Introduction
Dental implantation has been carried out for the
last 25 years and involves placing artificial tooth roots into the
jaw to hold a replacement tooth or bridge following tooth loss. The
success of long-term dental implant placement relies on essential
interactions between the jawbone and dental implant (1). However, insufficient bone volume has
a serious impact on these interactions. Furthermore, bone loss
following implantation is another major issue to address.
Consequently, bone regeneration may require stimulation prior to
implantation for a successful outcome (2). To resolve deficiencies in jawbone
regeneration and to prevent subsequent bone loss, researchers and
dentists have applied numerous strategies, including the use of
autografts, xenografts, allografts and alloplastic materials.
However, limitations exist for these approaches, including limited
availability of grafting material for autografts, and morbidity and
insufficient osteogenesis of xenografts, allografts and alloplastic
materials, due to an absence of cell populations (2,3).
Cell transplantation technologies may address the
limitations of bone transplantation (2). The application of stem or stromal
cells offers a promising approach for enhancing osseointegration
(1,4). Mesenchymal stem cells (MSCs) can
differentiate into bone, adipose, cartilage, muscle and ligament
cells. Bone marrow-derived MSCs (BMSCs) can be immunoselected from
bone marrow and culture-expanded. BMSCs are one of the most widely
applied stem cells for bone regeneration, including that of the
jawbone. However, the application of BMSCs for osteogenesis has
limitations since cell numbers are insufficiently high for several
clinical indications, and the steps for their harvesting and
expansion are complex (1). Adipose
tissue-derived stem cells (ADSCs) result in bone formation
comparable to that of BMSCs and address many of the limitations of
BMSCs. Adipose tissue is an attractive MSC source because it is
readily accessible by routine liposuction with minimal morbidity
(5). Furthermore, a higher number
of stem cells can be harvested from adipose tissue. ADSCs have been
reported to accelerate bone healing in combination with dental
implants (1); however, drawbacks
to cell-based therapies include risk of tumor and the formation of
emboli (6).
Recent evidence has suggested that the secreted
factors released by MSCs are more beneficial in tissue regeneration
than their direct tissue intercalation and differentiation
(7). Additionally, MSCs, including
ADSCs, produce exosomes that serve a role in several biological
functions. Exosomes are nanovesicles ranging between 30 and 100 nm
in size, which are derived from numerous cell types (8). They act as cell-to-cell messengers
and contain mRNA, microRNA (miRNA/miR), proteins and lipids, all of
which influence cell fate. Exosomes are considered a novel
alternative to stimulate bone regeneration with fewer safety
considerations, by resolving the risks of toxicity, emboli,
tumorigenicity and immunogenicity (9). Furthermore, they have powerful
pro-osteogenic potential (6) and
very high stability, in they can be maintained for ~6 months in
vitro at −20°C without loss of potency (10).
MSC-derived exosomes (MSC-exo) can induce naïve stem
cells to differentiate through the osteogenic lineage (11). Additionally, mineralized osteoblast
(12), dendritic (13) and monocyte cell-derived exosomes
have been reported to increase MSC osteogenic differentiation
(14). Over the past 3–4 years,
exosomes, in particular MSC-exo, have gained prominence in research
on bone regenerative medicine (11). However, MSC-exo have the same
limitations as MSCs with regards to resource and quality.
Although immortalization compromises the
differentiation potential of the MSCs, it does not affect the
production or quality of the exosomes for therapy (15). Consequently, ADSC-derived exosomes
(ADSC-exo) may represent a more promising tool for bone
regeneration. ADSC-exo has been reported to improve osteogenesis
via the promotion of vessel formation (16); however, the effects of these
exosomes on ADSC osteogenesis are unclear. The present study
identified a murine preadipocyte cell line, 3T3L1 cells, as having
MSC-like functions and an ability to differentiate into osteoblasts
in response to stimulation with differentiation factors.
Subsequently, 3T3L1 cell-derived exosomes (3T3L1-exo) were
generated, and were used to determine their effects on 3T3L1 cell
osteogenesis and to identify a possible mechanism of action. The
results revealed that 3T3L1-exo promoted 3T3L1 preadipocytes to
undergo osteogenic differentiation via reduced miR-223 expression.
Based on these findings, 3T3L1-exo may represent a useful tool for
investigating preadipocyte-induced bone regeneration.
Materials and methods
Cell culture
The 3T3L1 murine preadipocyte cell line was obtained
from the American Type Culture Collection (Manassas, VA, USA) and
cells were maintained in Dulbecco's modified Eagle's medium (DMEM:
high-glucose, 4,500 mg/l; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal calf serum (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 For
preparation of exosomes, 3T3L1 cells were seeded to 80% confluence
in 100-mm tissue culture dishes. Cells were then cultured for 2
days in the presence of medium containing 2% exosome-free serum
(obtained by ultracentrifugation of serum at 100,000 × g for 14 h
at 4°C).
Isolation of exosomes
In accordance with a previously reported method
(17), cell culture supernatants
were collected and centrifuged at 300 × g for 10 min, 1,200 × g for
20 min, and 10,000 × g for 30 min at 4°C. The supernatant from the
final centrifugation was then ultracentrifuged at 100,000 × g for 1
h at 4°C. After removing the supernatant, the exosome pellets were
washed in a large volume of ice-cold PBS and centrifuged at 100,000
× g for another 1 h at 4°C (17).
Detection of 3T3L1 cell proliferation
and apoptosis
To determine the effects of 3T3L1-exo on 3T3L1 cell
survival, cell apoptosis and proliferation assays were performed.
Briefly, cells were stimulated with 2 µg/ml 3T3L1-exo for 24 h
prior to analysis of the degree of apoptosis or proliferation at
37°C with 5% CO2. For the detection of apoptosis,
2×105 cells were stained with fluorescein
isothiocyanate-Annexin V (BD Pharmingen; BD Biosciences, San Diego,
CA, USA) and propidium iodide (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 5 min at 4°C in the dark. The cells were
then analyzed by fluorescence-activated cell sorting to identify
positively stained apoptotic cells, as described previously
(17).
