Introduction
Systemic lupus erythematosus (SLE) is a chronic
autoimmune disease characterized by the production of large
quantities of autoantibodies. These antibodies are deposited in the
vascular bed of target tissues and organs, including the glomeruli
and the renal microvasculature, leading to systemic inflammation
and lupus nephritis (LN) (1–6). LN
is the principal cause of morbidity and mortality in patients with
SLE (6), and as of resistance to
existing medications, the development of novel treatments is
required (7).
Sirolimus (also known as rapamycin) is an inhibitor
of the mammalian target of rapamycin (mTOR) (8,9), and
has been widely used in transplantation patients to prevention
allograft rejection (10,11). It has also been reported as an
effective treatment for pediatric tuberous sclerosis complex
(12,13), cirrhosis or hepatocellular
carcinoma accompanied with psoriasis following liver
transplantation (14), and tumor
recurrence in patients with hepatocellular carcinoma (15).
Transcriptomics analysis has been used to study the
therapeutic mechanisms of LN (16). Recently, sirolimus has been
administered as a treatment for patients with LN (17); however, its effects on patients
with LN, in addition to its mechanism of action, are yet to be
clarified. In the present study, the regulatory mechanism of
sirolimus in LN was elucidated using transcriptomics analysis,
which was performed using RNA-sequencing, differentially expressed
gene (DEG) identification and annotation, Gene Ontology (GO)
functional and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment.
Materials and methods
Animal experiments
Murphy Roths Large/lymphoproliferation strain
(MRL/lpr) mice an established model of LN, were selected as the
animal model for the present study. Specific pathogen free (SPF)
grade female MRL/lpr mice (n=28) weighing 16–20 g at 12 weeks of
age, were obtained from Shanghai SLAC Laboratory Animal Co., Ltd.
(Shanghai, China). Age and weight matched SPF female C57BL/6 mice
(n=7; Shanghai SLAC Laboratory Animal Co., Ltd.) were used as the
normal control group (NC). MRL/lpr mice were randomly divided into
the LN control group (LN, n=7), sirolimus low-dose treatment group
(SIRL, n=7), sirolimus medium-dose treatment group (SIRM, n=7) and
the sirolimus high-dose treatment group (SIRH, n=7), respectively.
Mice of the low, medium and high-dose treatment groups were
administered sirolimus (Shanghai Topbiochem Technology Co., Ltd.)
at a dose of 0.1, 0.3 and 1 mg/kg per day, respectively, by
intragastric administration for 4 weeks. Control groups, including
the NC and LN groups, received daily intragastric administration of
equal amounts of 1% sodium carboxymethylcellulose for 4 weeks. The
present study was approved by the Animal Care and Use Committee of
the Children's Hospital of Fudan University (Shanghai, China), and
complied with our institutional regulations. The survival rate in
each treatment group was calculated by dividing the number of mice
that survived until the end of the experiments by the amount of
animals at the start.
Sample collection
Mice were anesthetized with chloral hydrate (400
mg/kg) by intraperitoneal injection. Subsequently, the mice were
sacrificed by cervical dislocation and their kidneys were removed
and weighed. The kidney index, which represented the relative
kidney size, was the ratio between the average kidney weight and
body weight. The fresh kidneys were fixed in 4% paraformaldehyde at
room temperature for 24 h, and the residual kidney tissues were
snap frozen in liquid nitrogen, and stored at −80°C until use.
Masson trichome staining
Following fixation with paraformaldehyde, the
samples were embedded in paraffin and cut into 4-µm-thick sections.
Then, the sections were deparaffinized in xylene at room
temperature for 20 min, rehydrated in a descending ethanol series
(100 and 75% ethanol at room temperature for 5 min) and washed in
water prior to overnight incubation in potassium dichromate at room
temperature. After washing in water, the sections were stained with
hematoxylin iron solution for 3 min at room temperature, followed
by 0.5% acid alcohol, and washed in water once more. Samples were
stained with Ponceau S solution for 5 min at room temperature, and
subsequently rinsed in water. Finally, samples were stained using
phosphomolybdic acid for 1 min, followed by aniline blue solution
for 3 min, and washed with 1% acetic acid solution at room
temperature. The sections were dehydrated using 100% alcohol,
vitrified in xylene, and mounted with neutral gum. The accumulation
of collagen fibers (blue) was determined using Image-Pro Plus 6.0
image analysis software (Media Cybernetics, Inc., Rockville, MD,
USA).
Immunofluorescence
The 4 µm sections were deparaffinized in xylene,
rehydrated using a descending ethanol series (100, 85 and 75%
ethanol, respectively) and washed in water. Following antigen
retrieval, the spontaneous fluorescence quencher (Wuhan Servicebio
Technology Co., Ltd; cat. no. G1221) was added for 5 min at room
temperature, and the samples were rinsed with water for 10 min. The
sections were subsequently blocked using bovine serum albumin
(Wuhan Servicebio Technology Co., Ltd.) for 30 min at room
temperature, and incubated with antibodies against E-cadherin
(1:100; Wuhan Servicebio Technology Co., Ltd.; cat. no. GB12082),
α-smooth muscle actin (α-SMA, 1:500; Wuhan Servicebio Technology
Co., Ltd.; cat. no. GB13044) and vimentin (1:200, Wuhan Servicebio
Technology Co., Ltd.; cat. no. GB11192) at 4°C overnight. Following
the primary incubation, the sections were washed and incubated with
the corresponding conjugated secondary antibodies with Cy3 (1:300,
Wuhan Servicebio Technology Co., Ltd.; cat. no. GB21303) and Alexa
Fluor 488 (1:400, Wuhan Servicebio Technology Co., Ltd.; cat. no.
