Introduction
Uveal melanoma (UM), a rare subset of melanoma
accounting for only 5% of all melanomas, makes up 85–95% of all
ocular melanomas; however, UM ranks as the predominant intraocular
malignancy in adult patients (1–4).
Tumors located on the choroid are the most common, affecting ≤90%
of patients (5–7). In addition, ~50% of patients will
develop metastasis, >90% of which disseminates to the liver
(8,9). Once the tumor has metastasized, the
median overall survival (OS) is only 6–12 months (10–12).
The average age at diagnosis is 60-years-old, and UM affects both
sexes almost equally, with a slightly increased frequency in males
(13,14). Recent advances in the detection of
molecular pathology and a various immune-based therapies have
exhibited efficacy in treating UM. However, the molecular
biomarkers that may be effective in predicting the development and
prognosis of UM in patients are limited, which has led to the
unsatisfactory clinical management and targeted therapy of this
disease (15–17); thus, UM is associated with high
mortality. Therefore, there is an urgent need to identify ideal
prognostic biomarkers and treatment options for patients with
UM.
Recently, increasing evidence has indicated that
non-coding RNAs serve a vital regulatory role in tumorigenesis and
tumor progression, including small nucleolar RNAs (snoRNAs)
(18,19). snoRNAs, encoded by introns, exist
stably in the cellular environment and can regulate the expression
of specific genes (20–23). Numerous studies have proposed that
snoRNA expression profiles are dysregulated in various types of
tumors (24,25). Furthermore, numerous dysregulated
snoRNAs have also been identified to be associated with the
development and prognosis of cancer, including melanoma, which
suggests the potential of snoRNAs as prognostic predictors
(26–28). Unfortunately, the association
between snoRNAs and UM has rarely been reported.
In the present study, the expression landscape of
snoRNAs was integrated using the snoRNA in cancers (SNORic)
database and the clinical information of patients with UM in The
Cancer Genome Atlas (TCGA) database. snoRNas associated with
survival were systematically selected and a specific prognosis
index (PI) was proposed, which could be an ideal prognostic
signature to predict the prognosis of patients with UM. Further
bioinformatics analysis revealed survival-associated pathways in
UM. These findings may aid the identification of the novel
biological functions of snoRNAs in UM, and improve the clinical
management of this disease.
Materials and methods
General characteristics
The data of 80 patients with UM were submitted for
survival analysis. Among the 80 patients with UM, the histological
types included epithelioid cell (n=34) and spindle cell (n=46). A
total of 35 patients were female and 45 patients were male. The
average age was 61.65 years (ranging from 22–86 years).
Data acquisition
SnoRNA expression profiles were downloaded from the
SNORic (http://bioinfo.life.hust.edu.cn/SNORic) database,
which provides the quantified expression levels of snoRNAs in the
form of reads per kilobase per million (RPKM). Then, the original
values were calculated via log2 conversion. The corresponding
clinical information of UM patients was also downloaded from the
TCGA data portal (http://cancergenome.nih.gov/). To obtain more accurate
results, low-abundance snoRNAs with average log (RPKM+1) expression
levels in all samples were omitted.
Survival analysis
A univariate Cox regression analysis was performed
to select snoRNAs that were correlated with the overall survival
(OS) of patients with UM. Subsequently, multivariate Cox
proportional hazards regression was implemented to further identify
whether the prognostic snoRNAs were independent biomarkers.
Finally, the snoRNA-based prognostic signature was proposed
according to the linear multiplication of the expression profiles
of each prognostic snoRNA, weighted by their estimated regression
coefficients in the multivariate Cox analysis. All survival
analyses were conducted using the Survival package version 2.43-1
in R software version 3.4.4 (https://CRAN.R-project.org/package=survival). Then,
patients were separated into high- and low-risk groups according to
median risk score values. Kaplan-Meier (K-M) survival analysis
followed by a log-rank test was used to assess the value of the
snoRNAs in predicting prognosis. The number of patients in the low-
and high-risk groups was recorded every 500 days.
Bioinformatics procedure
To further investigate the potential pathways
affected in the high- and low-risk groups, the Limma package
version 3.38.2 in R software was used to identify the
differentially expressed genes between high- and low-risk groups
(29). The mRNA expression
profiles were downloaded from TCGA database; the raw count data of
mRNAs were submitted for analysis. The thresholds for definite
differentially expressed genes were set to |log fold change|>2
and false discovery rate <0.01. Subsequently, differentially
expressed genes were submitted to the DAVID online database for
gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG; http://www.genome.jp/kegg/) analysis.
GO analysis includes three categories: ‘Biological process’ (BP),
‘cellular component’ (CC) and ‘molecular function’ (MF).
Furthermore, a protein-protein interaction (PPI) network was also
generated to demonstrate the association between these genes. PPI
analysis was performed using STRING version 10.5 (https://string-db.org/). The results were visually
presented using the ggplot2 package version 3.1.0 in R software
(30). Providing that
BRCA1-associated protein-1 (BAP1) mutation is a critical indicator
for monitoring the poor progression of UM, the prognostic value of
BAP1 mutation was determined in the present study. Data for UM
patients, including BAP1 mutation status, were downloaded from the
cBioportal database (http://www.cbioportal.org/).
Results
Survival-associated snoRNAs
After removing the low-abundance snoRNAs, 380
snoRNAs were submitted for survival analysis. Among these, 64
snoRNAs were significantly associated with the OS of patients with
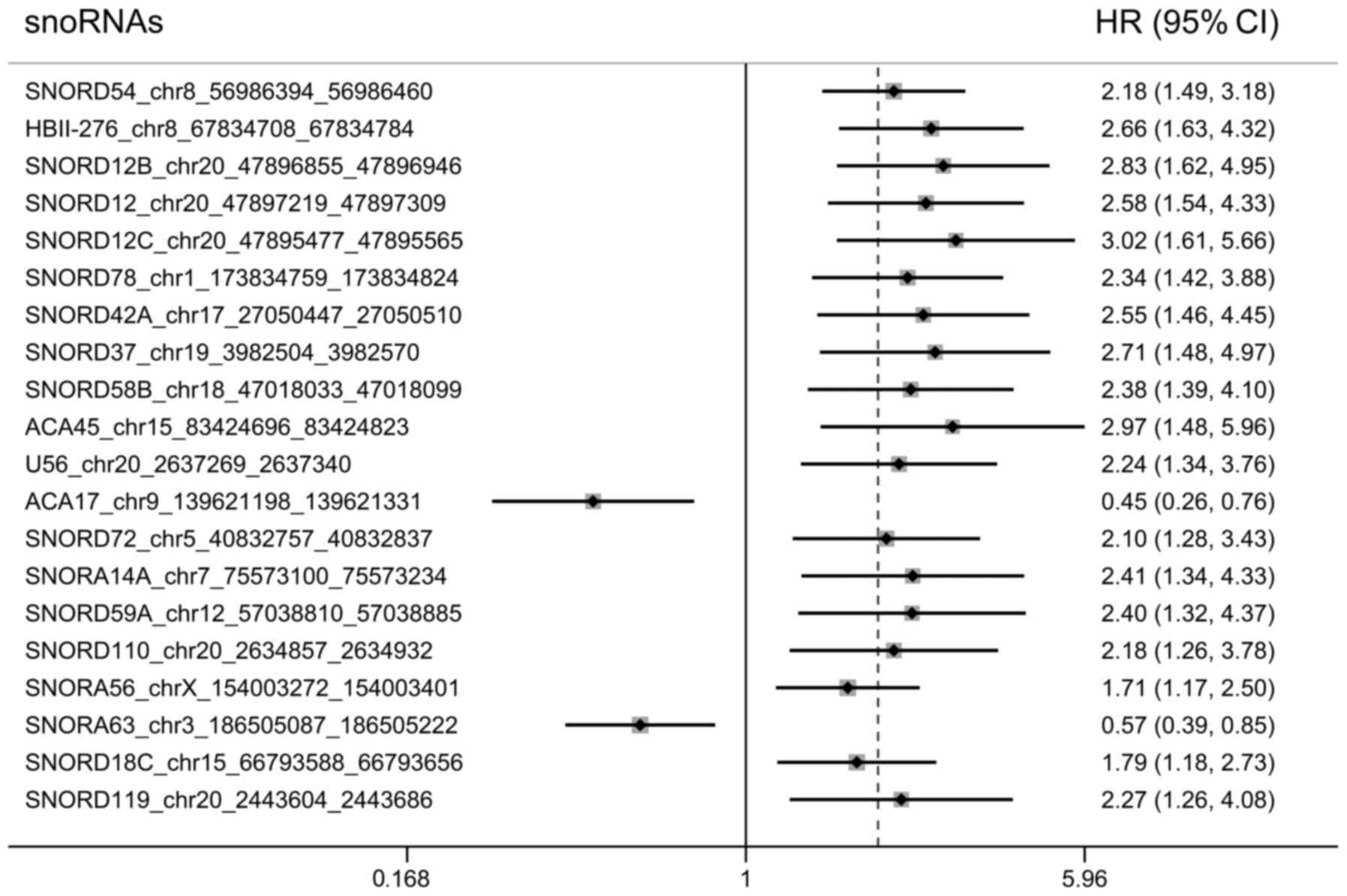
UM (P<0.05). The top 20 most significant survival-associated
snoRNAs were presented in Fig. 1.
