a single strand, the
slides were placed in a horizontal electrophoresis tank filled with
freshly prepared alkaline electrophoresis buffer. Electrophoresis
was run for 30 min at 25 V and 300 mA. Following this, the slides
were transferred to a neutralizing buffer for 10 min, and then
aspirated. This was repeated three times. Thereafter, the slides
were allowed to air dry, and 50 µl/well of 5 mg/ml PI was added to
each slide for 5 min in the dark at room temperature for DNA
staining. DNA migration was observed using a fluorescence
microscope (Olympus Corporation, Tokyo, Japan). For each sample,
200 cells were selected at random and were analyzed using CASP
software version 1.2.2 (Comet Assay Software Project, Beijing,
China).
Exposure of mononuclear cells from
patients with chemoresistant leukemia, to Olaparib combined with
ADR
To determine the potential clinical significance of
the cell line studies, the present study isolated mononuclear cells
from patients with chemoresistant leukemia, Each sample cells were
divided into two parts; one part at 1×105 cells/ml were
seeded in duplicate in a 96-well culture plate. The cells were
exposed to 0, 2, 4, 8, 16 and 32 µmol/l ADR at 37°C for 3 days.
Then 10 µl CCK-8 solution was added to each well and plates were
incubated at 37°C for 4 h. The absorbance at 570 nm was then
measured using a microplate reader (BioTek Instruments, Inc.,
Winooski, VT, USA). The other part of the sample was exposed to 2
µM ADR, 5 µM Olaparib, or a combination of the two drugs at 37°C
for 3 days. Subsequently, the cells were collected by
centrifugation (30,000 × g for 5 min at 4°C), washed with cold PBS,
and finally lysed with a cell lysis buffer (Cell Signaling
Technology, Inc.). Western blotting, as above was then performed to
analyze the levels of the selected proteins.
Statistical analysis
Statistical analyses was performed using GraphPad
Prism 6.02 version software (GraphPad Software, Inc., La Jolla, CA,
USA). The data were expressed as the mean ± standard deviation
(SD), and statistical comparisons were made using the two-tailed
Student's t-test and one-way analysis of variance followed by
Fisher's Least Significant Difference for multiple comparisons. All
