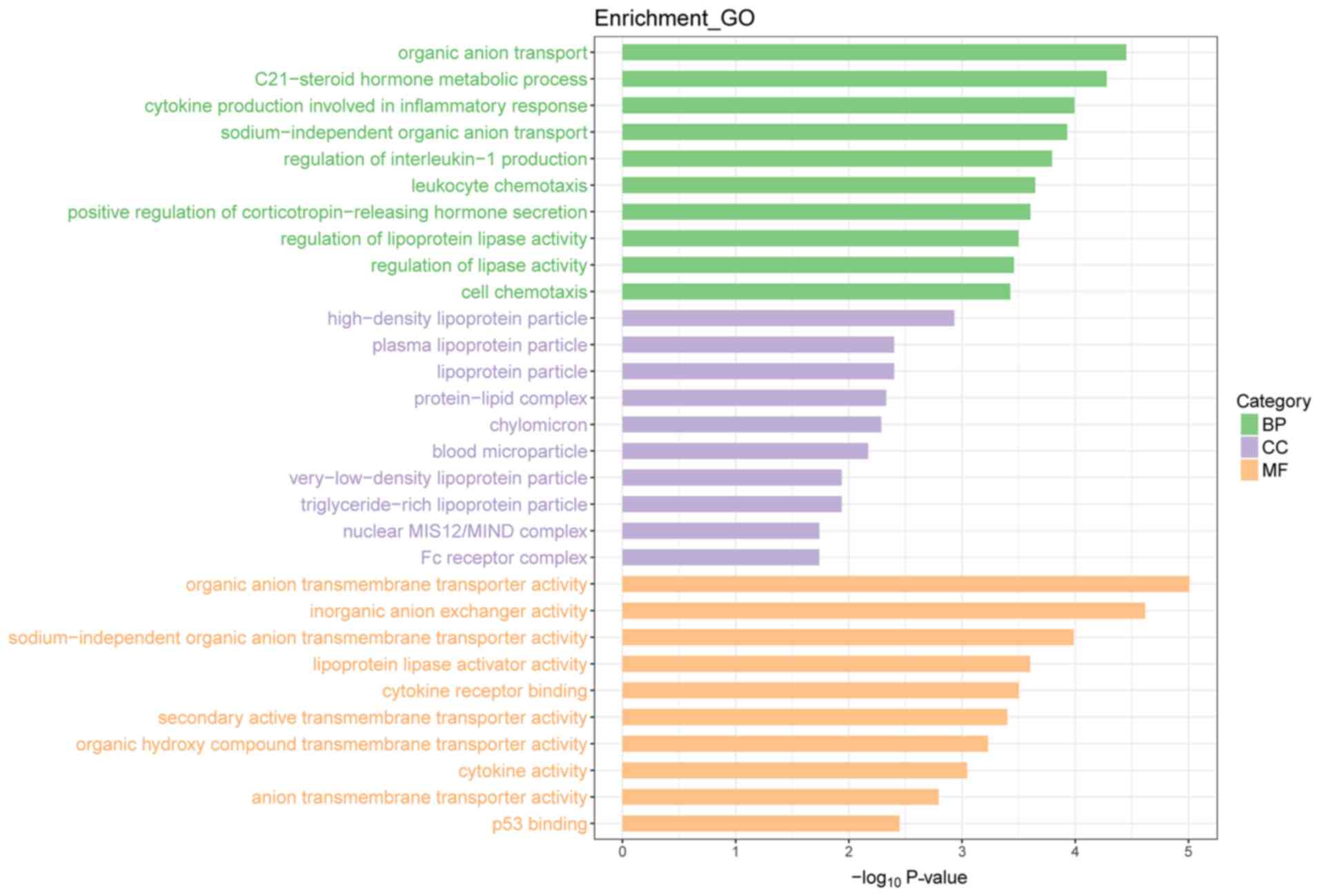
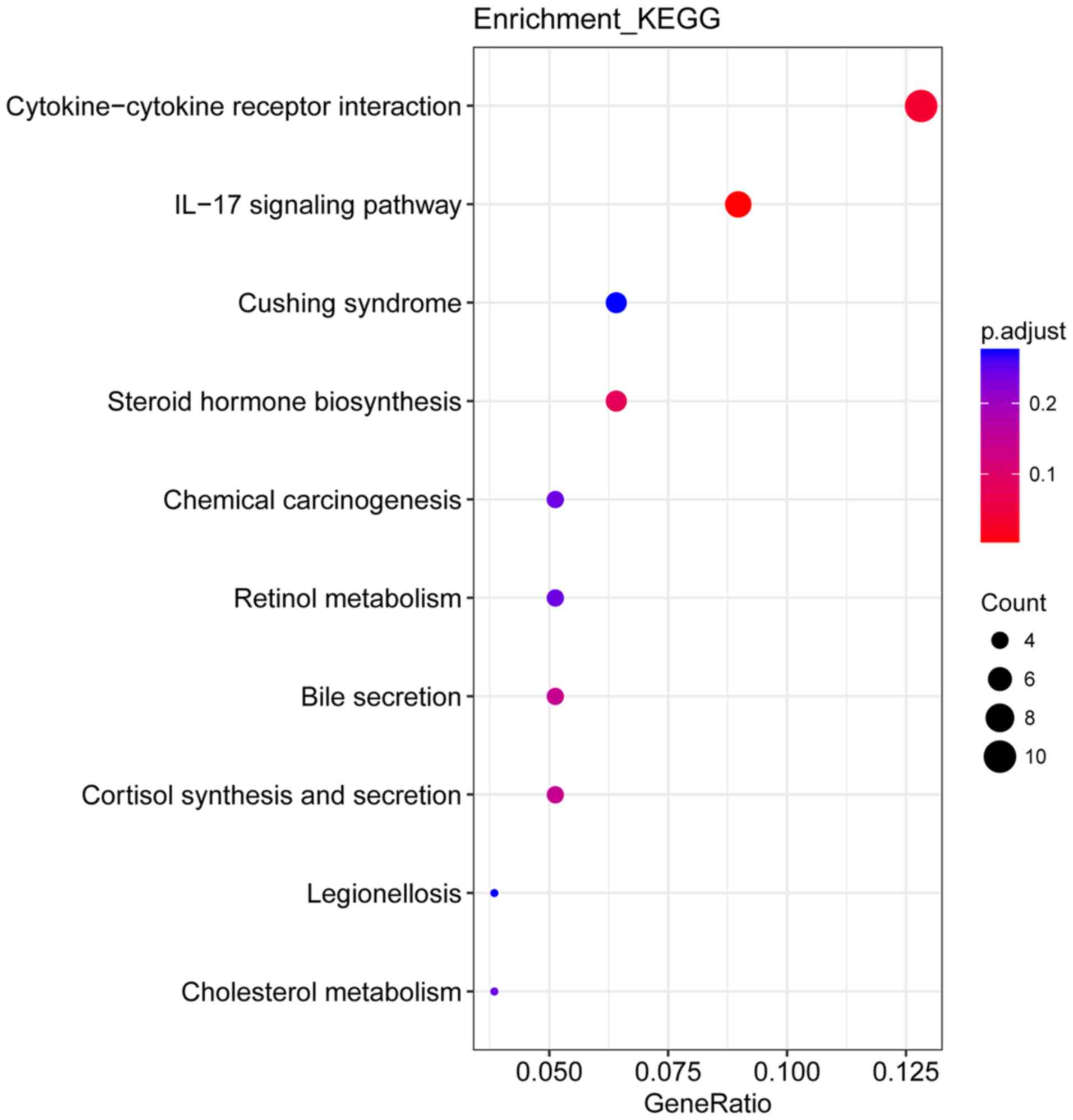
|
1
|
Wang DD, Lu JM, Li Q and Li ZP: Population
pharmacokinetics of tacrolimus in paediatric systemic lupus
erythematosus based on real-world study. J Clin Pharm Ther.