GB25301) at room temperature, in the dark room for 50 min. The
sections were stained with DAPI solution and incubated in the dark
at room temperature for 10 min, prior to quenching using an
anti-fluorescence quenching tablet. Images were acquired using a
fluorescence microscope (Nikon Eclipse C1; Nikon Corporation).
RNA library construction and
sequencing
Our preliminary analysis using various doses of
sirolimus revealed that medium-dose sirolimus treatment had a
therapeutic effect on LN. Therefore, the kidneys of mice in the LN
and SIRM groups were used to conduct transcriptome analysis. RNA
library construction and sequencing were performed in triplicate by
Suzhou Base Pair Biotechnology (Suzhou, China) using an Illumina
HiSeq X™ Ten Sequencing System (Illumina, Inc., San Diego, CA,
USA).
DEG identification and annotation
Raw reads were generated from image data and stored
in FASTQ format. Raw data were filtered to remove
adaptor-contaminated and low quality sequences, and to obtain clean
reads (18). The quality of the
clean reads was checked using FastQC (version 0.11.4; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
and aligned to the reference genome Ensemble-GRCM38 using Hisat2
(version 2.1.0) (19); ≤2 base
mismatches or read gaps were permitted in the alignment. Gene
coverage was calculated as the percentage of genes covered by
reads, and gene functional annotation was performed using the
ANNOVAR tool (20). DEGs were
identified using DEGseq panormalage (21) based on a negative binomial
distribution. Gene expression levels were measured based on
fragments per kilobase of transcript per million read pairs and
count values. The SIRM group reflected the ‘case’ and the LN group
was the ‘control’ for analysis. The results represented the case
gene expression relative to that of the control. Genes with an
adjusted |log2(fold change)|>1 and P<0.05 were identified as
DEGs. KEGG pathway and GO annotation of DEGs was performed using
Kobas with the KEGG (22), and GO
(23) databases, respectively.
Significantly enriched KEGG pathways and GO terms were identified
by P<0.05.
Results
Sirolimus treatment in mice with
LN
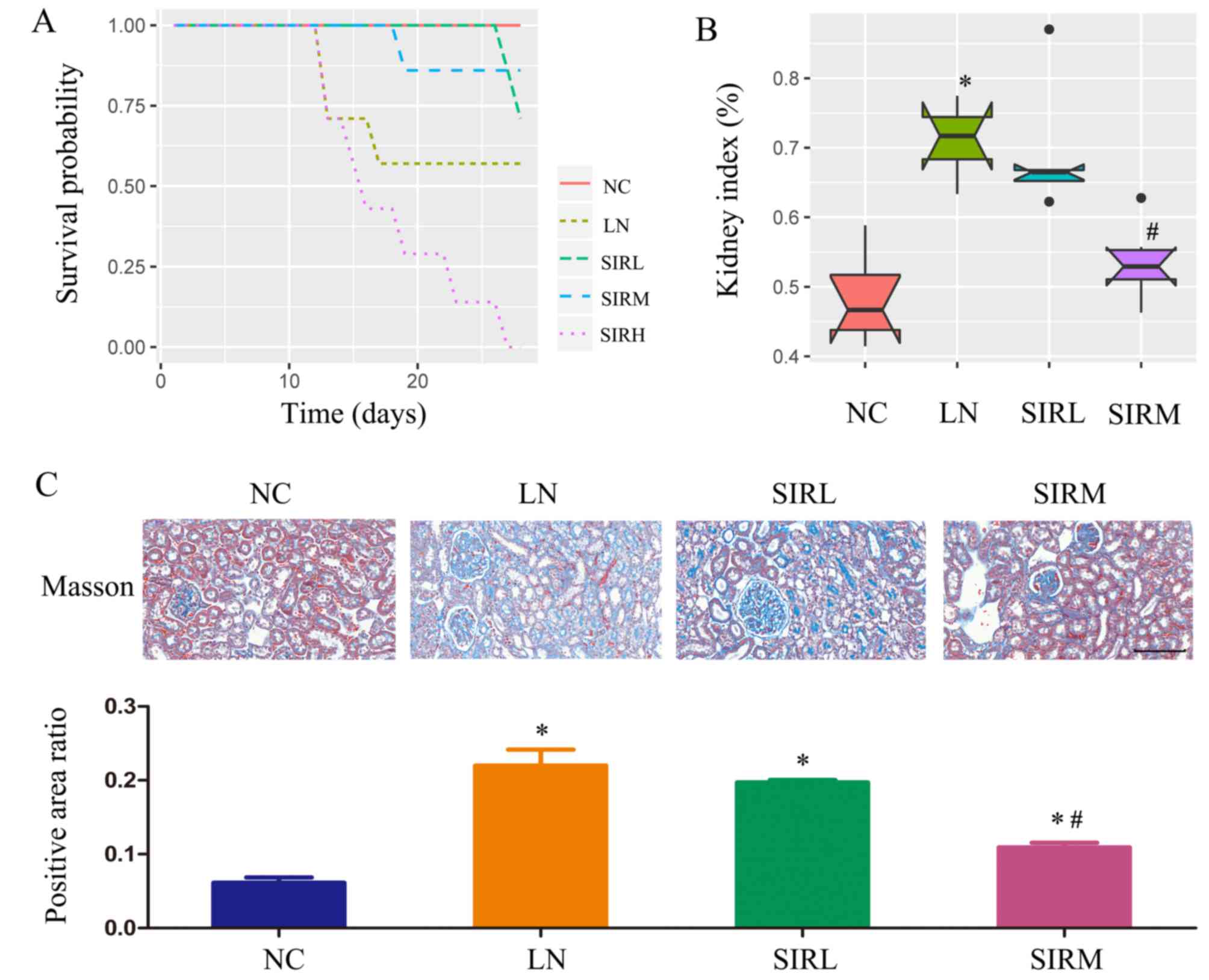
As demonstrated in Fig.