of the experiments were repeated at least three times as
independent experiments in order to be effective. P<0.05 was
considered to indicate a statistically significant difference.
Results
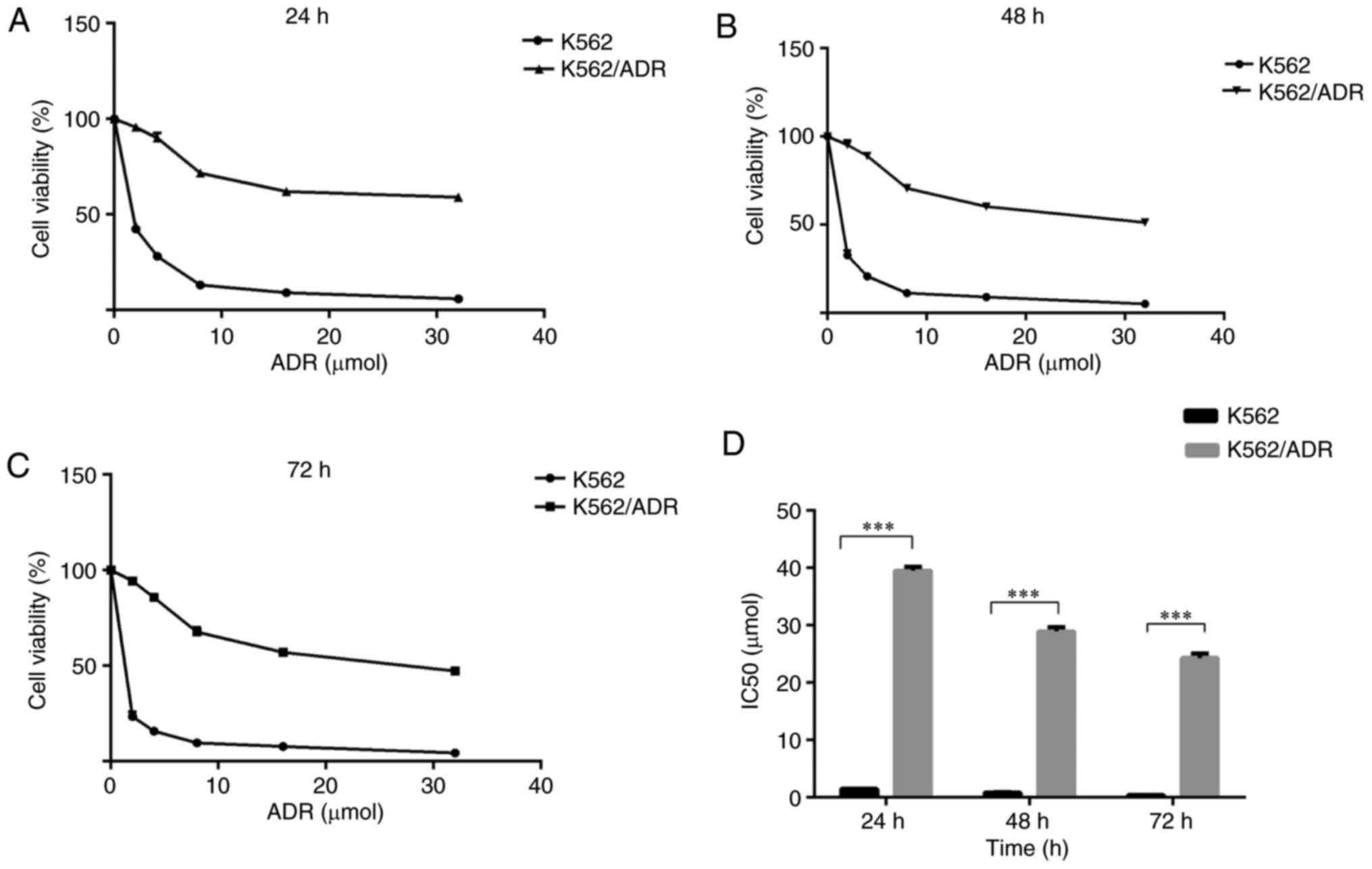
ADR inhibits the proliferation of K562
cells more than K562/ADR cells
To investigate the sensitivity of K562/ADR and K562
cells to chemotherapeutic drugs, the two cell lines were treated
with a gradually increasing concentration of ADR at different times
as indicated. The CCK-8 assay revealed that ADR inhibited the
growth of K562/ADR and K562 cells to varying degrees. ADR-induced
cytotoxicity was dose- and time-dependent in the two cell lines
(Fig. 1). The half-maximal
inhibitory concentration (IC50) values of ADR were
calculated for K562 and K62/ADR cells. The IC50 values
of ADR for K562 cells were 1.50±0.03, 0.82±0.09 and 0.40±0.05
µmol/l at 24, 48 and 72 h, respectively. In addition, the
corresponding values for the K562/ADR cells were 39.51±0.64,
28.93±0.74 and 24.31±0.78 µmol/l, respectively (Fig. 1D). The degree of ADR-resistance in
K562/ADR cells was 26.34–60.70 fold greater when compared with K562
cells, which suggests that K562/ADR cells may have greater
potential for resistance to ADR compared with K562 cells.
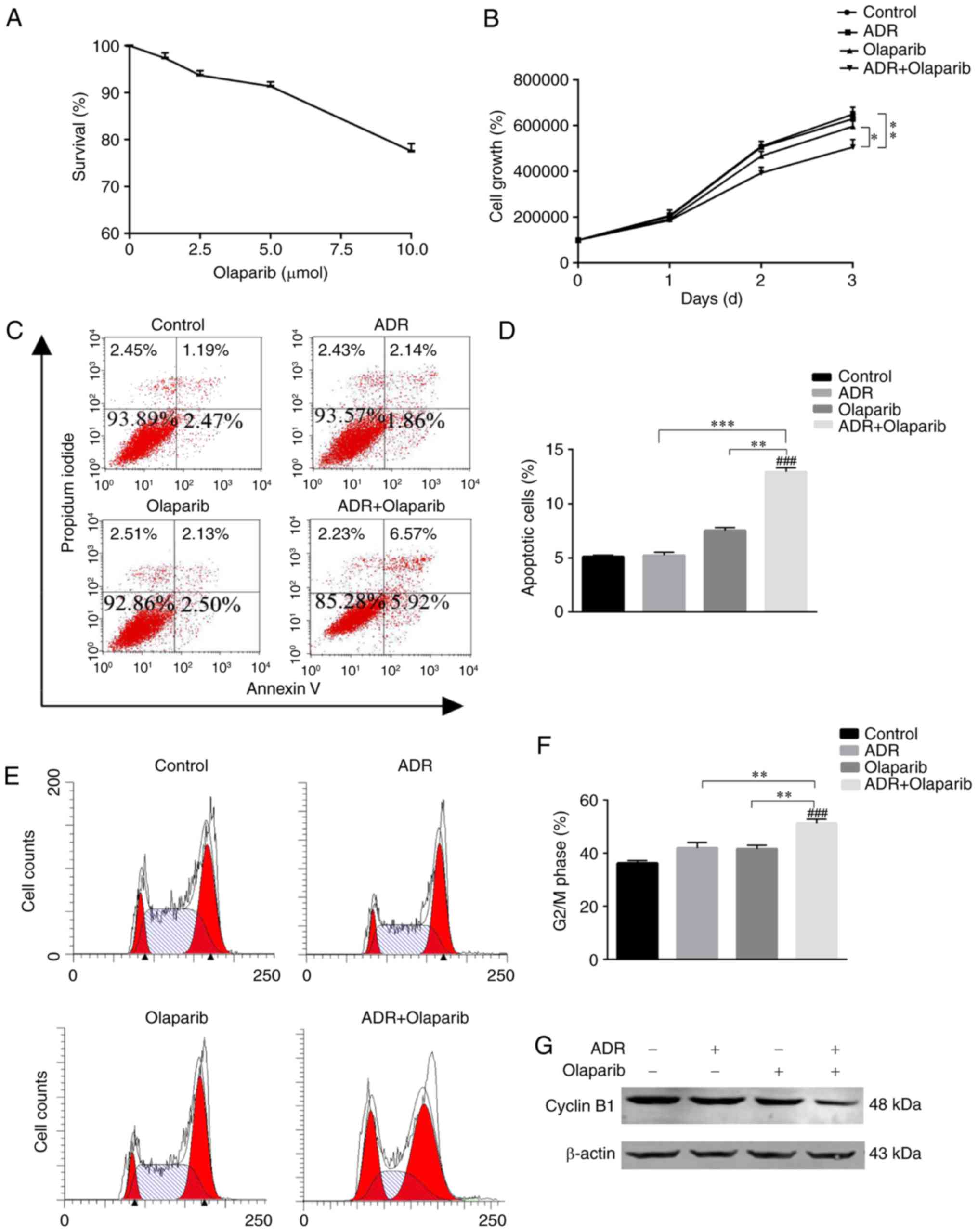
Olaparib enhances apoptosis and
arrests cell cycle progression in K562/ADR cells
In the present study, 2 µM ADR and 5 µM Olaparib
were selected for use in the experiments for the following reasons:
i) ADR at 2 µM consistently enhanced toxicity in the K562 cell
line, but not in the K562/ADR cell line (Fig. 1); and ii) Olaparib alone did not
exhibit a cytotoxic effect on the viability of K562/ADR cells at
doses up to 5 µM as determined by CCK-8 assays (Fig. 2A). The proliferation capability of
K562/ADR cells was inhibited and cell growth was slower in the
group treated with ADR+Olaparib for 3 days (Fig. 2B), which was significantly
different compared with the other control group. This indicated
that the cells treated with ADR+Olaparib had reduced proliferation
rates when compared with cells treated with Olaparib alone, or in
the untreated control cell group (P<0.01; Fig. 2B).