43:476–483. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takeuchi T, Tsuzaka K, Abe T, Yoshimoto K,
Shiraishi K, Kameda H and Amano K: T cell abnormalities in systemic
lupus erythematosus. Autoimmunity. 38:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rahman A and Isenberg DA: Systemic lupus
erythematosus. N Engl J Med. 358:929–939. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gurevitz SL, Snyder JA, Wessel EK, Frey J
and Williamson BA: Systemic lupus erythematosus: A review of the
disease and treatment options. Consult Pharm. 28:110–121. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
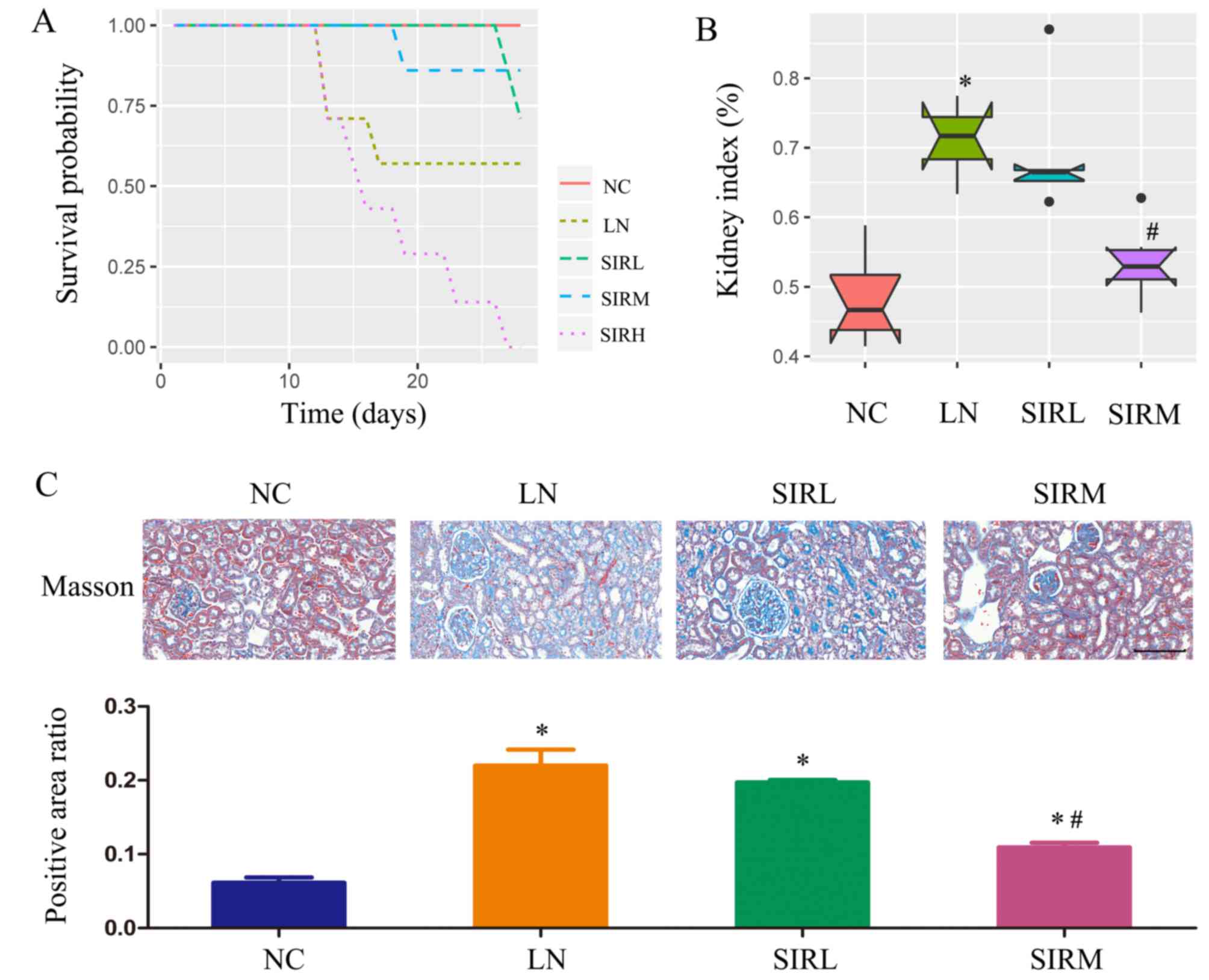
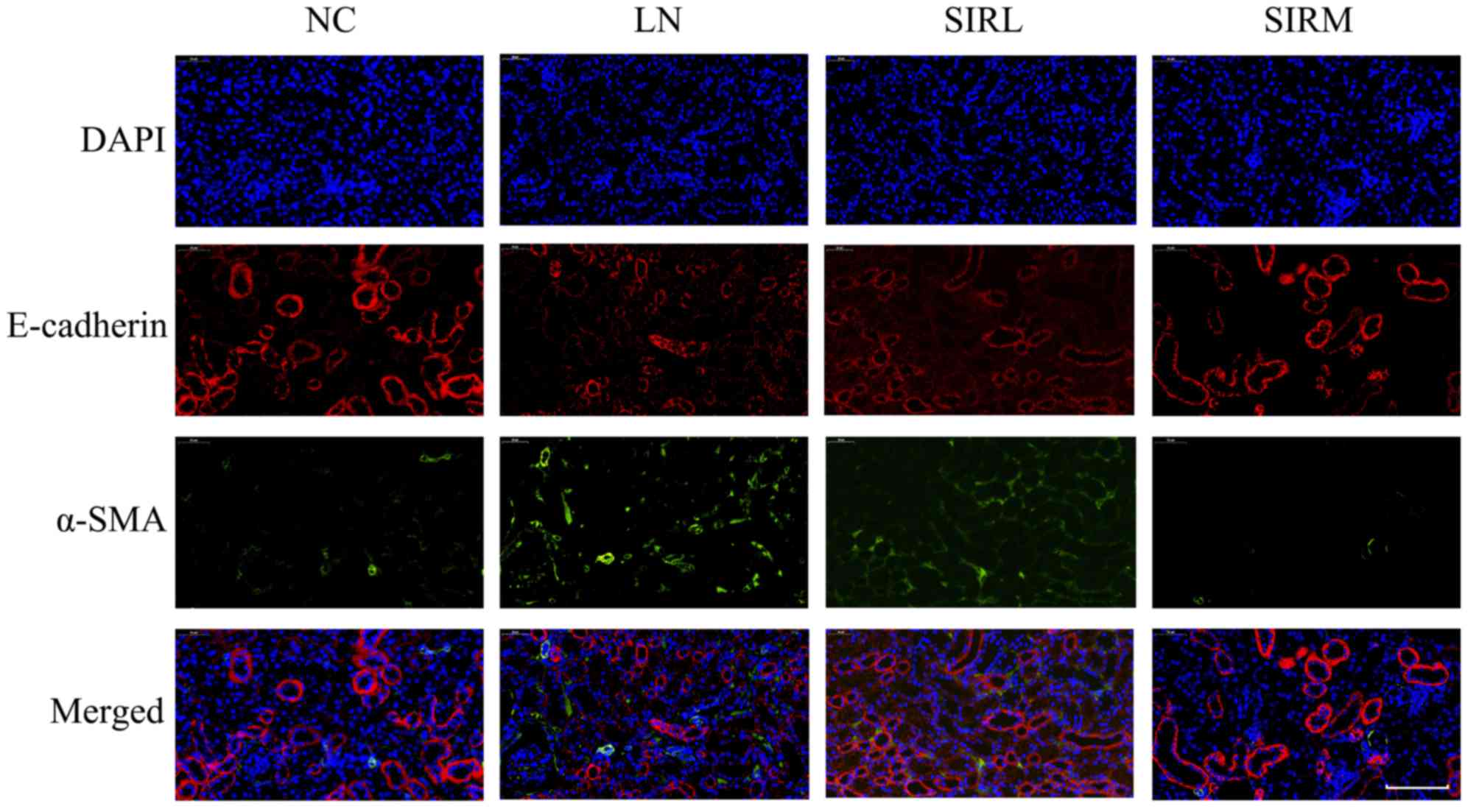
|
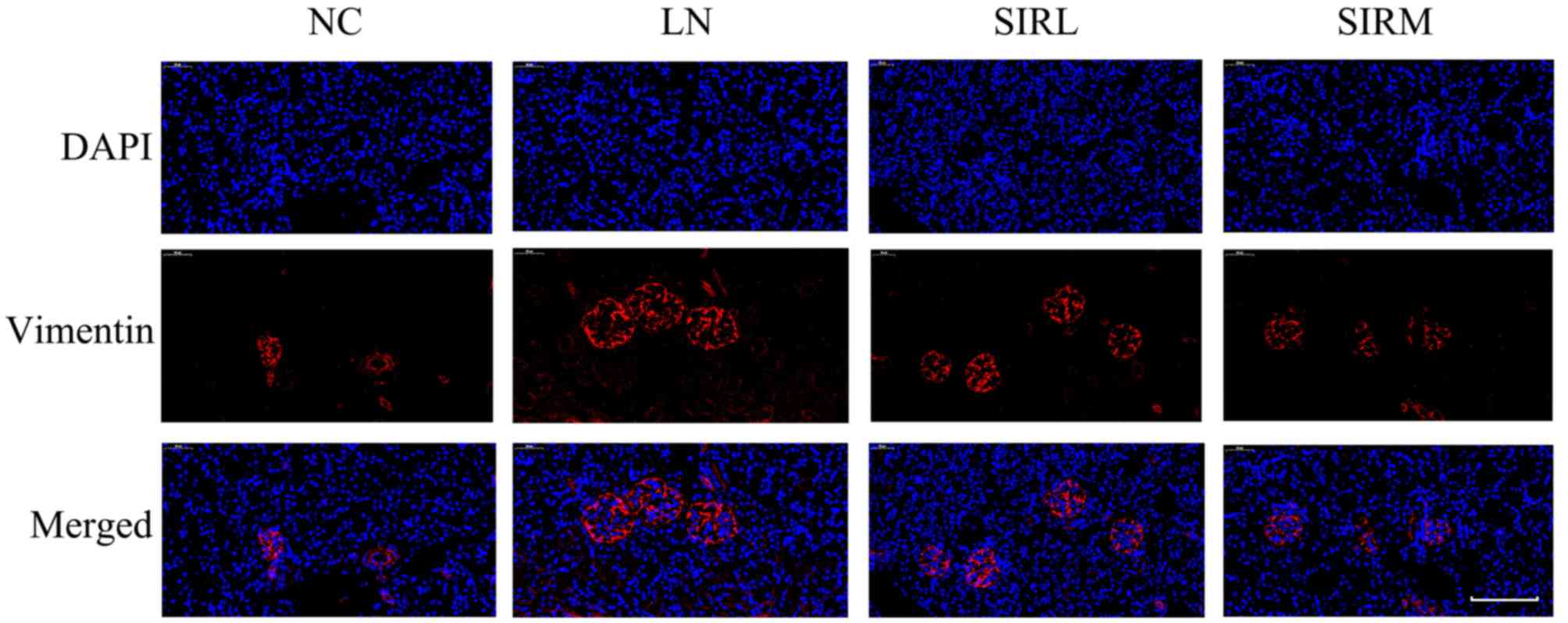
D'Cruz DP, Khamashta MA and Hughes GR:
Systemic lupus erythematosus. Lancet. 369:587–596. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liao R, Liu Q, Zheng Z, Fan J, Peng W,
Kong Q, He H, Yang S, Chen W, Tang X and Yu X: Tacrolimus protects