For the detection of proliferation, 10 µl Cell
Counting kit-8 reagent (7Sea Biotech, Shanghai, China) was added to
each well (2×104/well) in a 96-well plate for 4 h at
37°C, as described previously (17). Fluorescence intensity was
determined using a Bio-Rad microplate reader (450 nm; Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Osteogenic differentiation in
vitro
For osteogenic differentiation, 3T3L1 cells were
plated at a density of 5×104 cells/well in 12-well
plates for 24 h prior to induction. Osteogenic differentiation was
induced by culturing the 3T3L1 cells in osteogenic differentiation
medium (ODM, DMEM supplemented with 20 mM β-glycerol phosphate, 50
µg/ml ascorbic acid and 100 nM dexamethasone) for 21 days; the
medium was replaced every 3–4 days (18). In order to determine the effects of
the exosomes on osteogenic differentiation, Alizarin red staining
(ARS) and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) were carried out. In addition, 2 µg/ml 3T3L1-exo
was added to the 3T3L1 cells in ODM for 15, 30, 60 and 120 min to
determine its effects on differentiation.
RT-qPCR analysis of osteogenic gene
expression
RT-qPCR was used to determine the expression levels
of osteogenic differentiation-associated genes in 3T3L1 cells
stimulated by 3T3L1-exo or pretreated with 10 µM transforming
growth factor-β (TGF-β1) inhibitor (SB431542) at 37°C for 30 min,
and miRNA expression in 3T3L1-exo or 3T3L1 cells. Briefly, RNA was
isolated using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and cDNA was synthesized using
PrimeScript™ RT Reagent kits (Takara Biotechnology Co., Ltd.,
Dalian, China) prior to qPCR, according manufacturer's
protocol.
The expression levels of six pro-osteogenic genes
were analyzed. The genes and primers used in the present study are
listed in Table I. qPCR analysis
of mRNA expression was performed using SYBR Primer Ex Taq™ II kits
(Takara Biotechnology Co., Ltd.) under the following conditions:
One cycle at 95°C for 30 sec, followed by 40 cycles at 95°C for 5
sec and 60°C for 34 sec. Primers for miRNA were purchased from
iGeneBio (Guangzhou, China). qPCR analysis of miRNA expression was
performed using All-in-One™ miRNA qRT-PCR Detection kit
(GeneCopoeia, Inc., Rockville, MD, USA) under the following
conditions: One cycle at 95°C for 10 min, followed by 40 cycles at
95°C for 10 sec, 60°C for 20 sec and 72°C for 10 sec. Expression
levels were quantified using the 2−∆∆Cq method (19). Data are presented as mean fold
changes with respect to controls. Statistical significance was
determined using Student's t-test or one-way analysis of variance
(ANOVA) followed by Tukey's multiple comparisons test (11).
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|
| ALP |
GAGCGTCATCCCAGTGGAG |
TAGCGGTTACTGTAGACACCC |
| OCN |
GAGGGCAATAAGGTAGTGAA |
CATAGATGCGTTTGTAGGC |
| BSP |
CAGGGAGGCAGTGACTCTTC |
AGTGTGGAAAGTGTGGCGTT |
| RUNX2 |
ATGCTTCATTCGCCTCACAAA |
GCACTCACTGACTCGGTTGG |
| Osterix |
GGAAAGGAGGCACAAAGAAGC |
CCCCTTAGGCACTAGGAGC |
| Col I |
CCCTGCCTGCTTCGTGTA |
TTGAGTTTGGGTTGTTCGTC |
| β-actin |
CGTTGACATCCGTAAAGACC |
AACAGTCCGCCTAGAAGCAC |
Western blot analysis
In accordance with a previously reported method
(17), 10 µg exosomes or crude
proteins extracted from cell lysates were separated by 12% SDS-PAGE
and transferred onto polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MA, USA). Membranes were blocked with 5% milk
in PBS-Tween and were then incubated with the primary antibodies
(1:1,000) at 4°C overnight, followed by horseradish
peroxidase-conjugated secondary antibodies (cat. nos. 7074 and
7076; 1:5,000; Cell Signaling Technology, Inc., Danvers, MA, USA)
at room temperature for 1 h. The membranes were scanned using a
Tanon 4500 (Tanon Science and Technology Co., Ltd., Shanghai,
China), according to the manufacturer's protocol. The following
primary antibodies were used: Tumor susceptibility gene 101
(TSG101) mouse monoclonal antibody (mAb) (C-2; cat. no. sc-7964),
heat shock protein 90β family member 1 (GRP94) rabbit polyclonal
antibody (H-212; cat. no. sc-11402), heat shock protein 70 (HSP70)
mouse mAb (3A3; cat. no. sc-32239), β-actin (4E8H3; cat. no.
sc-130065) mouse mAb (Santa Cruz Biotechnology, Inc., Dallas, TX,
USA), SMAD family member 3 (smad3) rabbit mAb (C67H9),
phosphorylated (p)-smad3 rabbit mAb (Ser423/425) (C25A9), and
peroxisome proliferator-activated receptor-γ (PPAR-γ) rabbit mAb
(C26H12) (Cell Signaling Technology, Inc.).
Histological analysis
Osteogenic differentiation was detected using ARS on
day 21, in order to quantify mineralization. For mineralization
quantification, 40 mM ARS (Sigma-Aldrich; Merck KGaA) was prepared
in dH2O (pH 4.1). Cells (3T3L1; 1×105/well)
were cultured in ODM and 2 µg/ml 3T3L-exo for 21 days, rinsed three
times with PBS, and fixed in 10% (v/v) buffered neutral formalin
(Sigma-Aldrich; Merck KGaA) for 15 min at room temperature. The
cells were then rinsed three times with dH2O and
incubated at room temperature in ARS for 20 min with gentle
agitation. Following the aspiration of unincorporated ARS, cells
were rinsed four times with dH2O. Images of stained
cells were subsequently captured.
RNA sequencing of 3T3L1 exosomes and
3T3L1 cells
Total RNA was prepared from 3T3L1 cells and
3T3L1-exo using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA quantity was determined using an Agilent 2100 system
(Agilent Technologies, Inc., Santa Clara, CA, USA). cDNA sequence
libraries were established, sequenced and analyzed by Beijing
Genomics Institute (Beijing, China) using the BGISEQ-500 sequencing
technique (MGI Tech Co., Ltd., Shenzhen, China). Cluster analysis
was performed using pheatmap in R software version 3.1.1
(www.r-project.org).
Electron microscopy
In accordance with a previously described method
(17), exosome pellets were fixed
in 4% paraformaldehyde at 4°C for 1 h. The pellets were then loaded
onto electron microscopy grids coated with Formvar carbon,
contrasted, and embedded in a mixture of 2% uranyl acetate with
methylcellulose. Sections were observed using a Philips Tecnai-10
transmission electron microscope operating at 80 kV (Phillips
Electronic Instruments, Mahwah, NJ, USA).