The percentage of apoptotic cells was measured by
flow cytometry using Annexin V/PI staining. Notably, there was a
significantly increased population of apoptotic cells in the
combined treatment group (11.23±0.64%) when compared with the
untreated control group (3.72±0.24%; P<0.001), the ADR group
(4.30±0.31%; P<0.001) and the Olaparib group (4.42±0.66%;
P<0.001). Therefore, combined treatment with Olaparib
significantly improved the apoptotic percentage of K562/ADR cells
(Fig. 2C and D).
To investigate whether cell cycle arrest contributed
to growth inhibition, flow cytometry analysis was performed.
Treatment with ADR+Olaparib significantly increased the percentage
of K562/ADR cells in the G2/M phase. The percentage of cells at the
G2/M phase were 38.23±0.86 for the control cells, 41.81±1.94 for
the 2 µmol ADR-treated K562/ADR cells, 41.97±1.86 for the 5 µmol
Olaparib-treated K562/ADR cells and 50.93±1.53 for the
ADR+Olaparib-treated K562/ADR cells (Fig. 2E and F). K562/ADR cells treated
with ADR+Olaparib had a greater number of G2/M-phase
cells than the cells treated with ADR alone, Olaparib alone or the
untreated control cells (P<0.005, P<0.01 and P<0.005,
respectively). Cyclin B1 was markedly downregulated when the
K562/ADR cells were treated with ADR+Olaparib (Fig. 2G). Cyclin B1 is the regulatory
subunit of Cyclin-dependent kinase 1, and a reduction in the
expression of cyclin B1 can arrest cells in the G2/M phase of the
cell cycle, triggering cell death (19). These results indicate that
re-sensitization of ADR resistant leukemia cells may be partly
mediated by cell cycle arrest at the G2/M phase (Fig. 2E and F).
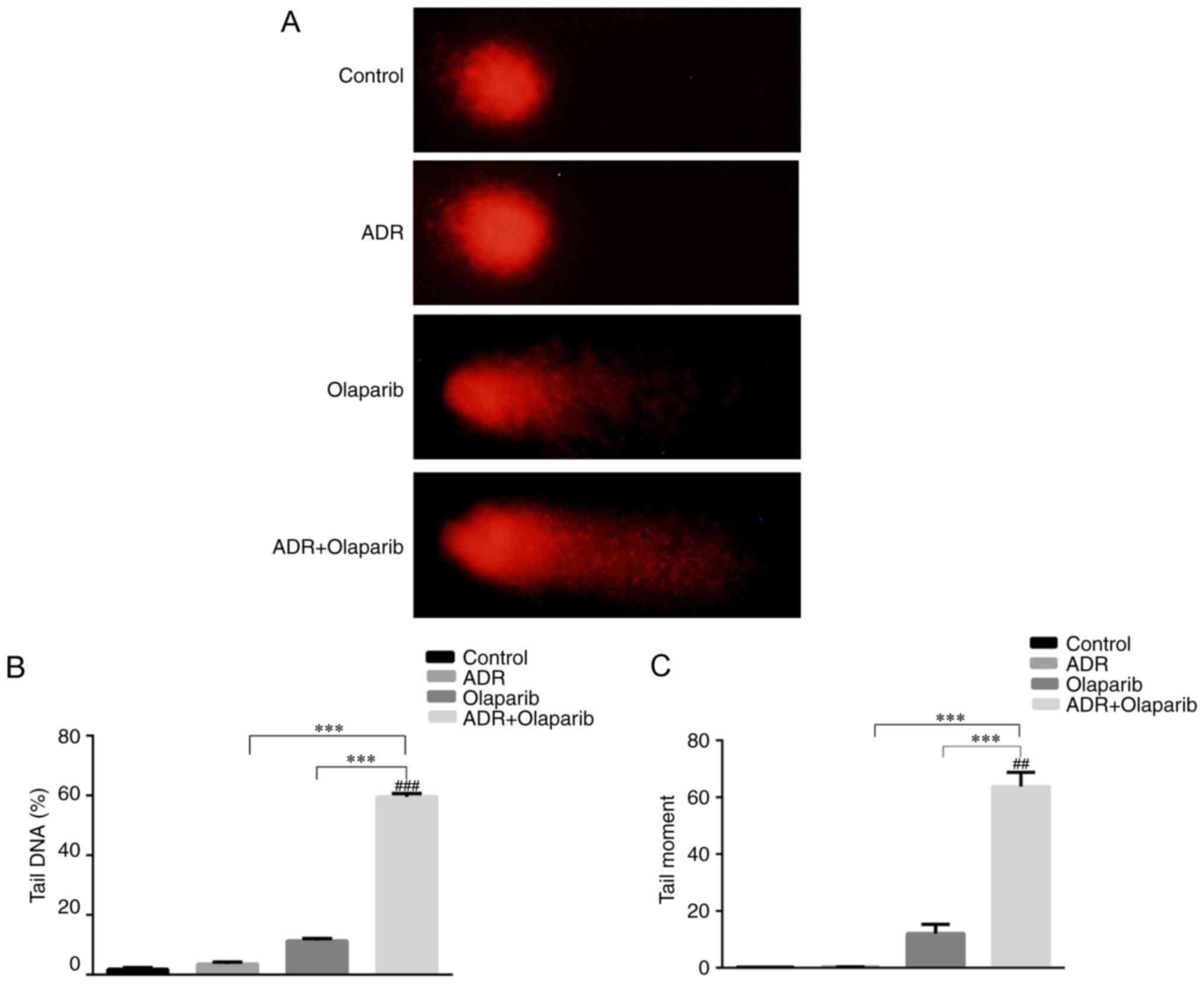
Combination of Olaparib and
ADR-induced DNA damage
The alkaline comet assay was conducted to
investigate the genotoxic potential of DNA damage. Olaparib
enhanced DNA damage in ADR treated cells as shown by the prevalence
of distinct comet tails (Fig. 3A).