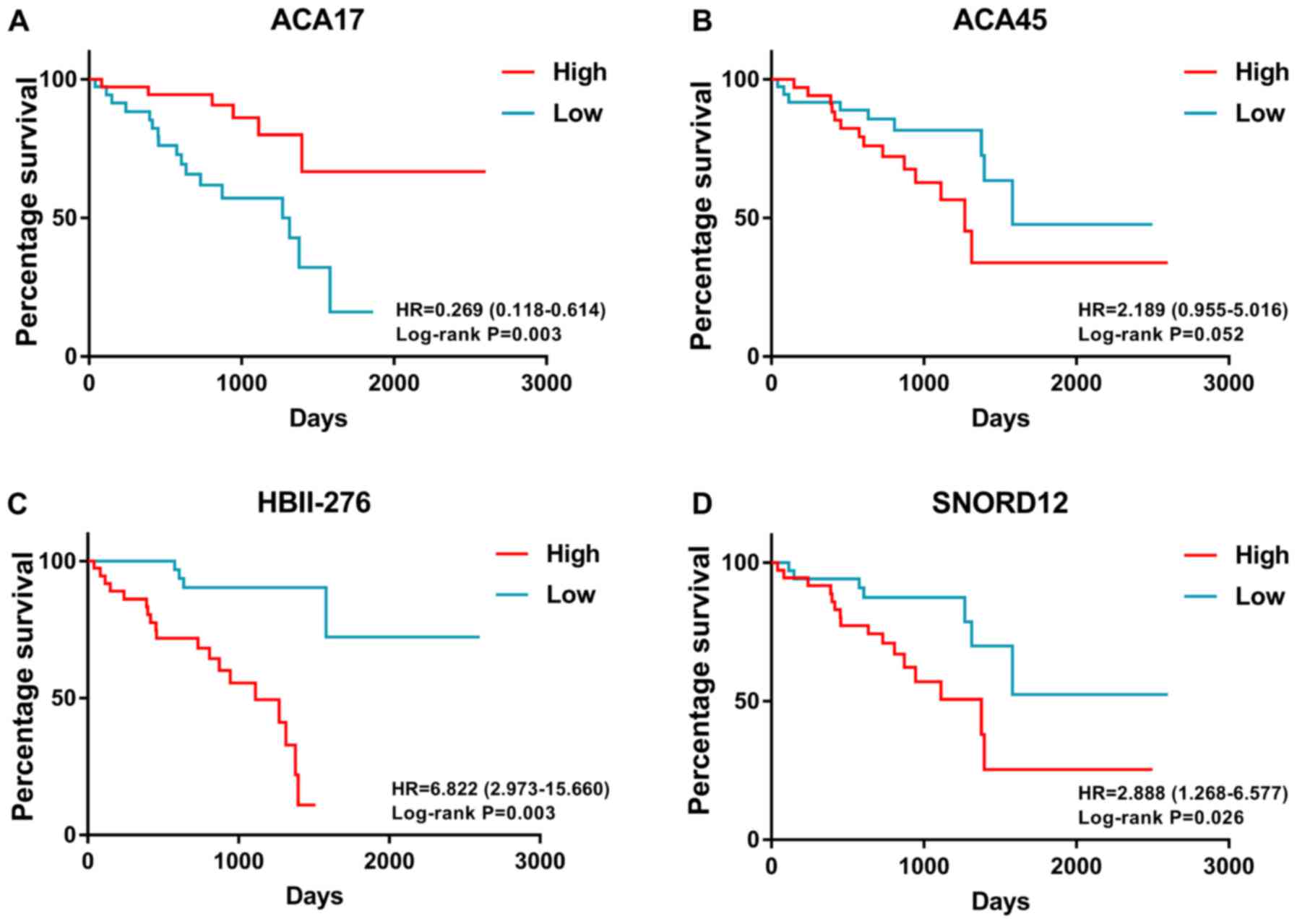
Then, multivariate Cox analysis was performed and four snoRNAs were
selected to construct a prognostic signature, including ACA45
(chr15: 83424696_83424823), ACA17 (chr9: 139621198_139621331),
HBII-276 (chr8: 67834708_67834784) and SNORD12 (chr20:
47897219_47897309). A survival-risk formula was constructed based
on the four snoRNAs as follows: ACA17 * (−1.602) + ACA45 * 0.803 +
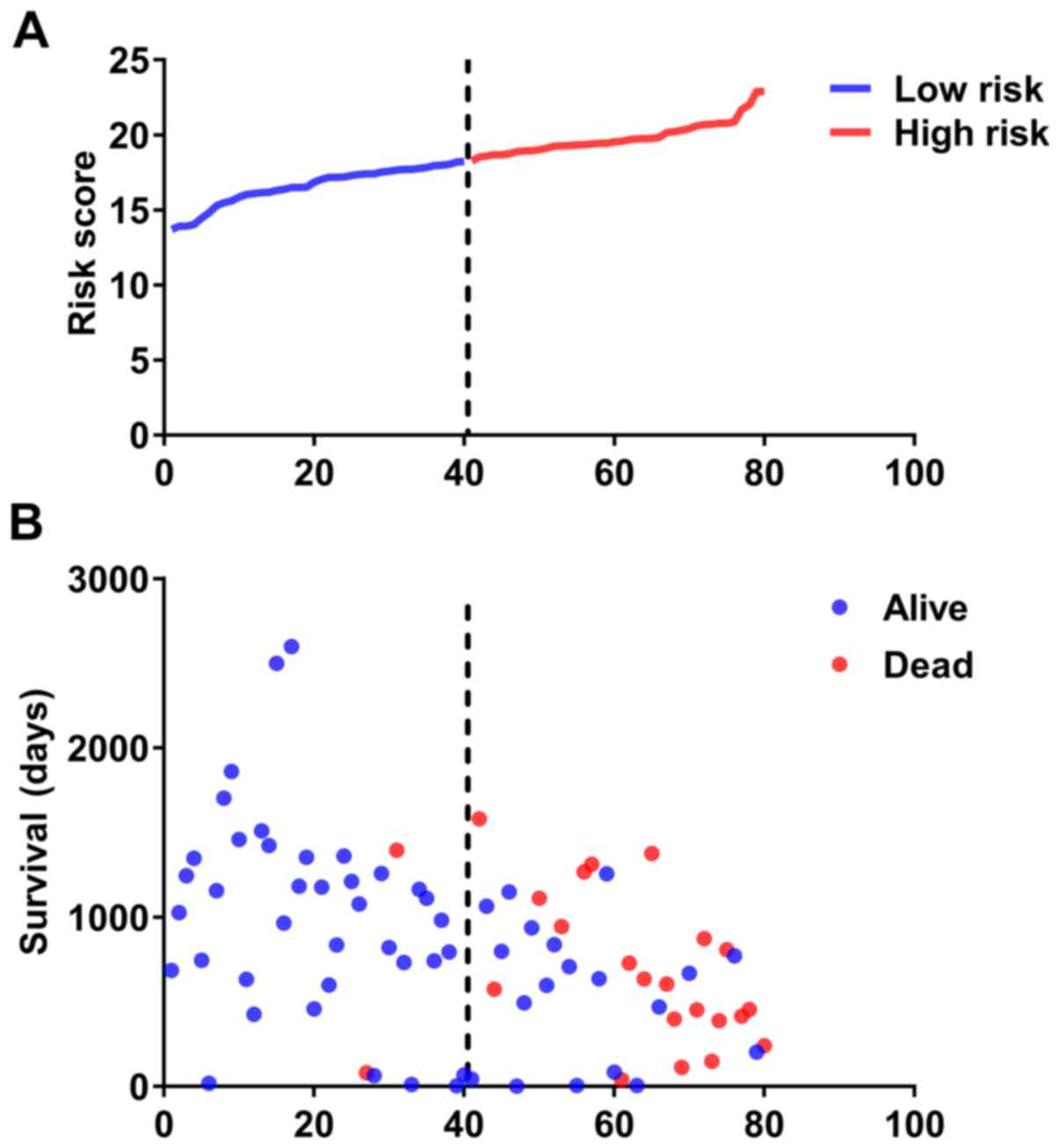