1A, the SIRM group revealed a high probability of survival,
while animals of the SIRH group had succumbed. As presented in
Fig. 1B, the LN group exhibited a
markedly higher kidney index compared with the NC group; data of
the SIRH group could not be obtained were thus emitted from
subsequent analysis. Of note, the SIRM group possessed a
significantly lower kidney index compared with the LN group.
Compared with the NC group, fibrosis was significantly promoted in
the LN group, as demonstrated by Masson Trichome staining. Compared
with the LN group, significantly reduced renal fibrosis was
observed in the SIRM group (Fig.
1C). Therefore, medium-dose sirolimus treatment was proposed to
possess notable therapeutic potential against LN.
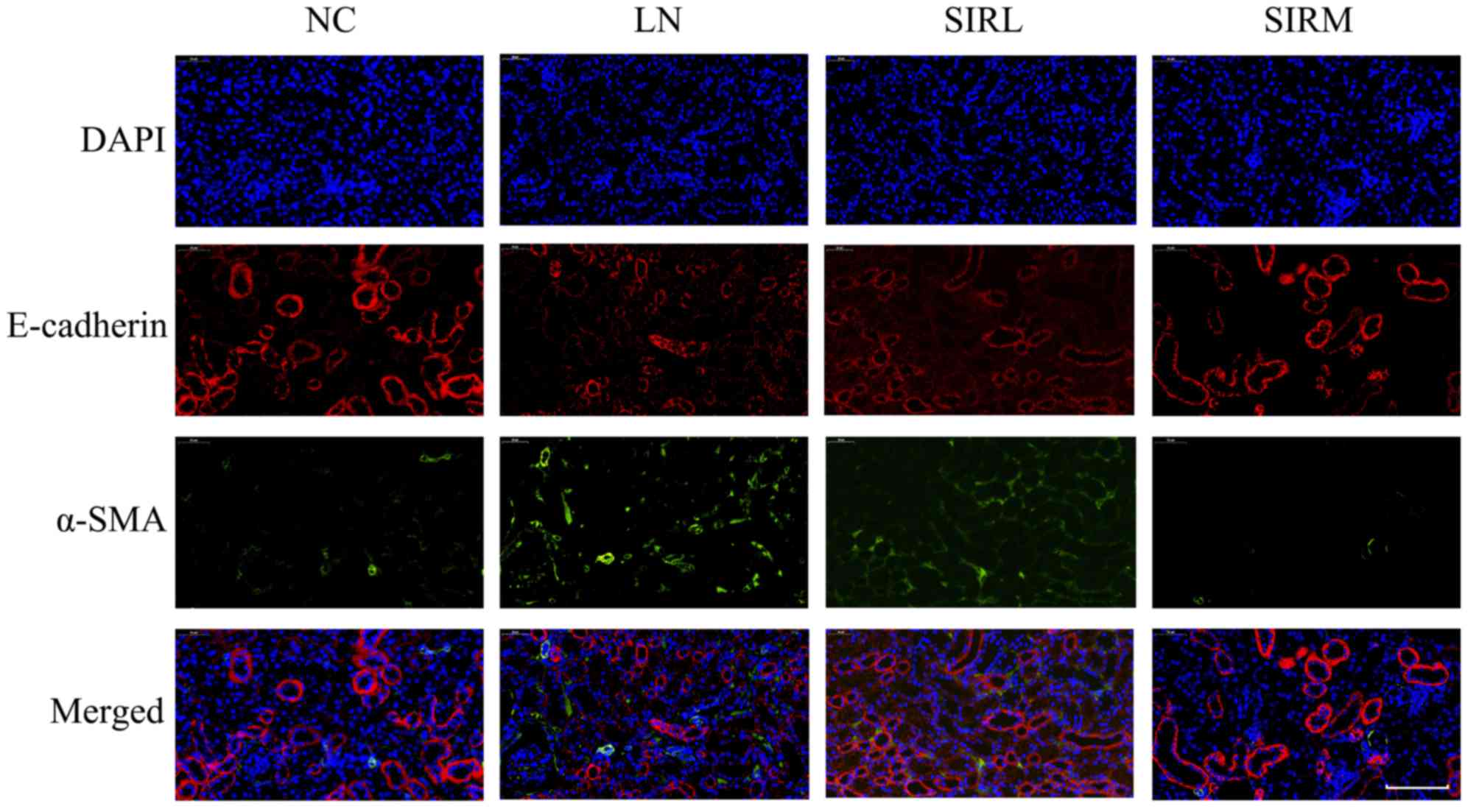
As presented in Fig.