The addition of Olaparib enhanced the average tail DNA by ~15-fold
when compared with the control group (P<0.001; Fig. 3B). In addition, the tail moment was
enhanced by ~150-fold P<0.01; Fig.
3C).
Olaparib and ADR induce apoptosis by
altering the expression of PARP1, cleaved PARP1, caspase 3, cleaved
caspase 3 and γ-H2AX in K562/ADR cells
The expression of γ-H2AX was significantly increased
in cells treated with Olaparib alone compared with the control
cells (Fig. 4). In addition, it
was revealed that PARP1 was significantly downregulated and
cleaved-PARP, caspase 3 and cleaved caspase 3 were markedly
upregulated in cells following Olaparib+ADR treatment, compared
with the control group (Fig. 4C and
D).
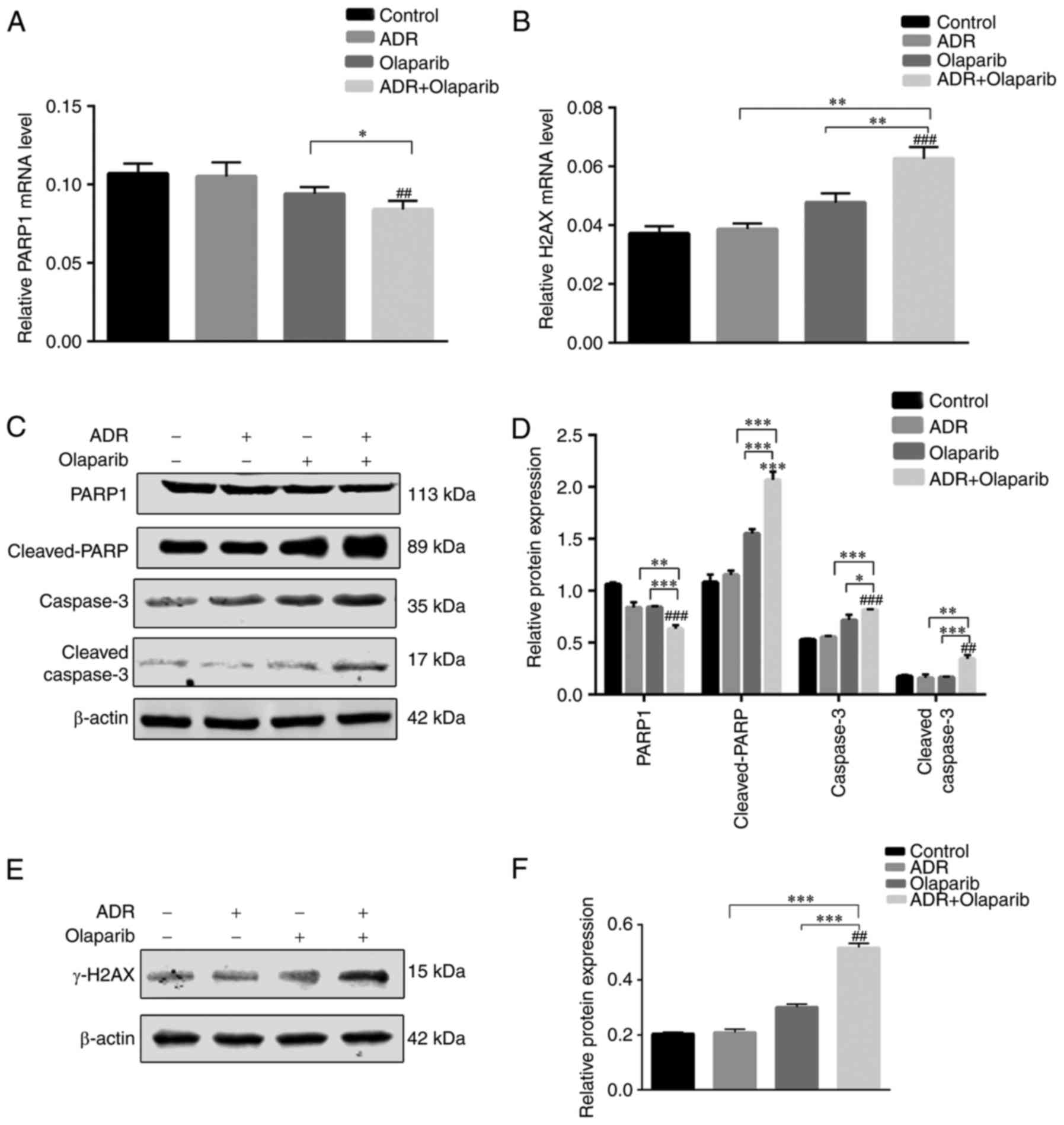 | Figure 4.ADR+Olaparib induces apoptosis by
altering the protein expression of γ-H2AX, PARP1, cleaved-PARP,
caspase 3 and cleaved-caspase 3 in K562/ADR cells. (A) mRNA
expression of PARP1 and (B) H2AX. (C) Western blotting and (D)
statistical analysis of PARP1, cleaved-PARP, Caspase 3 and
cleaved-Caspase 3 normalized to β-actin. (E) Western blotting and
(F) statistical analysis of γ-H2AX normalized to β-actin. Data are
presented as the mean ± standard deviation of three independent
experiments. *P<0.05, **P<0.01 and ***P<0.001, as
indicated; ##P<0.01 and ###P<0.001 vs.