Transient transfection of miR-223
mimics
To induce miR-223 overexpression, synthetic 100 nM
miRNA mimics were transfected into 3T3L1 cells
(2×105/well) using 3 µl INTERFERin® small
interfering RNA transfection reagent (Polyplus-transfection SA,
Illkirch, France) on a 24-well plate at 37°C for 24 h. The miR-223
mimic and NC mimic (Shanghai GenePharma Co., Ltd., Shanghai, China)
sequences were as follows: miR-223 mimic forward,
5′-UGUCAGUUUGUCAAAUACCCA-3′ and reverse,
5′-GGGUAUUUGACAAACUGACAUU-3′; NC mimics forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′.
Statistical analysis
Data are presented as the means ± standard error of
the mean. Data were analyzed by unpaired t-test or one-way analysis
of variance followed by Tukey's post-hoc test using GraphPad Prism
7 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Isolation and identification of
3T3L1-exo
Exosomes were isolated from 3T3L1 cells cultured
under normal growth conditions (3T3L1-exo). Exosome size and
morphology were determined using electron microscopy. Exosomes were
revealed to range between 50 and 100 nm in size, and exhibited a
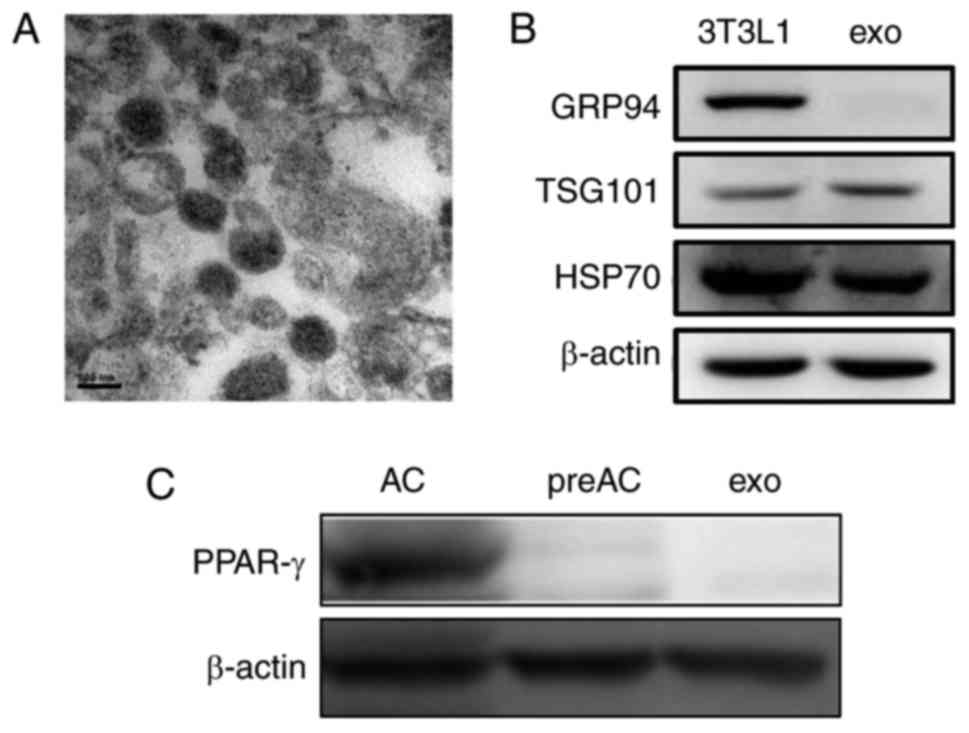
typical rounded shape (Fig. 1A).
Like their parent 3T3L1 cells, 3T3L1-exo was revealed to express
specific markers, including TSG101 and HSP70, whereas GRP94,
endoplasmic reticulum-residing protein, was absent in 3T3L1-exo
(Fig. 1B). Similar to their cell
counterparts, 3T3L1-exo did not express the adipogenic
transcription factor PPAR-γ; however, differentiated 3T3L1 cells
did express PPAR-γ (Fig. 1C).
These results confirmed the integrity of 3T3L1-exo.
3T3L1-exo has no effect on 3T3L1
preadipocyte proliferation and apoptosis
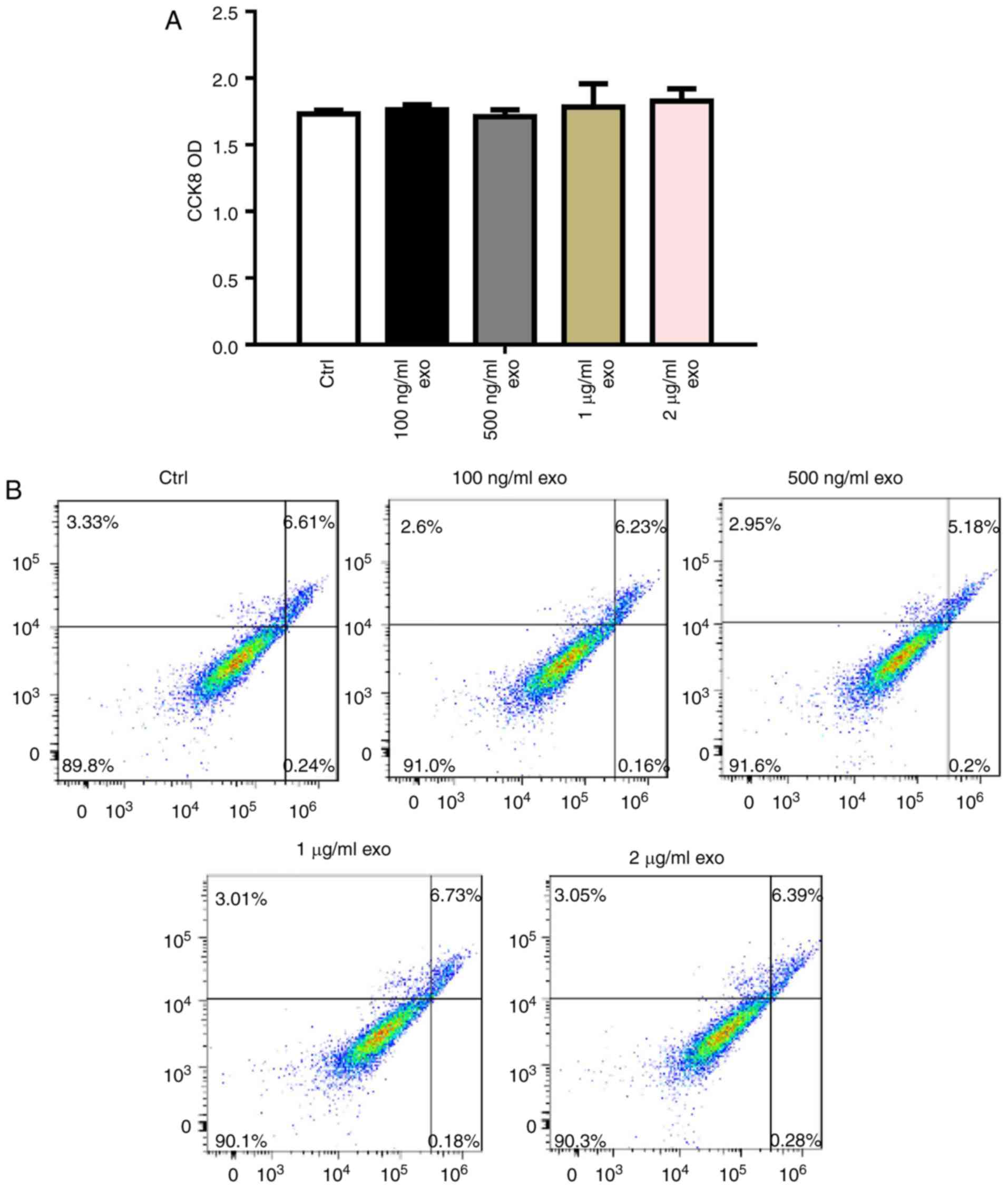
To determine the effects of 3T3L1-exo on 3T3L1 cell
survival, 3T3L1 preadipocyte proliferation and apoptosis were
detected following stimulation by 3T3L1-exo. Exosomes did not