HBII-276 * 0.603 + SNORD12 * 1.348. Then, patients were divided
into high- and low-risk groups according to median value of
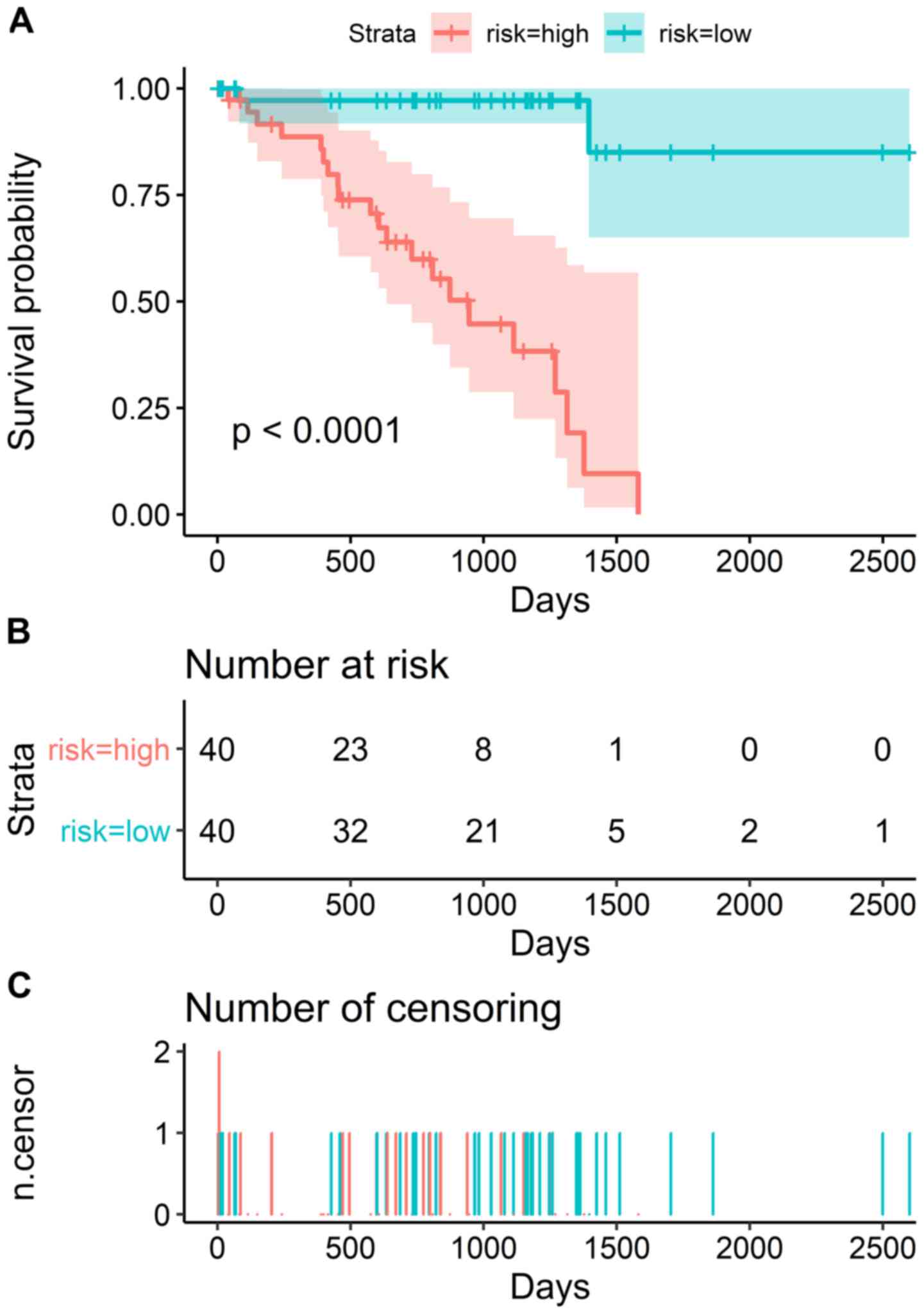
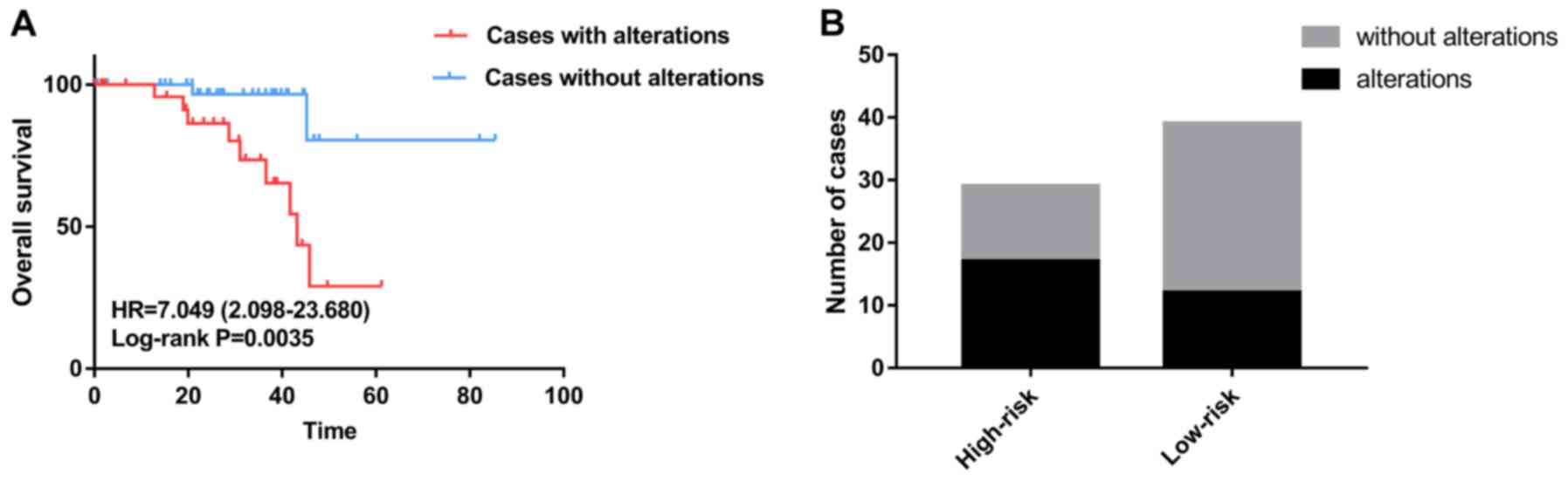
prognostic signature (Fig. 2). K-M
survival analysis revealed that patients in the high-risk group
exhibited shorter survival than those patients in the low-risk
group (Fig. 3A). The number of
patients in the high- and low-risk groups, and the number of
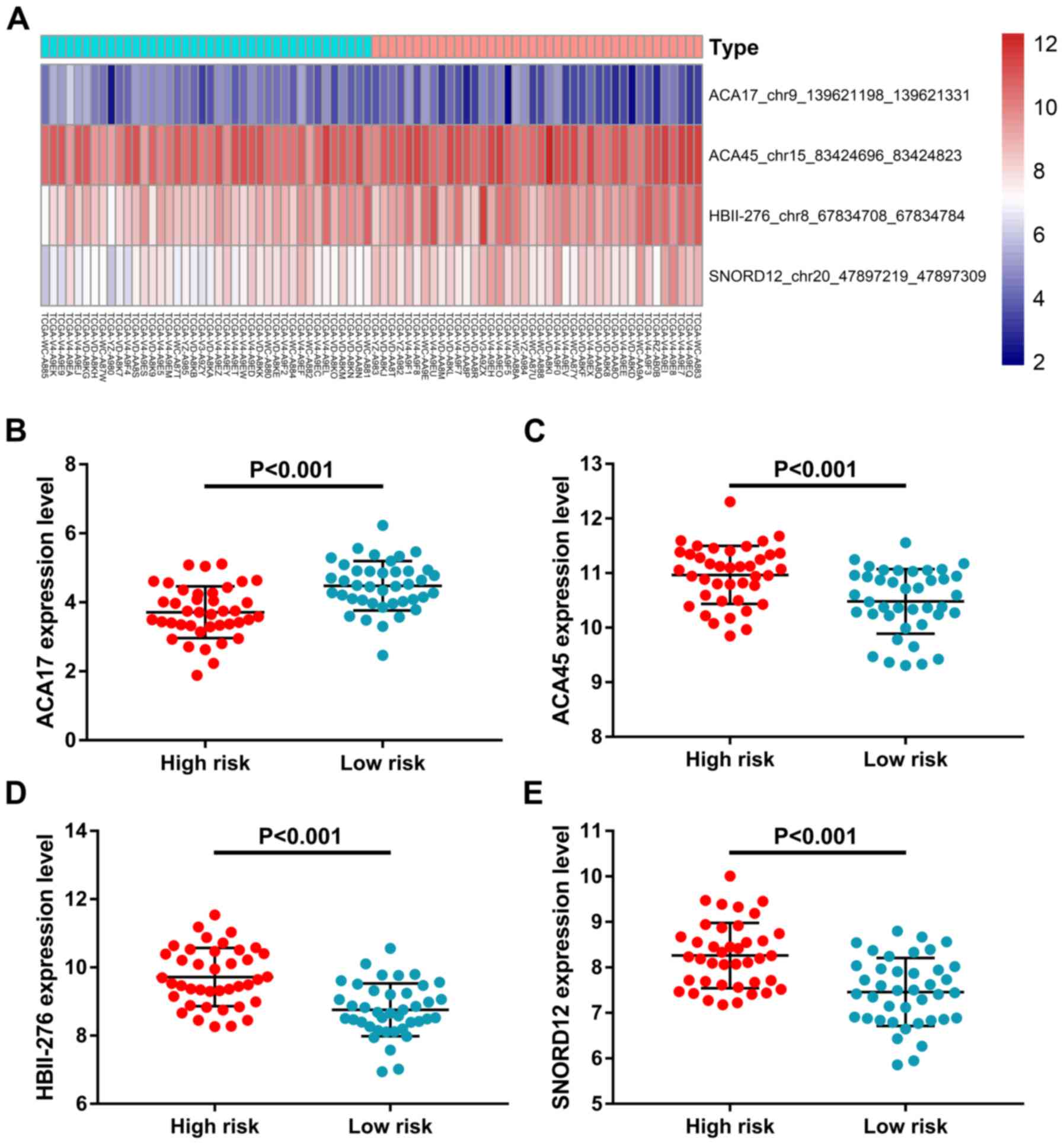
censored patients at different time points are shown in Fig. 3B and C. ACA17 was significantly
upregulated in the low-risk group, while ACA45, HBII-276 and
SNORD12 were significantly upregulated in the high-risk group
(Fig. 4). K-M survival plots were
also used to present the prognostic value of each snoRNA (Fig. 5). ACA17 upregulation indicated
better clinical outcome, while ACA45, HBII-276 and SNORD12
upregulation was associated with the poor survival of patients with
UM.