Control. ADR, Adriamycin; H2AX, H2A histone family member X; PARP,
poly (adenosine diphosphate-ribose) polymerase. |
Exposure of mononuclear cells from
patients with leukemia, to ADR+Olaparib activates the DNA-damage
response and apoptosis
To determine the potential clinical significance of
the aforementioned cell line studies, mononuclear cells were
isolated from the bone marrow of patients with chemoresistant
leukemia (their clinic pathological features are summarized in
Table I). These cells were exposed
to ADR, to test the degree of ADR resistance (Fig. 5A), as well as individual drugs or a
combination of the two. Increased levels of γ-H2AX were observed in
cells from the three leukemia patients exposed to ADR+Olaparib,
which indicated that the combined treatment activated a DNA damage
response. Cleaved PARP, caspase 3 and cleaved caspase 3 were
markedly upregulated in cells exposed to ADR+Olaparib treatment,
which indicated that apoptosis was activated (Fig. 5B). These results revealed that
there was drug synergism in cells derived from patients with
leukemia, involving mechanisms analogous to those observed in the
cultured cell lines.
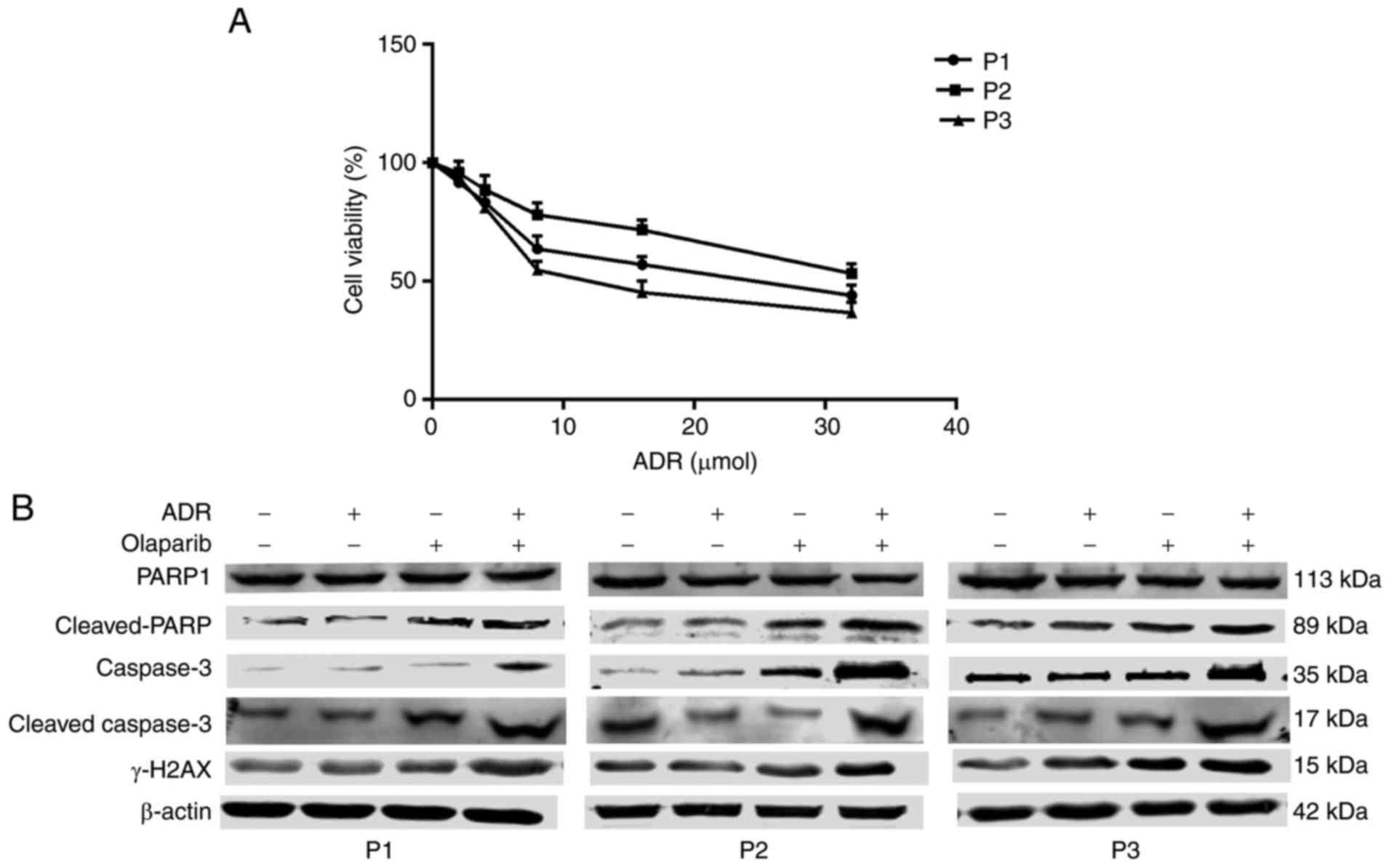 | Figure 5.Effect of patient cell sample
exposure to drugs on biomarkers of apoptosis and DNA-damage. (A)
Cells were exposed to 0, 2, 4, 8, 16 and 32 µmol/l ADR for 3 days,
and Cell Counting kit-8 assays were subsequently performed to
detect cell viability. Results are all presented as the mean ±
standard deviation of three separate experiments. (B) Cells were
exposed to ADR and Olaparib, alone or combined, or the
corresponding control for 3 days. Total cell extracts were analyzed
by western blotting. ADR, Adriamycin; H2AX, H2A histone family
member X; PARP, poly (adenosine diphosphate-ribose) polymerase; P,
patient. |
Discussion
ADR is a time- and dose-dependent antineoplastic
agent, which induces DNA damage in leukemia cells via topoisomerase
II (20). However, of leukemia
cell resistance to ADR reduces DNA damage through the FA/BRCA
signaling pathway in DNA inter-strand crosslink repair (21). In recent years, investigators have
performed numerous studies using PARP1 inhibitors (13), curcumin (22) and arsenic (23) to reverse multidrug resistance via
the FA/BRCA signaling pathway in tumor cells. These previous
studies have provided a strong rationale for the development of
PARP inhibitors for the treatment of ADR resistant leukemia.