2, the LN group possessed lower E-cadherin expression levels
and increased α-SMA expression levels compared with the NC group;
the SIRM group exhibited upregulated E-cadherin expression, and
reduced α-SMA expression when compared with the LN and low-dose
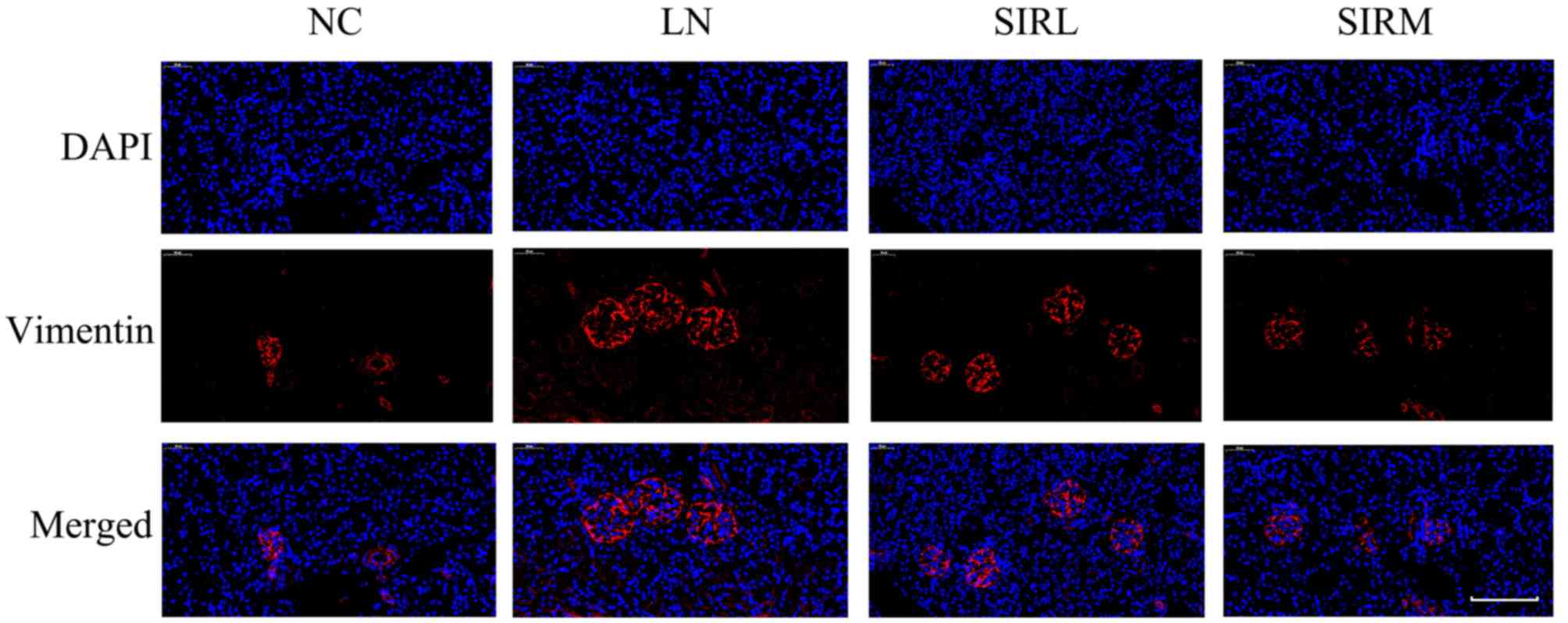
groups. The LN group exhibited increased vimentin expression
compared with NC group, however, downregulated vimentin expression
was reported compared with the LN and low-dose groups (Fig. 3). These results are consistent with
the findings presented in Fig. 1,
suggesting that medium-dose treatment was more effective, and that
the results could be used for future analysis of the
transcriptome.