Functional enrichment analysis
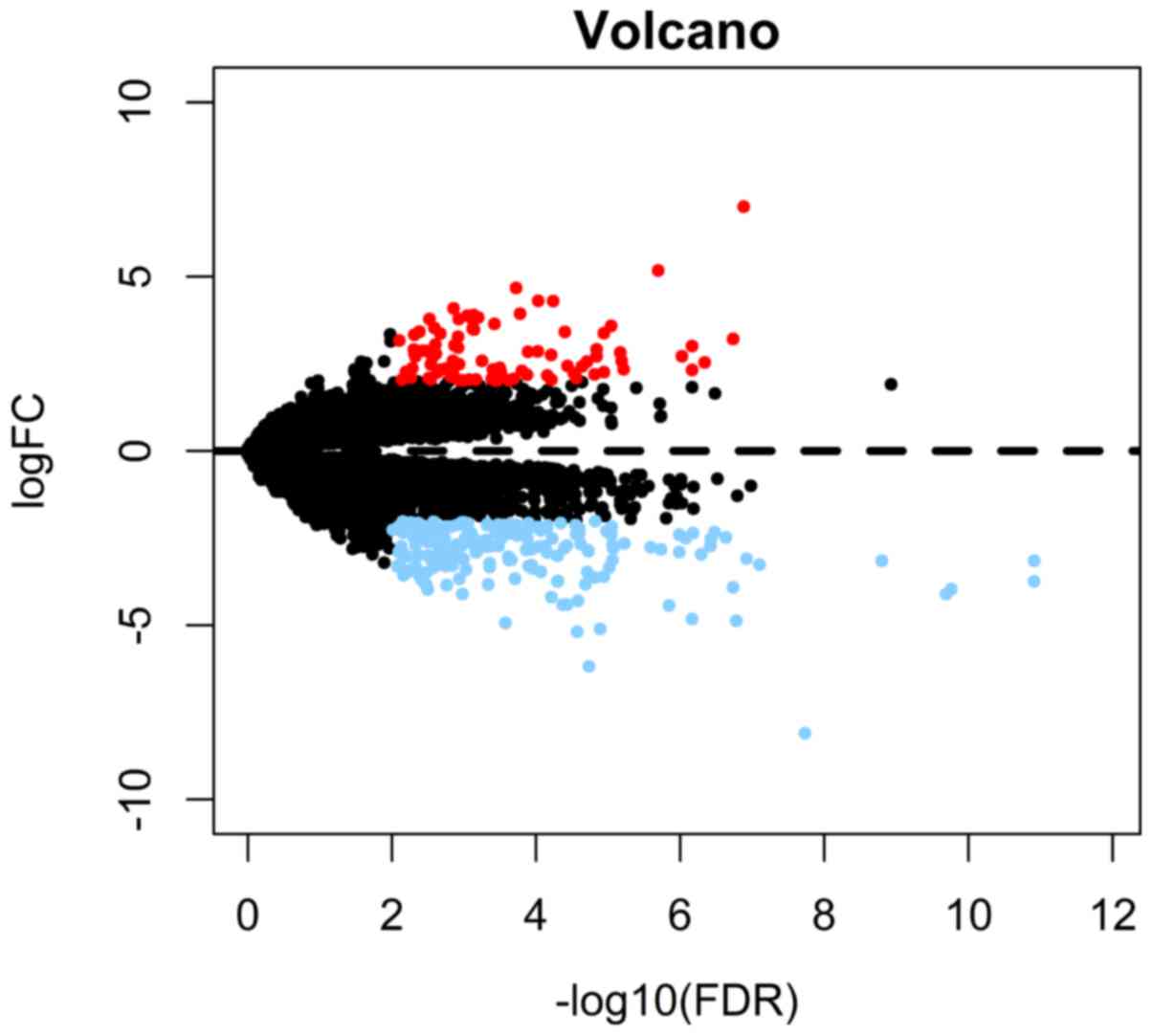
A total of 281 differentially expressed mRNAs
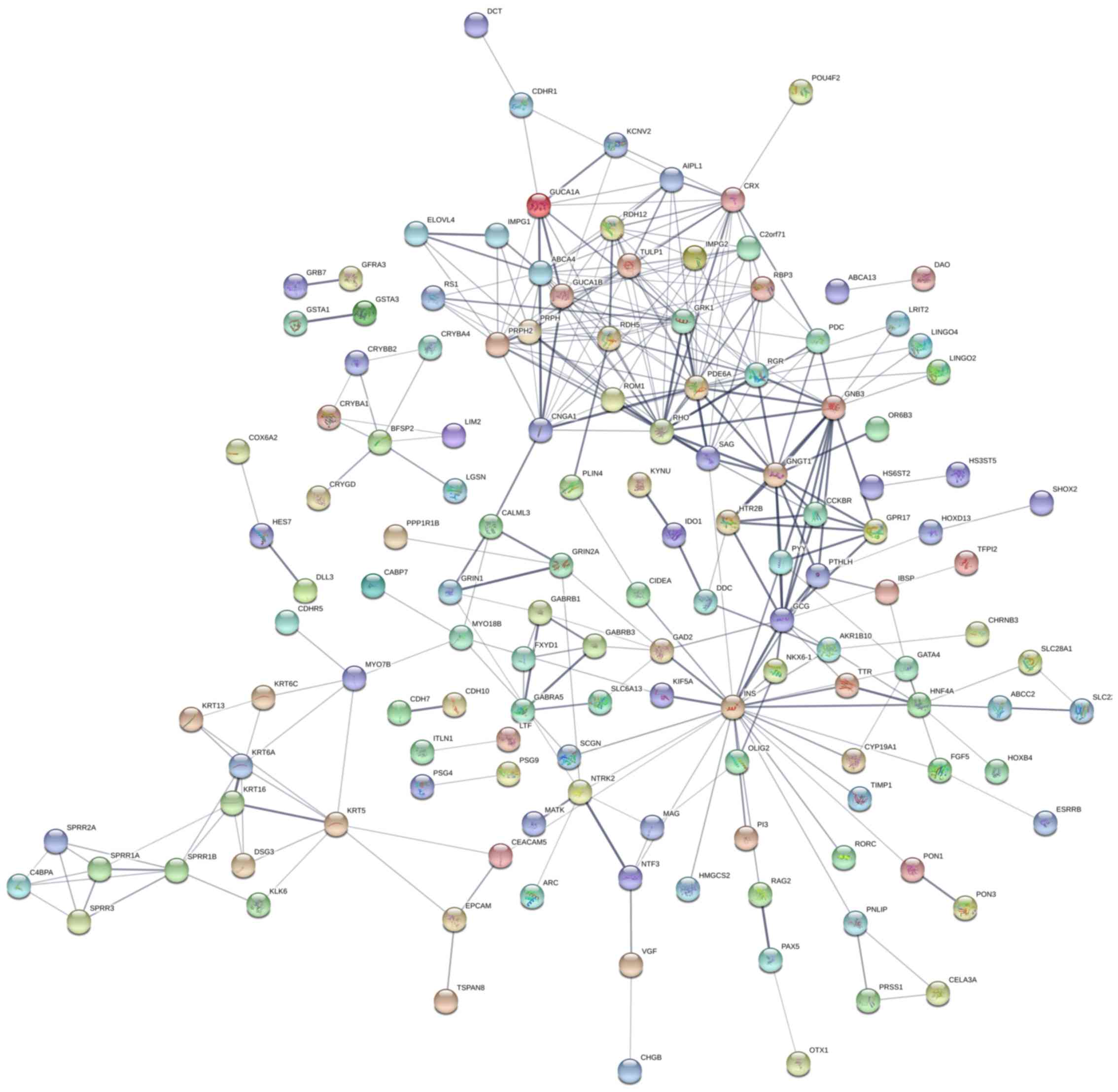
between the high- and low-risk groups were determined (Fig. 6). PPI network analysis suggested
that these genes notably interacted with each other (Fig. 7). Gene functional enrichment
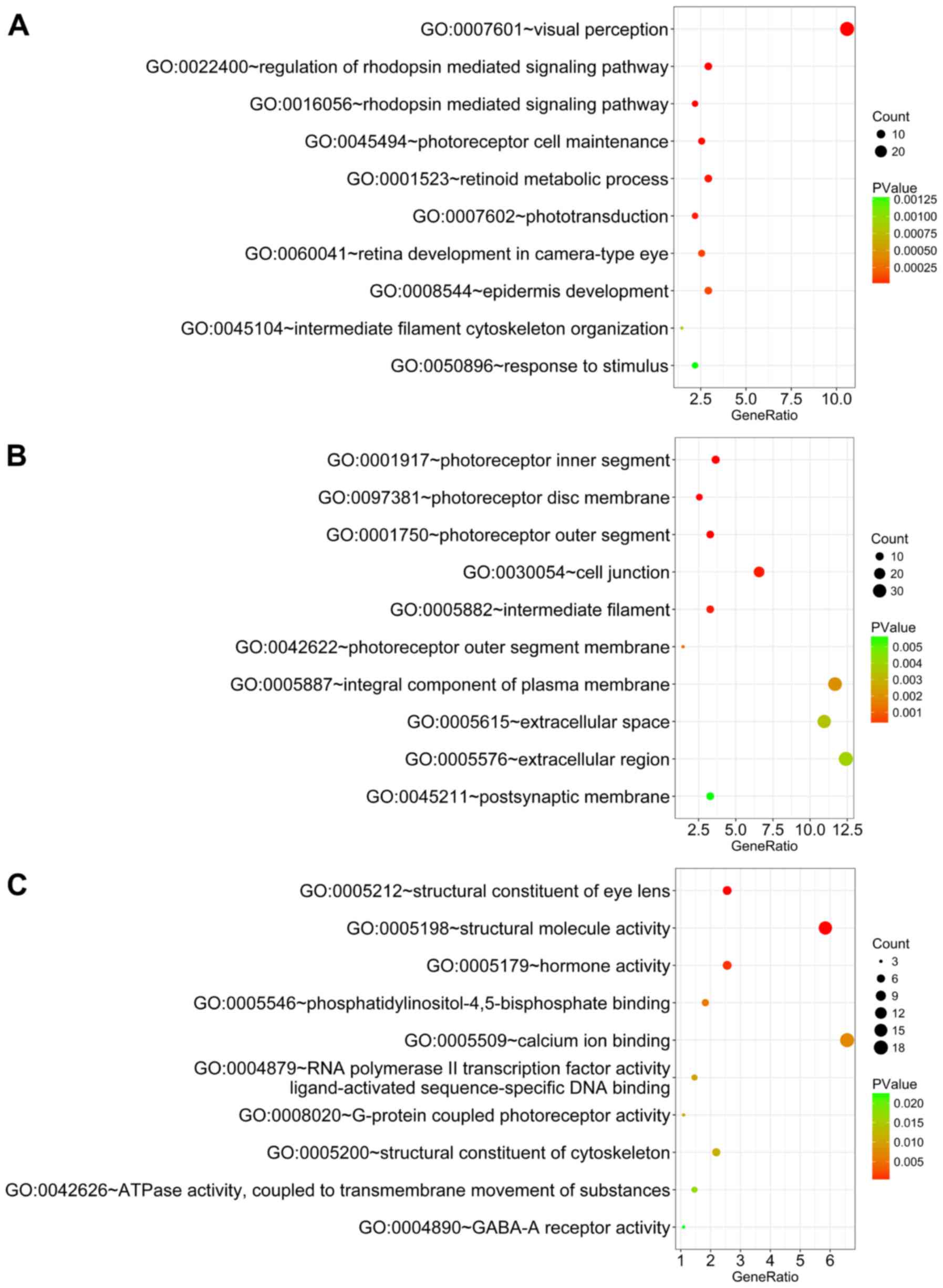
analysis revealed that these genes were significantly enriched in
several biological processes and pathways. For BP, the most notably
enriched functional terms were ‘visual perception’, ‘regulation of
rhodopsin-mediated signaling pathway’ and ‘rhodopsin-mediated
signaling pathway’ (Fig. 8A).
Regarding CC, genes were markedly enriched in ‘photoreceptor inner