podocytes from injury in lupus nephritis partly by stabilizing the
cytoskeleton and inhibiting podocyte apoptosis. PLoS One.
10:e01327242015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bertsias G, Ioannidis JP, Boletis J,
Bombardieri S, Cervera R, Dostal C, Font J, Gilboe IM, Houssiau F,
Huizinga T, et al: EULAR recommendations for the management of
systemic lupus erythematosus. Report of a task force of the eular
standing committee for international clinical studies including
therapeutics. Ann Rheum Dis. 67:195–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ji Y, Chen S, Xiang B, Li K, Xu Z, Yao W,
Lu G, Liu X, Xia C, Wang Q, et al: Sirolimus for the treatment of
progressive kaposiform hemangioendothelioma: A multicenter
retrospective study. Int J Cancer. 141:848–855. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jung KS, Lee J, Park SH, Park JO, Park YS,
Lim HY, Kang WK and Kim ST: Pilot study of sirolimus in patients
with PIK3CA mutant/amplified refractory solid cancer. Mol Clin
Oncol. 7:27–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Djebli N, Rousseau A, Hoizey G, Rerolle
JP, Toupance O, Le Meur Y and Marquet P: Sirolimus population
pharmacokinetic/pharmacogenetic analysis and bayesian modelling in
kidney transplant recipients. Clin Pharmacokinet. 45:1135–1148.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiao Z, Shi XJ, Li ZD and Zhong MK:
Population pharmacokinetics of sirolimus in de novo Chinese adult
renal transplant patients. Br J Clin Pharmacol. 68:47–60. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wataya-Kaneda M, Nakamura A, Tanaka M,
Hayashi M, Matsumoto S, Yamamoto K and Katayama I: Efficacy and
safety of topical sirolimus therapy for facial angiofibromas in the
tuberous sclerosis complex: A randomized clinical trial. JAMA
Dermatol. 153:39–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang DD, Lu JM, Li YZ, Li Q and Li ZP:
Population pharmacokinetics of sirolimus in pediatric tuberous
sclerosis complex: From real world study. Int J Clin Exp Med.