affect 3T3L1 cell proliferation, even with an increased
concentration (Fig. 2A).
Comparably, 3T3L1-exo had no effect on the apoptosis of 3T3L1 cells
(Fig. 2B).
3T3L1-exo mediates 3T3L1 preadipocyte
osteogenic differentiation
Since 3T3L1-exo had no effect on 3T3L1 cell
survival, and because MSC-exo has been reported to mediate
osteogenic differentiation of MSCs, preadipocytes and mature
osteoblasts (11), the present
study determined whether 3T3L1-exo promoted the osteogenic
differentiation of 3T3L1 cells (1). 3T3L1 cells were stimulated by
3T3L1-exo in ODM; subsequently, the cells were stained with ARS and
the expression levels of pro-osteogenic genes were analyzed by
RT-qPCR. As shown in Fig. 3A and
B, osteogenic differentiation was enhanced in 3T3L1 cells
stimulated by 3T3L1-exo in ODM. Furthermore, the expression levels
of pro-osteogenic genes, including alkaline phosphatase,
osteocalcin, bone sialoprotein, osterix, collagen-type I and
runt-related transcription factor 2 (RUNX2), were all increased in
3T3L1 cells exposed to ODM and 3T3L1-exo.
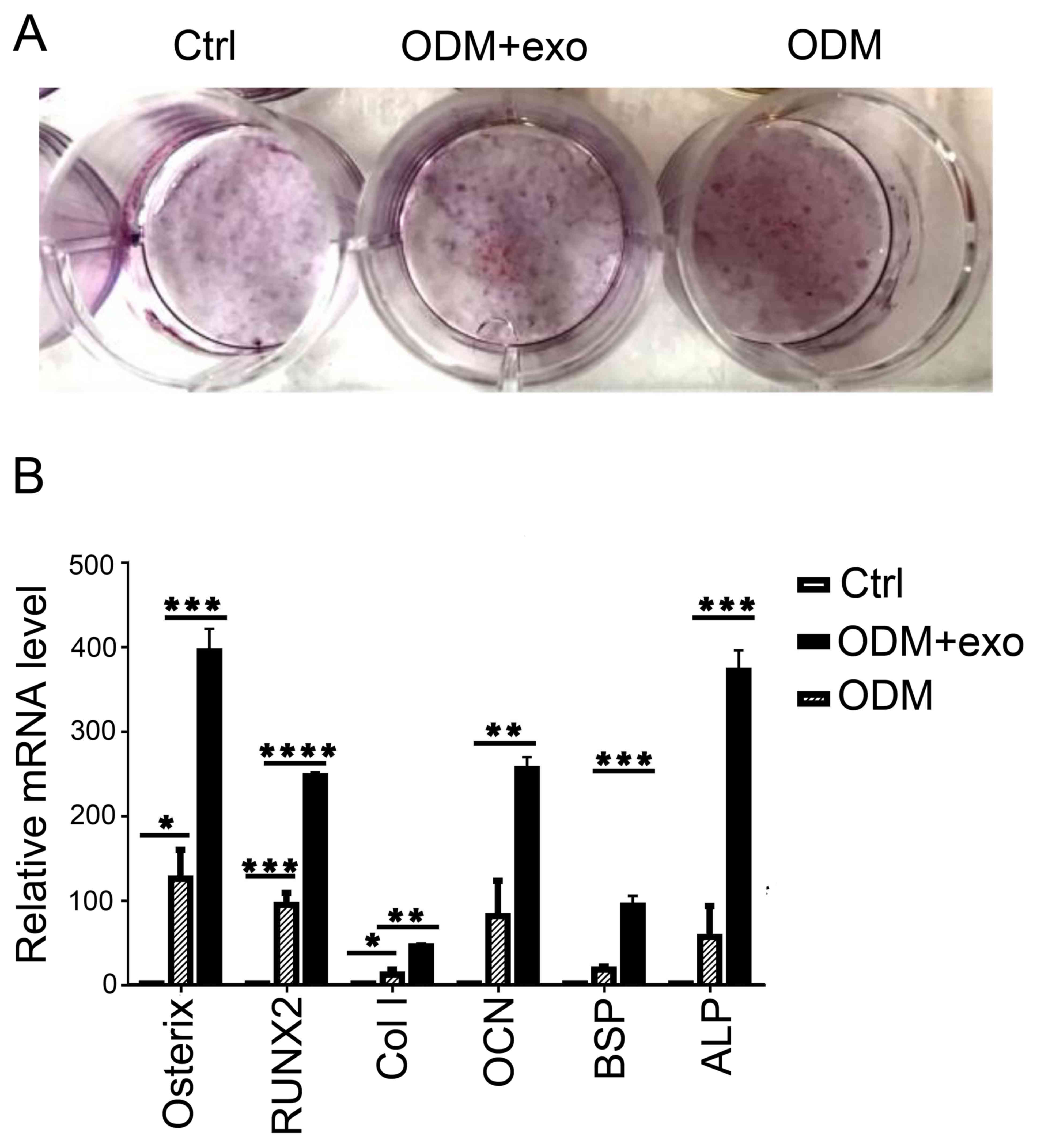 | Figure 3.3T3L1-exo mediates 3T3L1 preadipocyte
osteogenic differentiation. (A) ARS staining was conducted to
determine the effects of 3T3L1-exo on 3T3L1 preadipocyte osteogenic
differentiation. (B) Reverse transcription-quantitative polymerase
chain reaction was performed to determine the effects of 3T3L1-exo
on the expression of osteogenic differentiation-associated genes.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. (ODM + exo
vs. ODM: osterix, P=0.0007; RUNX2, P<0.0001; Col I, P=0.0022;
OCN, P=0.007; BSP, P=0.0008; ALP, P=0.0003, ctrl vs. ODM: osterix,
P=0.0298; RUNX2, P=0.0006; Col I, P=0.024; OCN, P=0.05; BSP,
P=0.2927; ALP, P=0.3391). ALP, alkaline phosphatase; BSP, bone
sialoprotein; Col I, collagen-type I; Ctrl, control; exo/3T3L1-exo,