Transcriptomics analysis of sirolimus
treatment in LN
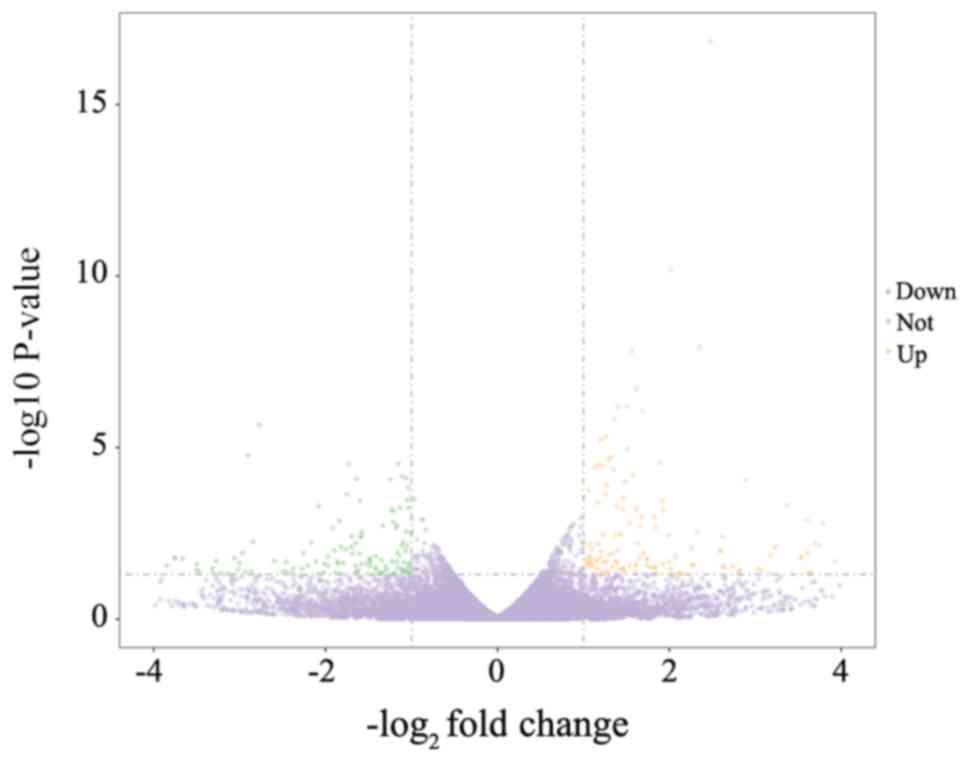
The results of the present study represent gene
expression in the SIRM group relative to that in the LN group.
Transcriptomics analysis indicated a total of 334 DEGs, of which
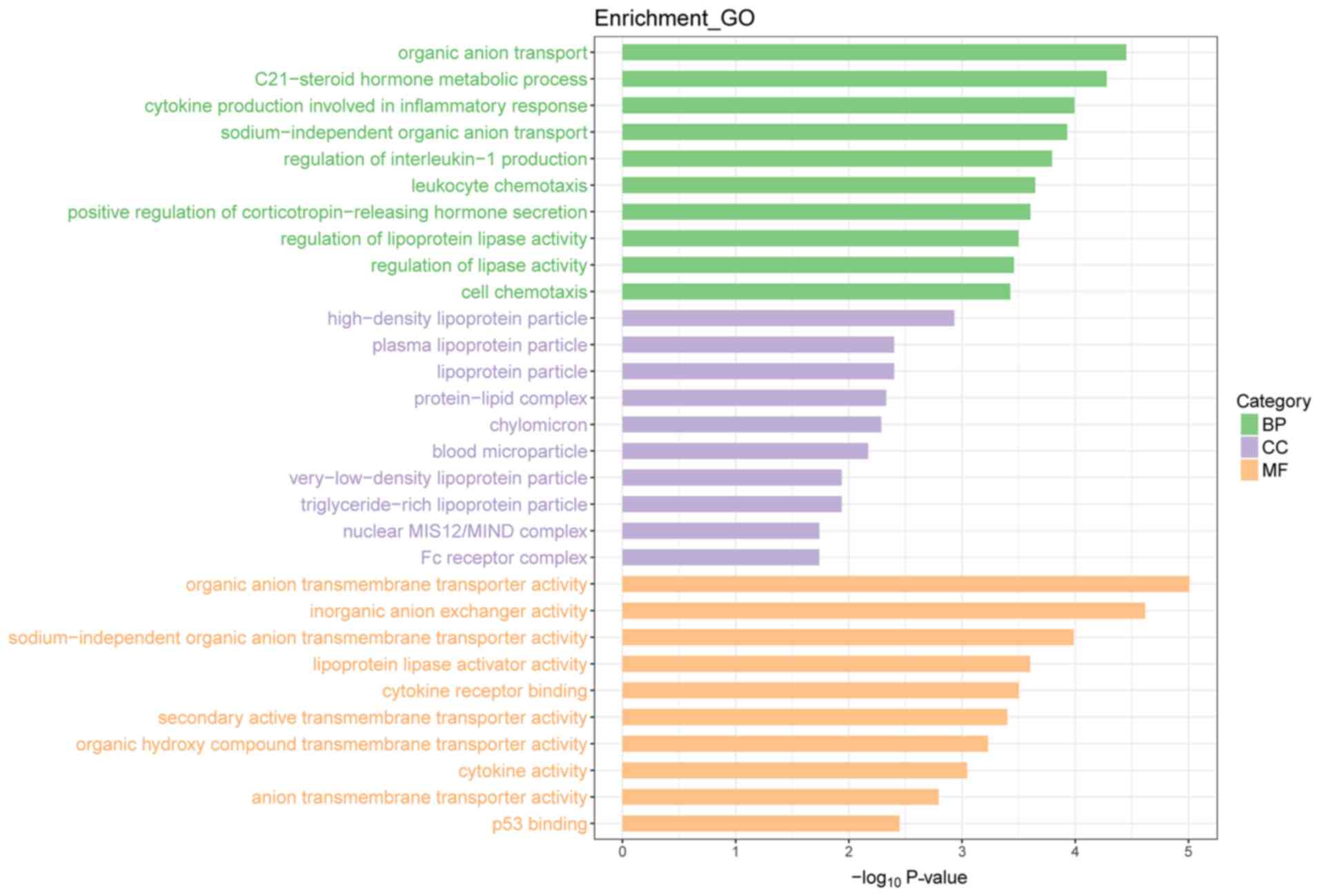
there were 176 upregulated and 158 downregulated genes (Fig. 4). Upon GO functional enrichment,