K562/ADR cells are multidrug-resistant cells, that
acquired resistance by exposing K562 cells to step-wise increased
concentrations of ADR. K562/ADR cells are resistant to ADR,
mitoxantrone, homoharringtonine, rubidomycin and etoposide
(24). K562/ADR cells appear to
have sophisticated cross drug-resistance to all of these
anti-cancer drugs, which have different structures and functions to
one another. The experiments performed in the present study
demonstrated that the IC50 of K562/ADR cells was
26.34–60.7 fold greater when compared with K562 cells, which
verified that K562/ADR cells were highly resistant to ADR-induced
proliferation inhibition and apoptosis.
The effect of Olaparib on the viability of K562/ADR
cells was investigated by CCK-8 assays. Olaparib alone did not
exhibit a cytotoxic effect on K562/ADR cells at doses up to 5 µM.
The concentrations of Olaparib selected for use within the present
study were below the peak concentration of Olaparib (24 µM) that
has been used in clinical trials (25). The concentrations used in
vitro can also be achieved in vivo. The ADR doses used
in vitro were dependent on the survival of the K562 and
K562/ADR cells. According to the results of previous experiments by
the authors, pre-treatment with ADR at 2 µM consistently enhanced
toxicity in K562 cell lines but not in K562/ADR cell lines
(11). Therefore, 2 µM ADR and 5
µM Olaparib were selected for use in further experiments.
Olaparib+ADR was capable of promoting ADR-mediated apoptosis in
K562/ADR cells.
Several previous studies have reported that PARP1
inhibitors can exert synergistic inhibitory effects in tumors with
various conventional chemotherapeutic agents, including doxorubicin
(26), temozolomide (7) and oxaliplatin (27). The results of the present study
demonstrated that treatment with Olaparib+ADR produced synergistic
effects and revealed a significant increase in the sensitivity of
ADR against K562/ADR cells. Cell cycle arrest at any phase will
inhibit cell proliferation (28).
The results revealed a synergistic effect in the treatment
combination of ADR and Olaparib; combined treatment induced G2/M
cell cycle arrest. In addition, the protein expression of Cyclin B1
was downregulated; the inhibition of cyclin B1 could lead to cell
cycle arrest in the G2/M phase (29). In conclusion, these results
suggested that the combined treatment of ADR and Olaparib may be
more effective than monotherapy in treating ADR resistant
leukemia.
Histone H2AX serves a critical role in the
regulation of DNA damage. H2AX phosphorylation is involved in DNA
damage, as well as apoptosis in chronic myelogenous leukemia cells
induced by imatinib (30).
Olaparib+ADR induced more DNA damage than Olaparib alone in the
present study. Olaparib may increase DNA damage induced by ADR by
inhibiting DNA damage repair.
To investigate the mechanism of PARP inhibitor
re-sensitization in ADR resistant leukemia, the effect of Olaparib
on apoptosis-associated proteins, such as cleaved caspase-3,
caspase-3 (31), cleaved PARP
(32) and PARP1 (33) was investigated. It was revealed
that apoptosis induced the upregulation of caspase-3, cleaved
caspase-3 and cleaved PARP protein expression, and downregulated
PARP1 expression. Caspase-3 is responsible for cleaving specific
cellular proteins during apoptosis (34). Cell death is accompanied by PARP
cleavage, a caspase-3 substrate (35). Caspase-3 is the most active
effector caspase in the intrinsic and extrinsic pathways where it
is processed and activated by caspase-9 and caspase-8, respectively
(36). A high level of caspase-3
activation and cleavage processing was observed in the present
study following ADR and Olaparib treatment of drug resistant
leukemia cells. PARP1 has a molecular weight of 113 kDa and is
located in the nucleus (37).