segment’, ‘photoreceptor disc membrane’ and ‘photoreceptor outer
segment’ (Fig. 8B). In regards to
MF, genes were notably enriched in ‘structural constituent of eye
lens’, ‘structural molecule activity’ and ‘hormone activity’
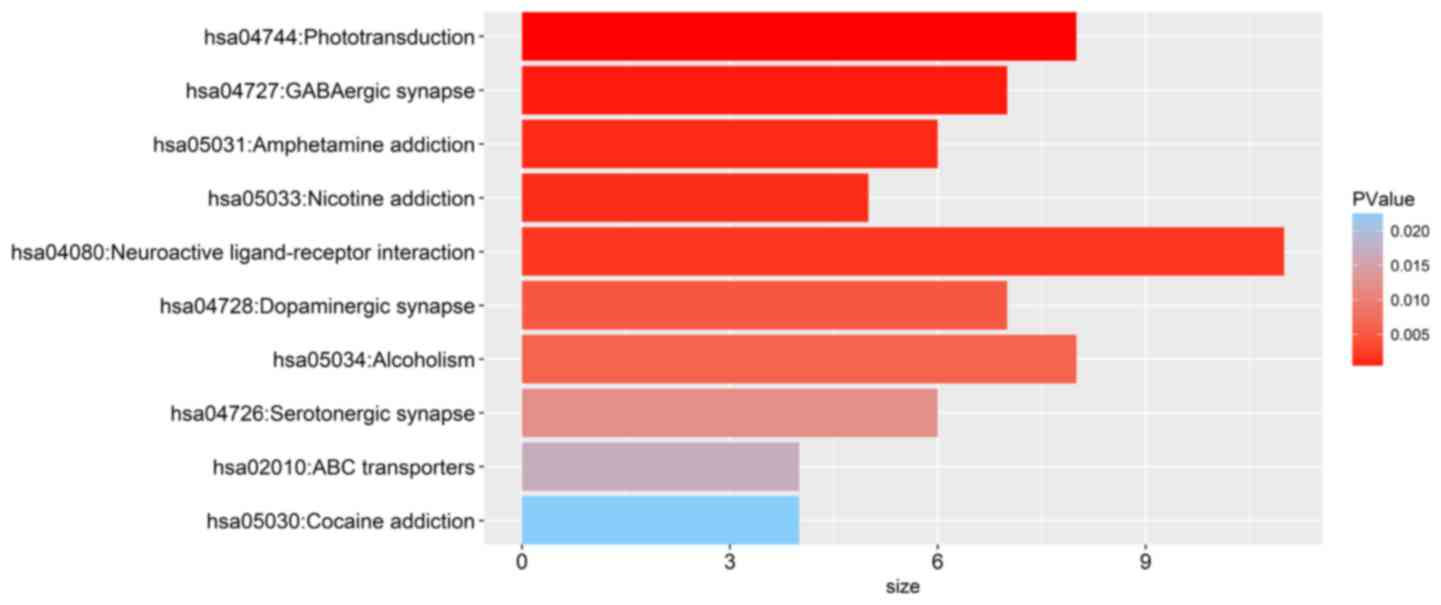
(Fig. 8C). Importantly, KEGG
enrichment analysis suggested that these genes were significantly
associated with ‘phototransduction’, ‘γ-aminobutyric acid (GABA)
ergic synapse’, ‘amphetamine (APA) addiction’, ‘nicotine addiction’
and ‘neuroactive ligand-receptor interaction’ (Fig. 9). Furthermore, the mutation status
of BAP1 may be useful in predicting the clinical outcome of
patients with UM; mutations in BAP1 demonstrated a significant
association with poorer overall survival compared with UM patients
without BAP1 mutations (Fig.