‘biological process’, ‘molecular function’ and ‘cellular component’
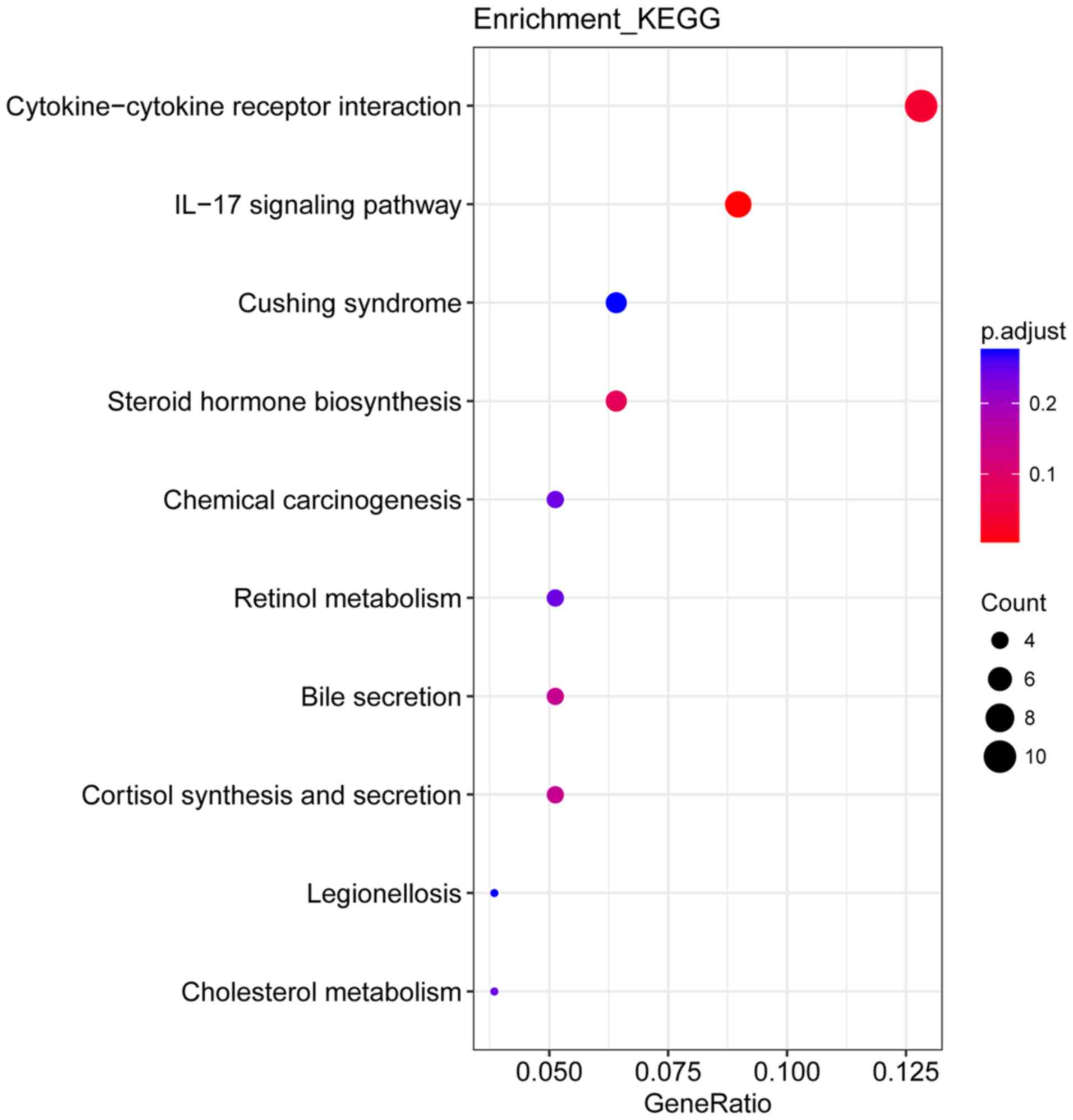
terms were identified (P<0.05) and can be found in Fig. 5. A total of 10 KEGG pathways were
enriched, of which ‘cytokine-cytokine receptor interaction’ and
‘interleukin (IL)-17 signaling pathway’ were significantly enriched
(P<0.05; Fig. 6).