3T3L1 cell derived-exosomes; OCN, osteocalcin; ODM, osteogenic
differentiation medium; RNX2, runt-related transcription factor
2. |
3T3L1-exo activates 3T3L1
preadipocytes to undergo osteogenic differentiation via TGF-β
signaling
The present findings suggested that 3T3L1-exo
promoted the osteogenic differentiation of 3T3L1 cells. RUNX2
activates and regulates osteogenesis and acts as a target gene for
numerous signaling pathways, including TGF-β1, bone morphogenetic
protein, Wnt, Hedgehog and Nel-like protein type-1 (5). The present study demonstrated that
exposure of cells to 3T3L1-exo induced Smad3 phosphorylation
(Fig. 4A). Subsequently, the mRNA
expression levels of RUNX2 were detected, and it was revealed that
there was no significant difference between the ODM + exo and ODM
groups when TGF-β was inhibited through a Smad3 inhibitor
(SB431542; Beyotime Institute of Biotechnology, Shanghai, China)
(Fig. 4B). These results indicated
that 3T3L1-exo may promote 3T3L1 cell osteogenic differentiation
via the TGF-β1 pathway.
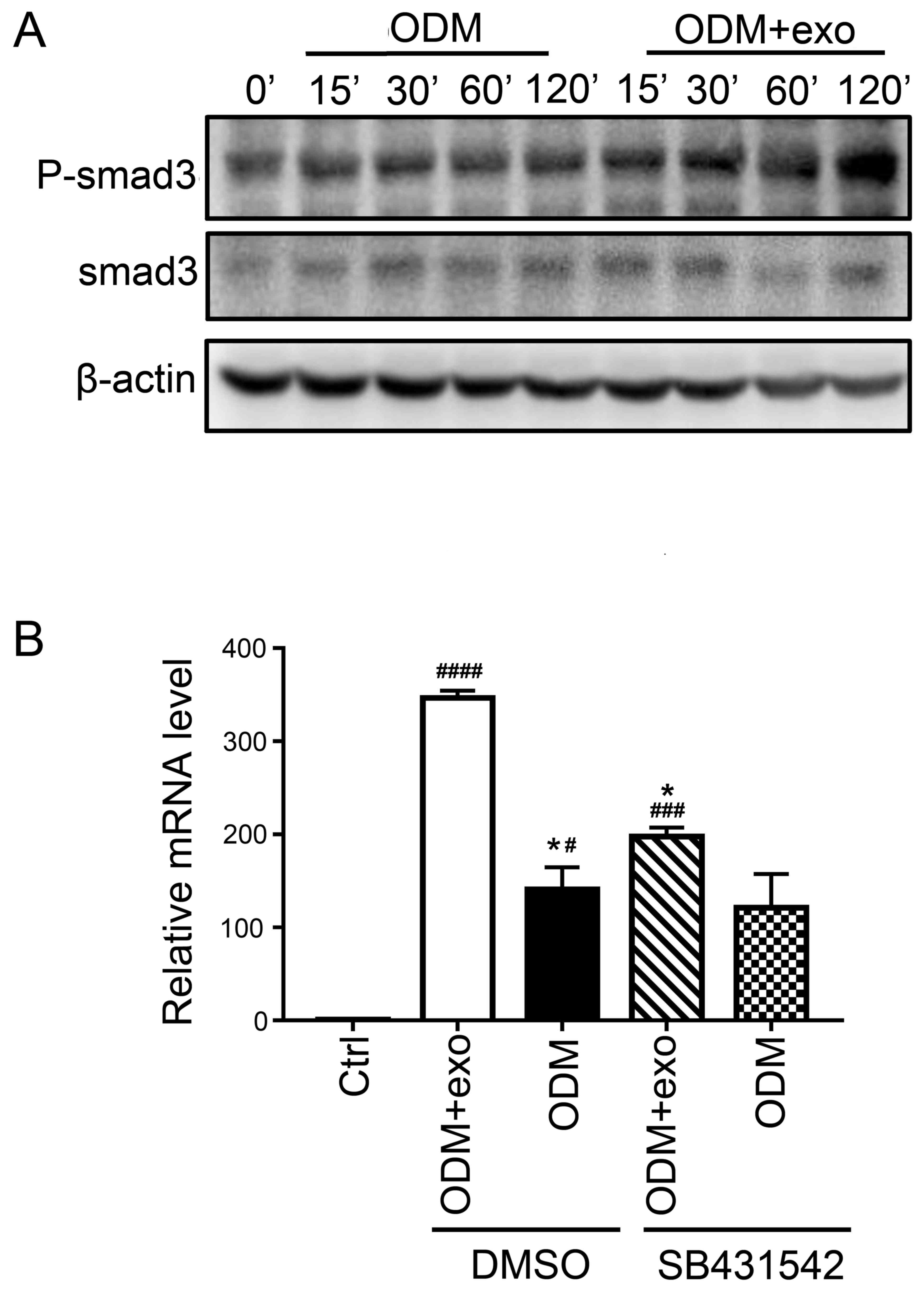 | Figure 4.3T3L1-exo activates 3T3L1
preadipocytes to undergo osteogenic differentiation via TGF-β
signaling. (A) Western blot analysis detected p-smad3 and smad3
expression in 3T3L1 cells following stimulation with 3T3L1-exo for
the indicated durations. (B) Reverse transcription-quantitative
polymerase chain reaction was conducted to determine the mRNA
expression levels of runt-related transcription factor 2 in 3T3L1
cells stimulated with ODM and/or 3T3L1 exo following treatment with
the transforming growth factor-β inhibitor (SB431542). *P<0.05
vs. DMSO+ODM+exo; #P<0.05, ###P<0.001,
####P<0.0001 vs. control. (ODM + exo vs. ODM: DMSO,
P=0.0204; SB431542, P=0.2204; Ctrl vs. DMSO+ODM+exo, P<0.0001;
Ctrl vs. DMSO+ODM, P=0.0285; Ctrl vs. SB431542+ODM+exo, P=0.0009;
Ctrl vs. SB431542+ODM, P=0.0883; DMSO+ODM+exo vs. SB431542+ODM+exo,