10A). Of note, the prognostic signature based on snoRNAs was
markedly associated high-risk patients with BAP1 mutations than
those without in the low-risk group (Fig. 10B).
Discussion
In the present study, survival-associated snoRNAs in
patients with UM were selected. Then, the snoRNAs that exhibited a
significant prognostic value were determined and identified by
multivariate analysis. Most importantly, a prognostic signature
comprising four novel snoRNAs was proposed: ACA17, ACA45, HBII276
and SNORD12. Notably, the prognostic signature of the present
study, based on these four novel snoRNAs, may be an ideal risk
model for patients with UM. To the best of our knowledge, the
present study is the first to propose a prognostic signature based
on snoRNAs. Furthermore, gene functional enrichment analysis
revealed that the optic nerve-associated pathways are dysregulated
most significantly between high-risk and low-risk groups. These
findings may provide novel insight into the clinical management and
molecular mechanisms underlying UM.
Previously, few studies have investigated the PI of
UM from TCGA database. Xu et al (31) reported that high plasmacytoma
variant translocation 1 (PVT1) expression was an independent
predictor in patients with UM; it was inferred that the expression
of PVT1 may act as a moderate and specific prognostic factor in
terms of OS in UM. Wan et al (32) identified 21 co-expression modules
that were analyzed based on 10,975 genes from 80 UM samples, by
using weighted correlation network analysis. It was revealed that
the hub genes solute carrier family 17 member 7, neurotrophic
receptor tyrosine kinase 2, ankyrin repeat and BTB domain
containing 1 and ADP-ribosylhydrolase like 1 may also serve a role
as potential diagnostic and prognostic biomarkers of UM. Robertson
et al (33) also analyzed
the genomic data of 80 patients with UM; the patients were divided
into several subgroups with different genomic aberrations,
transcriptional features and clinical outcomes. Field et al
(34) not only identified
preferentially expressed antigen in melanoma (PRAME) expression as
a biomarker for increased metastatic risk in class 1 UM; however,
PRAME expression was associated with poor prognosis among class 2
UM cases (35). These studies
provided novel and constructive methods of selecting prognostic
biomarkers for UM; however, the role of snoRNAs in the prognosis of
UM remains unclear. In the present study, a novel prognostic
signature based on snoRNAs was developed. More importantly, the
proposed risk score may attain satisfactory prognostic values for
UM. With the advantage of high-throughput RNA-sequencing, numerous
snoRNAs were identified; however, investigations into the clinical
significance of snoRNAs in UM are limited. The present study may
provide novel insight into the clinical management of UM.
Gene functional enrichment analysis revealed that
several biological processes and pathways were associated with
differentially expressed mRNAs between the high- and low-risk
groups; however, the biological mechanisms underlying this
association are unclear. The genes detected from KEGG enrichment
analysis were demonstrated to be associated with
‘phototransduction’, ‘GABAergic synapse’ and ‘APA addiction’ in
particular. Brown et al (36) revealed that brain-modulated
choroidal thickness has an unusual and well-established light
dependence. In addition, Wicks et al (37) reported that ultraviolet
phototransduction can evoke retina-dependent calcium flux in human
epidermal melanocytes, and increased cellular melanin content.
Furthermore, ocular melanocytosis is an important predisposing
factor for UM (38). Therefore, it
was speculated that phototransduction may be involved in the
formation of UM in the present study. GABAergic synapses are
important inhibitory neurotransmitters in the mammalian central
nervous system (CNS), as they serve to hyperpolarize the
postsynaptic neuron (39).
Interestingly, ‘APA addiction’, ‘nicotine addiction’ and
‘dopaminergic (DA) synapses’ were also associated with UM; however,
DA is an excitatory neurotransmitter in the CNS (40). APA promotes DA release in the CNS,
whilst also inducing other biogenic amine-releasing and inhibitory
neurons, and vesicle single-amine transporters (41). Monoamine oxidase can excite the CNS
(42). Nicotine initially inhibits
DA release, which is then enhanced by regulating GABAergic neurons
(43). This mechanism of action is
similar to that of APAs in the CNS. Thus, it may be inferred that
CNS activity-associated pathways may be notably dysregulated
between high-risk and low-risk groups.
However, several limitations should be considered.
An additional independent cohort was not used to validate the
performance of prognostic signature. Furthermore, the in
silico analysis for the molecular mechanisms require further
investigation.
Collectively, the results of the present study
demonstrated that snoRNAs may be notable prognostic markers for the
survival of patients with UM. To the best of our knowledge, the
present study is the first to demonstrate the prognostic value of
snoRNAs, and provides novel insight into the complex biological
functions underlying UM.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81160119), a funding
of Natural Science Foundation of Guangxi Zhuang Autonomous Region
(grant nos. 2017GXNSFAA198347 and 2016GXNsFAA380280).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
W-JZ made substantial contributions to the design of
the present study. QY and W-JZ conducted statistical analysis. QY
wrote the manuscript which was revised by W-JZ. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Álvarez-Rodríguez B, Latorre A, Posch C
and Somoza Á: Recent advances in uveal melanoma treatment. Med Res
Rev. 37:1350–1372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Komatsubara KM, Manson DK and Carvajal RD:
Selumetinib for the treatment of metastatic uveal melanoma: Past
and future perspectives. Future Oncol. 12:1331–1344. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reichstein D: New concepts in the
molecular understanding of uveal melanoma. Curr Opin Ophthalmol.
28:219–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vasalaki M, Fabian ID, Reddy MA, Cohen VM
and Sagoo MS: Ocular oncology: Advances in retinoblastoma, uveal
melanoma and conjunctival melanoma. Br Med Bull. 121:107–119. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kaliki S and Shields CL: Uveal melanoma:
Relatively rare but deadly cancer. Eye (Lond). 31:241–257. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shields CL, Kels JG and Shields JA:
Melanoma of the eye: Revealing hidden secrets, one at a time. Clin
Dermatol. 33:183–196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Helgadottir H and Höiom V: The genetics of
uveal melanoma: Current insights. Appl Clin Genet. 9:147–155. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Heppt MV, Steeb T, Schlager JG, Rosumeck
S, Dressler C, Ruzicka T, Nast A and Berking C: Immune checkpoint
blockade for unresectable or metastatic uveal melanoma: A
systematic review. Cancer Treat Rev. 60:44–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blum ES, Yang J, Komatsubara KM and
Carvajal RD: Clinical management of uveal and conjunctival
melanoma. Oncology (Williston Park). 30:29–32, 34-43, 48.
2016.PubMed/NCBI
|
|
10
|
Krantz BA, Dave N, Komatsubara KM, Marr BP
and Carvajal RD: Uveal melanoma: Epidemiology, etiology, and
treatment of primary disease. Clin Ophthalmol. 11:279–289. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou J, Jin B, Jin Y, Liu Y and Pan J: The
antihelminthic drug niclosamide effectively inhibits the malignant
phenotypes of uveal melanoma in vitro and in vivo. Theranostics.
7:1447–1462. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng H, Chua V, Liao C, Purwin TJ, Terai
M, Kageyama K, Davies MA, Sato T and Aplin AE: Co-targeting
HGF/cMET signaling with MEK inhibitors in metastatic uveal
melanoma. Mol Cancer Ther. 16:516–528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mahendraraj K, Lau CS, Lee I and
Chamberlain RS: Trends in incidence, survival, and management of
uveal melanoma: A population-based study of 7,516 patients from the
surveillance, epidemiology, and end results database (1973–2012).