Discussion
SLE is a systemic disease that can affect several
organs. During the course of the disease, there is a high incidence
of kidney damage, which is referred to as LN. Kidney damage is a
considerable cause of mortality and disability in patients with
SLE, and includes glomerular, tubular, renal interstitial and blood
vessel destruction (5,24).
Expansion and/or disruption of the intraglomerular
extracellular matrix (ECM) is a recognized phenomenon which occurs
during the development of LN, and may influence the deposition of
immune complexes in the renal system. The factors mediating this
process, and the structure and composition of the affected regions
require further investigation. Increased or altered synthesis of
ECM components and/or their accumulation could potentially serve a
role in LN, although the contribution made by each of these factors
remains unknown (25).
ECM accumulation results in mesangial expansion,
tubulointerstitial fibrosis and irreversible deterioration of renal
function (26–29). The epithelial-mesenchymal
transition (EMT) of renal tubular epithelial cells is one of the
underlying mechanisms of renal fibrosis, and encompasses a range of
events whereby epithelial cells no longer exhibit certain
epithelial traits, including E-cadherin expression. Instead, the
cells acquire characteristics typical of mesenchymal cells,
including the expression of α-SMA (30,31).
In addition, vimentin has also been reported as a potential novel
biomarker of renal fibrosis (32).
In the present study, the SIRM group exhibited a low
kidney index, reduced fibrosis, downregulated expression of
EMT-associated factors and vimentin, and improved survival in LN
mice. To reveal the molecular mechanism underlying the effects of
sirolimus in LN mice, transcriptomic analysis of sirolimus-treated
mice was conducted using RNA-sequencing and DEG identification and
annotation. Upon GO functional enrichment, terms associated with
‘biological process’, ‘molecular function’ and ‘cellular component’
were identified. A total of 10 KEGG pathways were enriched,
including ‘Cytokine-cytokine receptor interaction’ and ‘IL-17
signaling pathway’.
Based on the transcriptomics results of the present
study, future studies should aim to further investigate the
specific mechanism underlying the effects of sirolimus in LN and to
discover novel drug targets for the treatment of this disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Clinical
Pharmacy Key Specialty Construction Project of Shanghai (grant no.
YZ2017/5), The Young Medical Talents of Wuxi (grant no. QNRC020),
Young Project of Wuxi Health and Family Planning Research (grant
no. Q201706), Wuxi science and technology development guidance plan
(medical and health care; grant no. CSZON1744) and the AOSAIKANG
pharmaceutical foundation (grant no. A201826).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL made substantial contributions to the conception
and design of the present study. DW, XC and MF performed the
experiments. DW and XC wrote, reviewed and edited the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Use Committee of the Children's Hospital of Fudan University
(Shanghai, China), and complied with our institutional
regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang DD, Lu JM, Li Q and Li ZP: Population
pharmacokinetics of tacrolimus in paediatric systemic lupus
erythematosus based on real-world study. J Clin Pharm Ther.
43:476–483. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takeuchi T, Tsuzaka K, Abe T, Yoshimoto K,
Shiraishi K, Kameda H and Amano K: T cell abnormalities in systemic
lupus erythematosus. Autoimmunity. 38:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rahman A and Isenberg DA: Systemic lupus
erythematosus. N Engl J Med. 358:929–939. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gurevitz SL, Snyder JA, Wessel EK, Frey J
and Williamson BA: Systemic lupus erythematosus: A review of the
disease and treatment options. Consult Pharm. 28:110–121. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
D'Cruz DP, Khamashta MA and Hughes GR:
Systemic lupus erythematosus. Lancet. 369:587–596. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liao R, Liu Q, Zheng Z, Fan J, Peng W,
Kong Q, He H, Yang S, Chen W, Tang X and Yu X: Tacrolimus protects
podocytes from injury in lupus nephritis partly by stabilizing the
cytoskeleton and inhibiting podocyte apoptosis. PLoS One.
10:e01327242015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bertsias G, Ioannidis JP, Boletis J,
Bombardieri S, Cervera R, Dostal C, Font J, Gilboe IM, Houssiau F,
Huizinga T, et al: EULAR recommendations for the management of
systemic lupus erythematosus. Report of a task force of the eular
standing committee for international clinical studies including
therapeutics. Ann Rheum Dis. 67:195–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ji Y, Chen S, Xiang B, Li K, Xu Z, Yao W,
Lu G, Liu X, Xia C, Wang Q, et al: Sirolimus for the treatment of
progressive kaposiform hemangioendothelioma: A multicenter
retrospective study. Int J Cancer. 141:848–855. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jung KS, Lee J, Park SH, Park JO, Park YS,
Lim HY, Kang WK and Kim ST: Pilot study of sirolimus in patients
with PIK3CA mutant/amplified refractory solid cancer. Mol Clin
Oncol. 7:27–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Djebli N, Rousseau A, Hoizey G, Rerolle
JP, Toupance O, Le Meur Y and Marquet P: Sirolimus population
pharmacokinetic/pharmacogenetic analysis and bayesian modelling in
kidney transplant recipients. Clin Pharmacokinet. 45:1135–1148.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiao Z, Shi XJ, Li ZD and Zhong MK:
Population pharmacokinetics of sirolimus in de novo Chinese adult
renal transplant patients. Br J Clin Pharmacol. 68:47–60. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wataya-Kaneda M, Nakamura A, Tanaka M,
Hayashi M, Matsumoto S, Yamamoto K and Katayama I: Efficacy and
safety of topical sirolimus therapy for facial angiofibromas in the
tuberous sclerosis complex: A randomized clinical trial. JAMA
Dermatol. 153:39–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang DD, Lu JM, Li YZ, Li Q and Li ZP:
Population pharmacokinetics of sirolimus in pediatric tuberous
sclerosis complex: From real world study. Int J Clin Exp Med.