P=0.0133; DMSO+ODM vs. SB431542+ODM, P=0.9662). Ctrl, control;
DMSO, dimethyl sulfoxide; exo/3T3L1-exo, 3T3L1 cell
derived-exosomes; ODM, osteogenic differentiation medium; P,
phosphorylated; Smad3, SMAD family member 3. |
miR-223 in 3T3L1-exo may be involved
in enhanced osteogenic differentiation of 3T3L1 preadipocytes
To explore the mechanism by which 3T3L1-exo promoted
3T3L1 osteogenic differentiation, 3T3L1 cells and 3T3L1-exo were
sequenced to determine miRNA profiles, and a similarly wide
distribution profile of read lengths was confirmed (Fig. 5A), with predominant peaks at 20–24
nucleotides and 17–20 nucleotides, respectively. Since miRNAs are
critical regulators of signaling pathways, miRNA expression
patterns were compared between exosomes and cells, and it was
revealed that 427 miRNAs were upregulated and 573 were
downregulated in the exosomes compared with in the cells (Fig. 5B). To confirm the differences and
relationships of miRNAs derived from cells and exosomes, cluster
analysis using pheatmap in R software was performed (Fig. 5C).
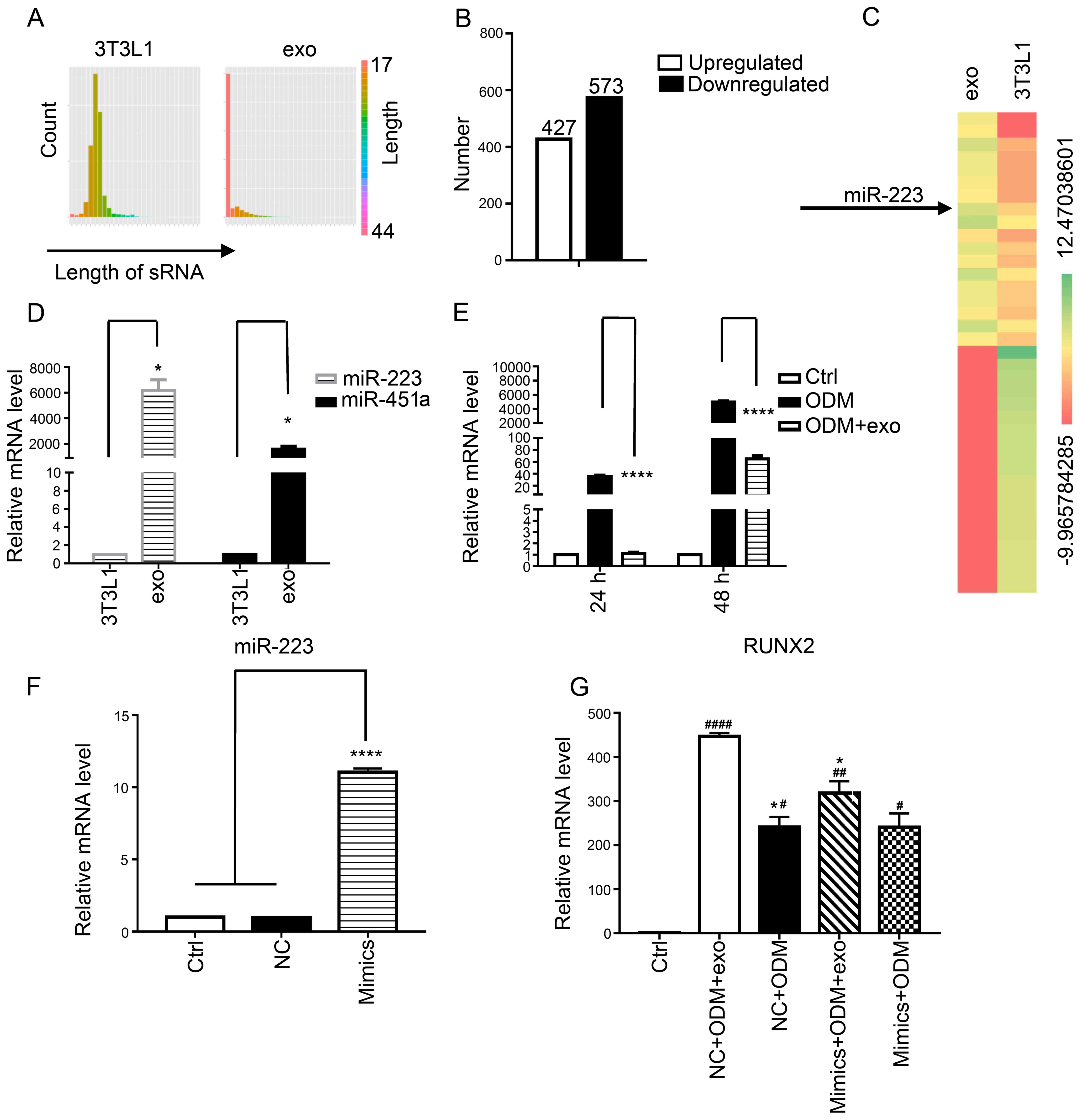 | Figure 5.3T3L1 cells and their exosomes differ
in miRNA composition. (A) Distribution of the length of miRNA reads
in 3T3L1 cells and 3T3L1-exo. (B) Differentially expressed miRNAs
(log2 ratio: 3T3L1-exo/3T3L1 cells). (C) Heat maps indicating the
expression of the top 20 upregulated (left) and downregulated
(right) miRNAs in 3T3L1-exo compared with in 3T3L1 cells. (D)
RT-qPCR analysis revealed differences in miR-223 and miR-451a
expression levels in 3T3L1 cells and 3T3L1-exo *P<0.05 vs. 3T3L1
cells. (3T3L1 cells vs. 3T3L1 exo: miR-223, P=0.0185; miR-451a,
P=0.0202). (E) RT-qPCR analysis was used to detect the expression
levels of miR-223 in 3T3L1 preadipocytes stimulated with ODM and/or
3T3L1-exo for 48 and 72 h. ****P<0.0001 vs. ODM. (ODM + exo vs.