Following treatment with Olaparib+ADR, caspase-3 was activated and
PARP1 was cleaved into its 89 kDa (cleaved PARP) and 24 kDa forms,
therefore the level of full-length PARP1 (113 kDa) was
significantly reduced. Xu et al (33) reported that caspase 3 activation
resulted in the cleavage of PARP1 and increased apoptosis, which is
consistent with the results observed in the present study. The
results demonstrated drug synergism between the cells derived from
patients with chemoresistant leukemia and the cultured cell lines,
through analogous mechanisms. Therefore, PARP inhibitor
re-sensitization of ADR resistant leukemia may be associated with
the PARP1-mediated signaling pathway of caspase-dependent
apoptosis. However, the apoptotic molecular mechanism of Olaparib
requires further investigation.
In conclusion, the present study provides evidence
of a number of associated mechanisms, that combine to generate DNA
damage and apoptosis in leukemia cell lines and patient-derived
samples. The present study had several limitations, such as the
lack of clinical samples, as well as only one cell line
demonstrating synergistic interactions between Olaparib and ADR in
ADR-resistant leukemia cells. Besides ideally γ-H2AX should be
normalized against the total level of H2AX, however, the remaining
protein of the present drug-resistant leukemia samples was not
enough to complete the H2AX western blot analysis. However, the
results can contribute to the design of clinical trials, which seek
to evaluate the efficacy of these drug combinations as components
of intensified induction therapy, or as a part of optimized
pre-transplant conditioning regimens for patients with
ADR-resistant leukemia.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81570155), Hunan
Provincial Health Committee project (grant no. B2015-06), Project
Fund for the Project of the Hunan Development and Reform Commission
(grant no. 42) and Health Department of Hunan Province (grant no.
B2014-035).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CY conceived and designed the experiments. JW wrote
the paper and SX revised it critically. JW, QL, WW and MY performed
the experiments. LH, SX, YO and GX performed data analysis. WW and
YO collected the patient's samples. SX, LH and GX supervised the
research group. All authors read and approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by Central South
University and written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang X, Ai Z, Chen J, Yi J, Liu Z, Zhao H
and Wei H: Glycometabolic adaptation mediates the insensitivity of
drug-resistant K562/ADM leukaemia cells to adriamycin via the
AKT-mTOR/c-Myc signalling pathway. Mol Med Rep. 15:1869–1876. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tabe Y, Konopleva M, Contractor R, Munsell
M, Schober WD, Jin L, Tsutsumi-Ishii Y, Nagaoka I, Igari J and
Andreeff M: Up-regulation of MDR1 and induction of doxorubicin
resistance by histone deacetylase inhibitor depsipeptide (FK228)
and ATRA in acute promyelocytic leukemia cells. Blood.
107:1546–1554. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vinod BS, Maliekal TT and Anto RJ:
Phytochemicals as chemosensitizers: From molecular mechanism to
clinical significance. Antioxid Redox Signal. 18:1307–1348. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tomicic MT and Kaina B: Topoisomerase
degradation, DSB repair, p53 and IAPs in cancer cell resistance to
camptothecin-like topoisomerase I inhibitors. Biochim Biophys Acta.
1835:11–27. 2013.PubMed/NCBI
|
|
5
|
Wang YQ, Wang PY, Wang YT, Yang GF, Zhang
A and Miao ZH: An Update on Poly(ADP-ribose)polymerase-1 (PARP-1)
Inhibitors: Opportunities and challenges in cancer therapy. J Med
Chem. 59:9575–9598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Neri P, Ren L, Gratton K, Stebner E,
Johnson J, Klimowicz A, Duggan P, Tassone P, Mansoor A, Stewart DA,
et al: Bortezomib-induced ‘BRCAness’ sensitizes multiple myeloma
cells to PARP inhibitors. Blood. 118:6368–6379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gill SJ, Travers J, Pshenichnaya I, Kogera
FA, Barthorpe S, Mironenko T, Richardson L, Benes CH, Stratton MR,
McDermott U, et al: Combinations of PARP inhibitors with
temozolomide drive PARP1 trapping and apoptosis in ewing's sarcoma.
PloS one. 10:e01409882015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim G, Ison G, McKee AE, Zhang H, Tang S,
Gwise T, Sridhara R, Lee E, Tzou A, Philip R, et al: FDA approval
summary: Olaparib monotherapy in patients with deleterious germline
BRCA-mutated advanced ovarian cancer treated with three or more
lines of chemotherapy. Clin Cancer Res. 21:4257–4261. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mateo J, Carreira S, Sandhu S, Miranda S,
Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A,
Tunariu N, et al: DNA-repair defects and olaparib in metastatic
prostate cancer. N Engl J Med. 373:1697–1708. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Helleday T: PARP inhibitor receives FDA
breakthrough therapy designation in castration resistant prostate
cancer: Beyond germline BRCA mutations. Ann Oncol. 27:755–757.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yao C, Du W, Chen H, Xiao S, Huang L and
Chen FP: Involvement of fanconi anemia genes FANCD2 and FANCF in
the molecular basis of drug resistance in leukemia. Mol Med Rep.
11:4605–4610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mouw KW and D'Andrea AD: Crosstalk between
the nucleotide excision repair and Fanconi anemia/BRCA pathways.