11:12302–12309. 2018.
|
|
14
|
Zhou L, Du GS, Pan LC, Zheng YG, Liu ZJ,
Shi HD, Yang SZ, Shi XJ, Xuan M, Feng LK and Zhu ZD: Sirolimus
treatment for cirrhosis or hepatocellular carcinoma patients
accompanied by psoriasis after liver transplantation: A single
center experience. Oncol Lett. 14:7817–7824. 2017.PubMed/NCBI
|
|
15
|
Zhou L, Pan LC, Zheng YG, Du GS, Fu XQ,
Zhu ZD, Song JY, Liu ZJ, Su XZ, Chen W, et al: Novel strategy of
sirolimus plus thymalfasin and huaier granule on tumor recurrence
of hepatocellular carcinoma beyond the UCSF criteria following
liver transplantation: A single center experience. Oncol Lett.
16:4407–4417. 2018.PubMed/NCBI
|
|
16
|
Fu J, Wang Z, Lee K, Wei C, Liu Z, Zhang
M, Zhou M, Cai M, Zhang W, Chuang PY, et al: Transcriptomic
analysis uncovers novel synergistic mechanisms in combination
therapy for lupus nephritis. Kidney Int. 93:416–429. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yap DYH, Tang C, Chan GCW, Kwan LPY, Ma
MKM, Mok MMY and Chan TM: Longterm data on sirolimus treatment in
patients with lupus nephritis. J Rheumatol. 45:1663–1670. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu J, Yang L, Hou Y, Soteyome T, Zeng B,
Su J, Li L, Li B, Chen D, Li Y, et al: Transcriptomics study on
staphylococcus aureus biofilm under low concentration of
ampicillin. Front Microbiol. 9:24132018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Burden CJ, Qureshi SE and Wilson SR: Error
estimates for the analysis of differential expression from RNA-seq
count data. PeerJ. 2:e5762014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seshan SV and Jennette JC: Renal disease
in systemic lupus erythematosus with emphasis on classification of
lupus glomerulonephritis: Advances and implications. Arch Pathol
Lab Med. 133:233–248. 2009.PubMed/NCBI
|
|
25
|
Tveita A, Rekvig OP and Zykova SN:
Glomerular matrix metalloproteinases and their regulators in the
pathogenesis of lupus nephritis. Arthritis Res Ther. 10:2292008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kolset SO, Reinholt FP and Jenssen T:
Diabetic nephropathy and extracellular matrix. J Histochem
Cytochem. 60:976–986. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang D, Zhang G, Chen X, Wei T, Liu C,
Chen C, Gong Y and Wei Q: Sitagliptin ameliorates diabetic
nephropathy by blocking TGF-beta1/Smad signaling pathway. Int J Mol
Med. 41:2784–2792. 2018.PubMed/NCBI
|
|
28
|
Chen X, Wang DD, Wei T, He SM, Zhang GY
and Wei QL: Effects of astragalosides from radix astragali on high
glucose-induced proliferation and extracellular matrix accumulation
in glomerular mesangial cells. Exp Ther Med. 11:2561–2566. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang GY, Wang DD, Cao Z, Wei T, Liu CX
and Wei QL: Sitagliptin ameliorates high glucose-induced cell
proliferation and expression of the extracellular matrix in
glomerular mesangial cells. Exp Ther Med. 14:3862–3867. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen X, Yang Y, Liu C, Chen Z and Wang D:
Astragaloside IV ameliorates high glucoseinduced renal tubular
epithelialmesenchymal transition by blocking mTORC1/p70S6K
signaling in HK2 cells. Int J Mol Med. 43:709–716. 2019.PubMed/NCBI
|
|
31
|
Badid C, Desmouliere A, Babici D,
Hadj-Aissa A, McGregor B, Lefrancois N, Touraine JL and Laville M:
Interstitial expression of alpha-SMA: An early marker of chronic
renal allograft dysfunction. Nephrol Dial Transplant. 17:1993–1998.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao YH, Lv LL, Zhang X, Hu H, Ding LH, Yin
D, Zhang YZ, Ni HF, Chen PS and Liu BC: Urinary vimentin mRNA as a
potential novel biomarker of renal fibrosis. Am J Physiol Renal
Physiol. 309:F514–F522. 2015. View Article : Google Scholar : PubMed/NCBI
|