ODM: 48 h, P<0.0001; 72 h, P<0.0001). (F) RT-qPCR was used to
test the efficacy of miR-223 mimic transfection (mimics vs. ctrl:
P<0.0001; mimics vs. NC: P<0.0001). (G) RT-qPCR was used to
detect the mRNA expression levels of runt-related transcription
factor 2 in 3T3L1 cells stimulated with ODM and/or 3T3L1 exo
post-transfection with miR-223 mimics. *P<0.05 vs. NC+ODM+exo;
#P<0.05, ##P<0.01,
####P<0.0001 vs. control. (ODM+exo vs. ODM: NC,
P=0.0200; mimics, P=0.1578; NC+ODM+exo vs. mimics+ODM+exo,
P=0.0322; NC+ODM vs. mimics+ODM, P>0.9999; Ctrl vs. NC+ODM+exo,
P<0.0001; Ctrl vs. NC+ODM, P=0.01; Ctrl vs. mimics+ODM+exo,
P=0.0079; ctrl vs mimics+ODM, P=0.0173). Ctrl, control;
exo/3T3L1-exo, 3T3L1 cell derived-exosomes; miR/miRNA, microRNA;
NC, negative control; ODM, osteogenic differentiation medium; sRNA,
small RNA. |
In the present study, miR-223 was highlighted as a
potential candidate target; this miRNA is a key regulatory factor
in osteoclast and osteoblast differentiation (20). Therefore, RT-qPCR was performed to
confirm the expression of miR-223 in the 3T3L1-exo and 3T3L1 cells;
miR-223 and miR-451a was revealed to be upregulated in 3T3L1-exo
compared with in 3T3L1 cells (Fig.
5D). Furthermore, miR-223 expression was detected in 3T3L1
cells stimulated with ODM and/or 3T3L1-exo for 48 and 72 h.
Notably, miR-223 was downregulated in ODM + 3T3L1-exo stimulated
cells compared with in cells stimulated with ODM alone (Fig. 5E). It was therefore hypothesized
that 3T3L1-exo may suppress miR-223 expression in 3T3L1
preadipocytes through a competitive mechanism or via another miRNA,
and that other factors may regulate the decreased levels of
miR-223. The results of a miR-223 RNA interference experiment
(Fig. 5F) demonstrated that
miR-223 mimics weakened the increased expression of RUNX2 in 3T3L1
cells exposed to ODM and 3T3L1-exo (Fig. 5G). These results suggested that
miR-223 in 3T3L1-exo may be involved in enhanced osteogenic
differentiation of 3T3L1 preadipocytes.
Discussion
Clinical bone implants often require bone fillers or
enhanced regeneration due to a shortage of bone. In dentistry, a
significant proportion of patients that need implants require
increased bone volume prior to implant placement. The clinical gold
standard for bone grafting is an autograft; however, this method
has limitations, including donor-site morbidity, limited
availability of grafting material and compromised bone quality in
patients with osteoporosis (4).
Aside from autografts, guided bone regeneration using a specially
selected bovine source is the most straightforward procedure for
bone transplantation; however, this method can result in rejection
and insufficient osteogenesis (3).
Bone regeneration requires the migration of specific
cells to the healing site to proliferate there and to provide a
biological substrate for new tissue growth. BMSCs have the ability
to form bone; therefore, bone marrow transplantation is used
clinically in combination with osteoconductive materials to augment
bone healing (21). However, BMSCs
are limited with regards to insufficiency in numbers. Research has
indicated that ADSCs are a more suitable tissue source (1), since they are also capable of
undergoing osteogenic differentiation but are more readily
obtained.
Because the numbers of stem cells are limited,
agents that promote their differentiation are required. To promote
bone formation, specific growth factors are often applied. However,
research has suggested that the secreted trophic factors are more
important than the process of stem cell differentiation in
mediating therapeutic efficacy. The exosome, a secreted membrane
vesicle, is therefore an active therapeutic factor in the process
of MSC secretion (22).
In the present study, 3T3L1-exo was revealed to
promote 3T3L1 preadipocyte osteogenic differentiation via the TGF-β
pathway. Notably, the TGF-β pathway can activate RUNX2 and further
induce the osteogenic differentiation of cells (5). Furthermore, miRNA sequences were
detected in 3T3L1 cells and 3T3L1-exo, and it was revealed that, in
some cases, miRNAs were comparable between cells and exosomes;
however, in other cases, miRNAs were expressed at a higher level in
exosomes.
MSCs can promote TGF-β expression in murine renal
tubular epithelial cells via miR-223 (23). Furthermore, bone marrow-derived
miR-223 has an effect on vascular endothelial cells as an endocrine
genetic signal, and is involved in vascular injury by targeting
insulin-like growth factor 1 receptor (24). In addition, Notch/miR-223 has been
reported to modify the osteogenic potential of bone marrow stromal
cells (25). Consequently, miR-223
may serve a regulatory role in 3T3L1-exo by enhancing the
osteogenic differentiation of 3T3L1 cells.
The present study confirmed that the expression of
miR-223 was increased in 3T3L1-exo compared with in 3T3L1 cells.