Clin Ophthalmol. 10:2113–2119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Andreoli MT, Mieler WF and Leiderman YI:
Epidemiological trends in uveal melanoma. Br J Ophthalmol.
99:1550–1553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chandran SS, Somerville RPT, Yang JC,
Sherry RM, Klebanoff CA, Goff SL, Wunderlich JR, Danforth DN, Zlott
D, Paria BC, et al: Treatment of metastatic uveal melanoma with
adoptive transfer of tumour-infiltrating lymphocytes: A
single-centre, two-stage, single-arm, phase 2 study. Lancet Oncol.
18:792–802. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Komatsubara KM and Carvajal RD:
Immunotherapy for the treatment of uveal melanoma: Current status
and emerging therapies. Curr Oncol Rep. 19:452017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Spagnolo F, Picasso V, Spano L, Tanda E,
Venzano C and Queirolo P: Update on metastatic uveal melanoma:
Progress and challenges. BioDrugs. 30:161–172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McMahon M, Contreras A and Ruggero D:
Small RNAs with big implications: New insights into H/ACA snoRNA
function and their role in human disease. Wiley Interdiscip Rev
RNA. 6:173–189. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou H, Xu Q, Ni C, Ye S, Xu X, Hu X,
Jiang J, Hong Y, Huang D and Yang L: Prospects of noncoding RNAs in
hepatocellular carcinoma. Biomed Res Int. 2018:65794362018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koduru SV, Leberfinger AN and Ravnic DJ:
Small non-coding RNA abundance in adrenocortical carcinoma: A
footprint of a rare cancer. J Genomics. 5:99–118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siprashvili Z, Webster DE, Johnston D,
Shenoy RM, Ungewickell AJ, Bhaduri A, Flockhart R, Zarnegar BJ, Che
Y, Meschi F, et al: The noncoding RNAs SNORD50A and SNORD50B bind
K-Ras and are recurrently deleted in human cancer. Nat Genet.
48:53–58. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Xu C, Gu D, Wu M, Yan B, Xu Z,
Wang Y and Liu H: H/ACA box small nucleolar RNA 7A promotes the
self-renewal of human umbilical cord mesenchymal stem cells. Stem
Cells. 35:222–235. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang C, Shi J, Guo Y, Huang W, Huang S,
Ming S, Wu X, Zhang R, Ding J, Zhao W, et al: A snoRNA modulates
mRNA 3′ end processing and regulates the expression of a subset of
mRNAs. Nucleic Acids Res. 45:8647–8660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou F, Liu Y, Rohde C, Pauli C, Gerloff
D, Köhn M, Misiak D, Bäumer N, Cui C, Göllner S, et al: AML1-ETO
requires enhanced C/D box snoRNA/RNP formation to induce
self-renewal and leukaemia. Nat Cell Biol. 19:844–855. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu B, Ye MH, Lv SG, Wang QX, Wu MJ, Xiao
B, Kang CS and Zhu XG: SNORD47, a box C/D snoRNA, suppresses
tumorigenesis in glioblastoma. Oncotarget. 8:43953–43966.
2017.PubMed/NCBI
|
|
26
|
Patterson DG, Roberts JT, King VM,
Houserova D, Barnhill EC, Crucello A, Polska CJ, Brantley LW,
Kaufman GC, Nguyen M, et al: Human snoRNA-93 is processed into a
microRNA-like RNA that promotes breast cancer cell invasion. NPJ
Breast Cancer. 3:252017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshida K, Toden S, Weng W, Shigeyasu K,
Miyoshi J, Turner J, Nagasaka T, Ma Y, Takayama T, Fujiwara T and
Goel A: SNORA21-an oncogenic small nucleolar RNA, with a prognostic
biomarker potential in human colorectal cancer. EBioMedicine.
22:68–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu L, Ziegelbauer J, Wang R, Wu WW, Shen
RF, Juhl H, Zhang Y and Rosenberg A: Distinct profiles for
mitochondrial t-RNAs and small nucleolar RNAs in locally invasive
and metastatic colorectal cancer. Clin Cancer Res. 22:773–784.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wickham H: ggplot2: Elegant graphics for
data analysis. Springer Publishing Company, Incorporated. 2009.
|
|
31
|
Xu H, Gong J and Liu H: High expression of
lncRNA PVT1 independently predicts poor overall survival in
patients with primary uveal melanoma. PLoS One. 12:e01896752017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wan Q, Tang J, Han Y and Wang D:
Co-expression modules construction by WGCNA and identify potential
prognostic markers of uveal melanoma. Exp Eye Res. 166:13–20. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Robertson AG, Shih J, Yau C, Gibb EA, Oba
J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al:
Integrative analysis identifies four molecular and clinical subsets
in uveal melanoma. Cancer Cell. 32:1512018. View Article : Google Scholar
|
|
34
|
Field MG, Decatur CL, Kurtenbach S, Gezgin
G, van der Velden PA, Jager MJ, Kozak KN and Harbour JW: PRAME as
an independent biomarker for metastasis in uveal melanoma. Clin
Cancer Res. 22:1234–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Field MG, Durante MA, Decatur CL, Tarlan
B, Oelschlager KM, Stone JF, Kuznetsov J, Bowcock AM, Kurtenbach S
and Harbour JW: Epigenetic reprogramming and aberrant expression of
PRAME are associated with increased metastatic risk in Class 1 and
Class 2 uveal melanomas. Oncotarget. 7:59209–59219. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brown JS, Flitcroft DI, Ying GS, Francis
EL, Schmid GF, Quinn GE and Stone RA: In vivo human choroidal
thickness measurements: Evidence for diurnal fluctuations. Invest
Ophthalmol Vis Sci. 50:5–12. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wicks NL, Chan JW, Najera JA, Ciriello JM
and Oancea E: UVA phototransduction drives early melanin synthesis
in human melanocytes. Curr Biol. 21:1906–1911. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Szuścik I, Romanowska-Dixon B, Jakubowska
B and Orlowska-Heitzman J: Uveal melanoma in patients with ocular
or oculodermal melanocytosis. Klin Oczna. 110:380–383. 2008.(In
Polish). PubMed/NCBI
|
|
39
|
Kuzirian MS and Paradis S: Emerging themes
in GABAergic synapse development. Prog Neurobiol. 95:68–87. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zieger E, Lacalli TC, Pestarino M,
Schubert M and Candiani S: The origin of dopaminergic systems in
chordate brains: Insights from amphioxus. Int J Dev Biol.
61:749–761. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vicente-Rodríguez M, Rojo Gonzalez L,
Gramage E, Fernández-Calle R, Chen Y, Pérez-García C, Ferrer-Alcón
M, Uribarri M, Bailey A and Herradón G: Pleiotrophin overexpression
regulates amphetamine-induced reward and striatal dopaminergic
denervation without changing the expression of dopamine D1 and D2
receptors: Implications for neuroinflammation. Eur
Neuropsychopharmacol. 26:1794–1805. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Baig AM: DARK side of amphetamine and
analogues: pharmacology, syndromic manifestation, and management of
amphetamine addiction. ACS Chem Neurosci. 9:2299–2303. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ortells MO and Arias HR: Neuronal networks
of nicotine addiction. Int J Biochem Cell Biol. 42:1931–1935. 2010.
View Article : Google Scholar : PubMed/NCBI
|