11:12302–12309. 2018.
|
|
14
|
Zhou L, Du GS, Pan LC, Zheng YG, Liu ZJ,
Shi HD, Yang SZ, Shi XJ, Xuan M, Feng LK and Zhu ZD: Sirolimus
treatment for cirrhosis or hepatocellular carcinoma patients
accompanied by psoriasis after liver transplantation: A single
center experience. Oncol Lett. 14:7817–7824. 2017.PubMed/NCBI
|
|
15
|
Zhou L, Pan LC, Zheng YG, Du GS, Fu XQ,
Zhu ZD, Song JY, Liu ZJ, Su XZ, Chen W, et al: Novel strategy of
sirolimus plus thymalfasin and huaier granule on tumor recurrence
of hepatocellular carcinoma beyond the UCSF criteria following
liver transplantation: A single center experience. Oncol Lett.
16:4407–4417. 2018.PubMed/NCBI
|
|
16
|
Fu J, Wang Z, Lee K, Wei C, Liu Z, Zhang
M, Zhou M, Cai M, Zhang W, Chuang PY, et al: Transcriptomic
analysis uncovers novel synergistic mechanisms in combination
therapy for lupus nephritis. Kidney Int. 93:416–429. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yap DYH, Tang C, Chan GCW, Kwan LPY, Ma
MKM, Mok MMY and Chan TM: Longterm data on sirolimus treatment in
patients with lupus nephritis. J Rheumatol. 45:1663–1670. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu J, Yang L, Hou Y, Soteyome T, Zeng B,
Su J, Li L, Li B, Chen D, Li Y, et al: Transcriptomics study on
staphylococcus aureus biofilm under low concentration of
ampicillin. Front Microbiol. 9:24132018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Burden CJ, Qureshi SE and Wilson SR: Error
estimates for the analysis of differential expression from RNA-seq
count data. PeerJ. 2:e5762014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seshan SV and Jennette JC: Renal disease
in systemic lupus erythematosus with emphasis on classification of
lupus glomerulonephritis: Advances and implications. Arch Pathol
Lab Med. 133:233–248. 2009.PubMed/NCBI
|
|
25
|
Tveita A, Rekvig OP and Zykova SN:
Glomerular matrix metalloproteinases and their regulators in the
pathogenesis of lupus nephritis. Arthritis Res Ther. 10:2292008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kolset SO, Reinholt FP and Jenssen T:
Diabetic nephropathy and extracellular matrix. J Histochem
Cytochem. 60:976–986. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang D, Zhang G, Chen X, Wei T, Liu C,
Chen C, Gong Y and Wei Q: Sitagliptin ameliorates diabetic
nephropathy by blocking TGF-beta1/Smad signaling pathway. Int J Mol
Med. 41:2784–2792. 2018.PubMed/NCBI
|
|
28
|
Chen X, Wang DD, Wei T, He SM, Zhang GY
and Wei QL: Effects of astragalosides from radix astragali on high
glucose-induced proliferation and extracellular matrix accumulation
in glomerular mesangial cells. Exp Ther Med. 11:2561–2566. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang GY, Wang DD, Cao Z, Wei T, Liu CX
and Wei QL: Sitagliptin ameliorates high glucose-induced cell
proliferation and expression of the extracellular matrix in
glomerular mesangial cells. Exp Ther Med. 14:3862–3867. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen X, Yang Y, Liu C, Chen Z and Wang D:
Astragaloside IV ameliorates high glucoseinduced renal tubular
epithelialmesenchymal transition by blocking mTORC1/p70S6K
signaling in HK2 cells. Int J Mol Med. 43:709–716. 2019.PubMed/NCBI
|
|
31
|
Badid C, Desmouliere A, Babici D,
Hadj-Aissa A, McGregor B, Lefrancois N, Touraine JL and Laville M:
Interstitial expression of alpha-SMA: An early marker of chronic
renal allograft dysfunction. Nephrol Dial Transplant. 17:1993–1998.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao YH, Lv LL, Zhang X, Hu H, Ding LH, Yin
D, Zhang YZ, Ni HF, Chen PS and Liu BC: Urinary vimentin mRNA as a
potential novel biomarker of renal fibrosis. Am J Physiol Renal
Physiol. 309:F514–F522. 2015. View Article : Google Scholar : PubMed/NCBI
|