Notably, the expression levels of miR-223 were decreased in 3T3L1
preadipocytes cultured in ODM and stimulated by 3T3L1-exo compared
with in cells without exosome stimulation. It may be hypothesized
that 3T3L1-exo suppresses the expression of miR-223 in 3T3L1
preadipocytes through a competitive mechanism, or by another miRNA,
or a factor regulated by decreased miR-223. However, these
competitive mechanisms or other regulated mechanisms require
further investigation In conclusion, the application of 3T3L1-exo
may be useful for investigating preadipocyte-induced bone
regeneration.
Acknowledgments
The authors would like to thank Dr Elizabeth Finnie
for editing the English text of a draft of this manuscript.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no.
81700972).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW conceived and designed the study; WD, LS and NZ
performed the experiments; and WD and LS completed the draft. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bressan E, Botticelli D, Sivolella S,
Bengazi F, Guazzo R, Sbricoli L, Ricci S, Ferroni L, Gardin C,
Velez JU and Zavan B: Adipose-derived stem cells as a tool for
dental implant osseointegration: An experimental study in the dog.
Int J Mol Cell Med. 4:197–208. 2015.PubMed/NCBI
|
|
2
|
Padial-Molina M, O'Valle F, Lanis A, Mesa
F, Dohan Ehrenfest DM, Wang HL and Galindo-Moreno P: Clinical
application of mesenchymal stem cells and novel supportive
therapies for oral bone regeneration. Biomed Res Int.
2015:3413272015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grayson WL, Bunnell BA, Martin E, Frazier
T, Hung BP and Gimble JM: Stromal cells and stem cells in clinical
bone regeneration. Nat Rev Endocrinol. 11:140–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
James AW: Review of signaling pathways
governing MSC osteogenic and adipogenic differentiation.
Scientifica (Cairo). 2013:6847362013.PubMed/NCBI
|
|
6
|
Qin Y, Sun R, Wu C, Wang L and Zhang C:
Exosome: A novel approach to stimulate bone regeneration through
regulation of osteogenesis and angiogenesis. Int J Mol Sci.
17:E7122016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baglio SR, Rooijers K, Koppers-Lalic D,
Verweij FJ, Pérez Lanzón M, Zini N, Naaijkens B, Perut F, Niessen
HW, Baldini N and Pegtel DM: Human bone marrow- and
adipose-mesenchymal stem cells secrete exosomes enriched in
distinctive miRNA and tRNA species. Stem Cell Res Ther. 6:1272015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gernapudi R, Yao Y, Zhang Y, Wolfson B,
Roy S, Duru N, Eades G, Yang P and Zhou Q: Targeting exosomes from
preadipocytes inhibits preadipocyte to cancer stem cell signaling
in early-stage breast cancer. Breast Cancer Res Treat. 150:685–695.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fleury A, Martinez MC and Le Lay S:
Extracellular vesicles as therapeutic tools in cardiovascular
diseases. Front Immunol. 5:3702014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu B, Zhang X and Li X: Exosomes derived
from mesenchymal stem cells. Int J Mol Sci. 15:4142–4157. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Narayanan R, Huang CC and Ravindran S:
Hijacking the cellular mail: Exosome mediated differentiation of
mesenchymal stem cells. Stem Cells Int. 2016:38086742016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui Y, Luan J, Li H, Zhou X and Han J:
Exosomes derived from mineralizing osteoblasts promote ST2 cell
osteogenic differentiation by alteration of microRNA expression.
FEBS Lett. 590:185–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Ding L, Zheng XL, Wang HX and Yan
HM: DC-derived exosomes induce osteogenic differentiation of
mesenchymal stem cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
22:600–604. 2014.(In Chinese). PubMed/NCBI
|
|
14
|
Ekström K, Omar O, Granéli C, Wang X,
Vazirisani F and Thomsen P: Monocyte exosomes stimulate the
osteogenic gene expression of mesenchymal stem cells. PLoS One.
8:e752272013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y,
Teh BJ and Lim SK: Mesenchymal stem cell: An efficient mass
producer of exosomes for drug delivery. Adv Drug Deliv Rev.
65:336–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lopatina T, Bruno S, Tetta C, Kalinina N,
Porta M and Camussi G: Platelet-derived growth factor regulates the
secretion of extracellular vesicles by adipose mesenchymal stem
cells and enhances their angiogenic potential. Cell Commun Signal.
12:262014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cai Z, Yang F, Yu L, Yu Z, Jiang L, Wang
Q, Yang Y, Wang L, Cao X and Wang J: Activated T cell exosomes
promote tumor invasion via Fas signaling pathway. J Immunol.
188:5954–5961. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan DC, Shi YY, Nacamuli RP, Quarto N,
Lyons KM and Longaker MT: Osteogenic differentiation of mouse
adipose-derived adult stromal cells requires retinoic acid and bone
morphogenetic protein receptor type IB signaling. Proc Natl Acad
Sci USA. 103:12335–12340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guan X, Gao Y, Zhou J, Wang J, Zheng F,
Guo F, Chang A, Li X and Wang B: miR-223 regulates adipogenic and
osteogenic differentiation of mesenchymal stem cells through a
C/EBPs/miR-223/FGFR2 regulatory feedback loop. Stem Cells.
33:1589–1600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marolt D, Knezevic M and Novakovic GV:
Bone tissue engineering with human stem cells. Stem Cell Res Ther.
1:102010. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lai RC, Yeo RW, Tan SS, Zhang B, Yin Y,
Kwan Sze NS, Choo A and Lim SK: Mesenchymal stem cell exosomes: The
future MSC-based therapy? Mesench Stem Cell Ther. 39–61. 2012.
|
|
23
|
Yuan X, Wang X, Chen C, Zhou J and Han M:
Bone mesenchymal stem cells ameliorate ischemia/reperfusion-induced
damage in renal epithelial cells via microRNA-223. Stem Cell Res
Ther. 8:1462017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chu M, Wu R, Qin S, Hua W, Shan Z, Rong X,
Zeng J, Hong L, Sun Y, Liu Y, et al: Bone marrow-derived
MicroRNA-223 works as an endocrine genetic signal in vascular
endothelial cells and participates in vascular injury from kawasaki
disease. J Am Heart Assoc. 6:e0048782017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berenstein R, Nogai A, Waechter M, Blau O,
Kuehnel A, Schmidt-Hieber M, Kunitz A, Pezzutto A, Dörken B and
Blau IW: Multiple myeloma cells modify VEGF/IL-6 levels and
osteogenic potential of bone marrow stromal cells via
Notch/miR-223. Mol Carcinog. 55:1927–1939. 2016. View Article : Google Scholar : PubMed/NCBI
|