DNA Repair (Amst). 19:130–134. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiong T, Wei H, Chen X and Xiao H: PJ34, a
poly(ADP-ribose) polymerase (PARP) inhibitor, reverses
melphalan-resistance and inhibits repair of DNA double-strand
breaks by targeting the FA/BRCA pathway in multidrug resistant
multiple myeloma cell line RPMI8226/R. Int J Oncol. 46:223–232.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
D'Andrea AD: Susceptibility pathways in
fanconi's anemia and breast cancer. N Engl J Med. 362:1909–1919.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McCabe N, Turner NC, Lord CJ, Kluzek K,
Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka
MZ, et al: Deficiency in the repair of DNA damage by homologous
recombination and sensitivity to poly(ADP-ribose) polymerase
inhibition. Cancer Res. 66:8109–8115. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pacher P, Liaudet L, Bai P, Virag L,
Mabley JG, Haskó G and Szabó C: Activation of poly(ADP-ribose)
polymerase contributes to development of doxorubicin-induced heart
failure. J Pharmacol Exp Ther. 300:862–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xia B, Tian C, Guo S, Zhang L, Zhao D, Qu
F, Zhao W, Wang Y, Wu X, Da W, et al: c-Myc plays part in drug
resistance mediated by bone marrow stromal cells in acute myeloid
leukemia. Leuk Res. 39:92–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang L, Zhang Y, Pan H, Luo Q, Zhu XM,
Dong MY, Leung PC, Sheng JZ and Huang HF: Involvement of cyclin B1
in progesterone-mediated cell growth inhibition, G2/M cell cycle
arrest, and apoptosis in human endometrial cell. Reprod Biol
Endocrinol. 7:1442009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Menear KA, Adcock C, Boulter R, Cockcroft
XL, Copsey L, Cranston A, Dillon KJ, Drzewiecki J, Garman S, Gomez
S, et al:
4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluoroben
zyl]-2H-phthalazin-1-one: A novel bioavailable inhibitor of
poly(ADP-ribose) polymerase-1. J Med Chem. 51:6581–6591. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao C, Du W, Chen H, Xiao S, Huang L and
Chen F: The Fanconi anemia/BRCA pathway is involved in DNA
interstrand cross-link repair of adriamycin-resistant leukemia
cells. Leuk Lymphoma. 56:755–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen P, Li J, Jiang HG, Lan T and Chen YC:
Curcumin reverses cisplatin resistance in cisplatin-resistant lung
caner cells by inhibiting FA/BRCA pathway. Tumour Biol.
36:3591–3599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peremarti J, Ramos F, Marcos R and
Hernandez A: Arsenic exposure disrupts the normal function of the
FA/BRCA repair pathway. Toxicol Sci. 142:93–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li GY, Liu JZ, Zhang B, Wang LX, Wang CB
and Chen SG: Cyclosporine diminishes multidrug resistance in
K562/ADM cells and improves complete remission in patients with
acute myeloid leukemia. Biomed Pharmacother. 63:566–570. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fong PC, Boss DS, Yap TA, Tutt A, Wu P,
Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et
al: Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA
mutation carriers. N Engl J Med. 361:123–134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mariano G, Ricciardi MR, Trisciuoglio D,
Zampieri M, Ciccarone F, Guastafierro T, Calabrese R, Valentini E,
Tafuri A, Del Bufalo D, et al: PARP inhibitor ABT-888 affects
response of MDA-MB-231 cells to doxorubicin treatment, targeting
snail expression. Oncotarget. 6:15008–15021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu K, Chen Z, Cui Y, Qin C, He Y and Song
X: Combined olaparib and oxaliplatin inhibits tumor proliferation
and induces G2/M arrest and gamma-H2AX foci formation in colorectal
cancer. OncoTargets Ther. 8:3047–3054. 2015. View Article : Google Scholar
|
|
28
|
Ujiki MB, Ding XZ, Salabat MR, Bentrem DJ,
Golkar L, Milam B, Talamonti MS, Bell RH Jr, Iwamura T and Adrian
TE: Apigenin inhibits pancreatic cancer cell proliferation through
G2/M cell cycle arrest. Mol Cancer. 5:762006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yadav N, Kumar P, Chhikara A and Chopra M:
Development of 1,3,4-oxadiazole thione based novel anticancer
agents: Design, synthesis and in-vitro studies. Biomed
Pharmacother. 95:721–730. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dong Y, Xiong M, Duan L, Liu Z, Niu T, Luo
Y, Wu X, Xu C and Lu C: H2AX phosphorylation regulated by p38 is
involved in bim expression and apoptosis in chronic myelogenous
leukemia cells induced by imatinib. Apoptosis. 19:1281–1292. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu B, Jian Z, Li Q, Li K, Wang Z, Liu L,
Tang L, Yi X, Wang H, Li C and Gao T: Baicalein protects human
melanocytes from H(2)O(2)-induced apoptosis via inhibiting
mitochondria-dependent caspase activation and the p38 MAPK pathway.
Free Radic Biol Med. 53:183–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
D'Amours D, Desnoyers S, D'Silva I and
Poirier GG: Poly(ADP-ribosyl)ation reactions in the regulation of
nuclear functions. Biochem J. 342:249–268. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu P, Cai X, Zhang W, Li Y, Qiu P, Lu D
and He X: Flavonoids of rosa roxburghii tratt exhibit
radioprotection and anti-apoptosis properties via the
Bcl-2(Ca(2+))/Caspase-3/PARP-1 pathway. Apoptosis. 21:1125–1143.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cohen GM: Caspases: The executioners of
apoptosis. Biochem J. 326:1–16. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nicholson DW and Thornberry NA: Caspases:
Killer proteases. Trends Biochem Sci. 22:299–306. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nicolini F, Burmistrova O, Marrero MT,
Torres F, Hernández C, Quintana J and Estévez F: Induction of G2/M
phase arrest and apoptosis by the flavonoid tamarixetin on human
leukemia cells. Mol Carcinog. 53:939–950. 2014.PubMed/NCBI
|
|
37
|
Uchida K and Miwa M: Poly(ADP-ribose)
polymerase: Structural conservation among different classes of
animals and its implications. Mol Cell Biochem. 138:25–32. 1994.
View Article : Google Scholar : PubMed/NCBI
|