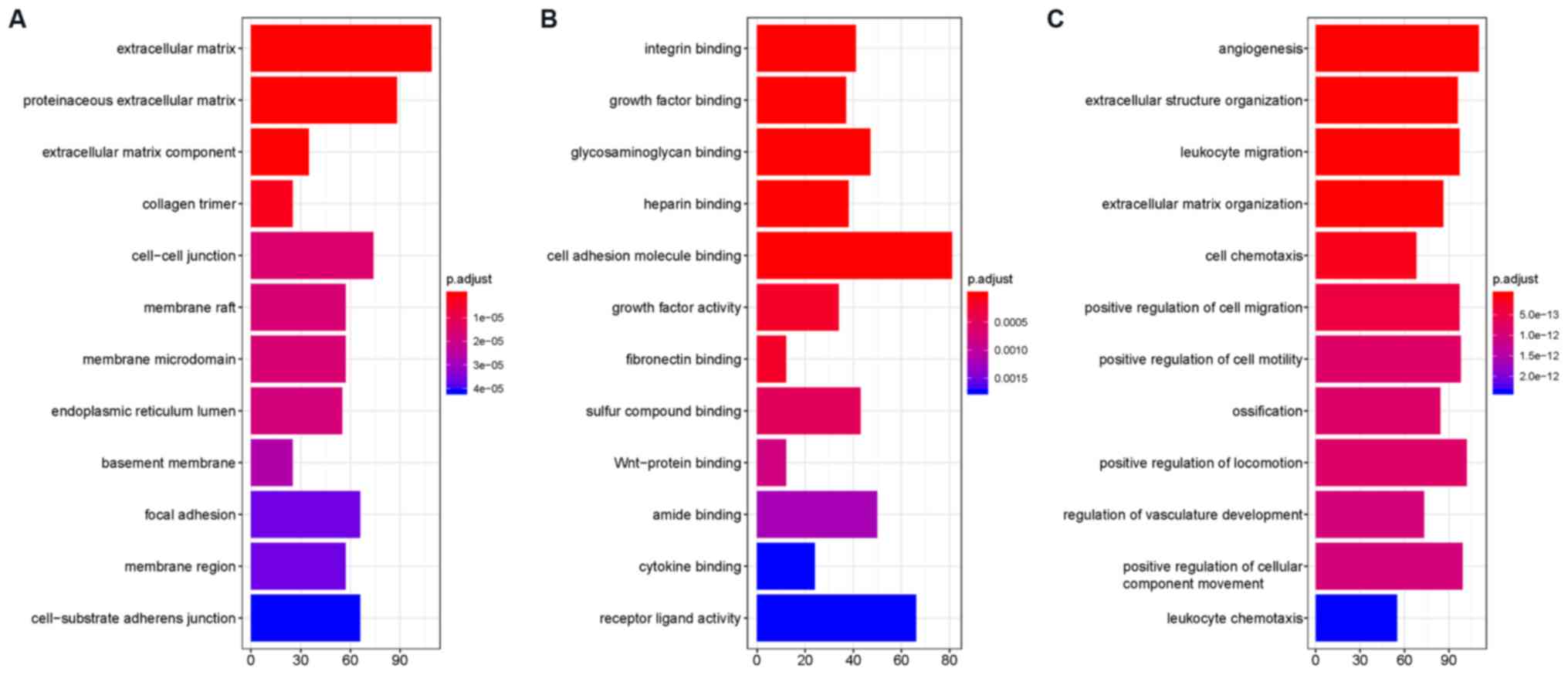
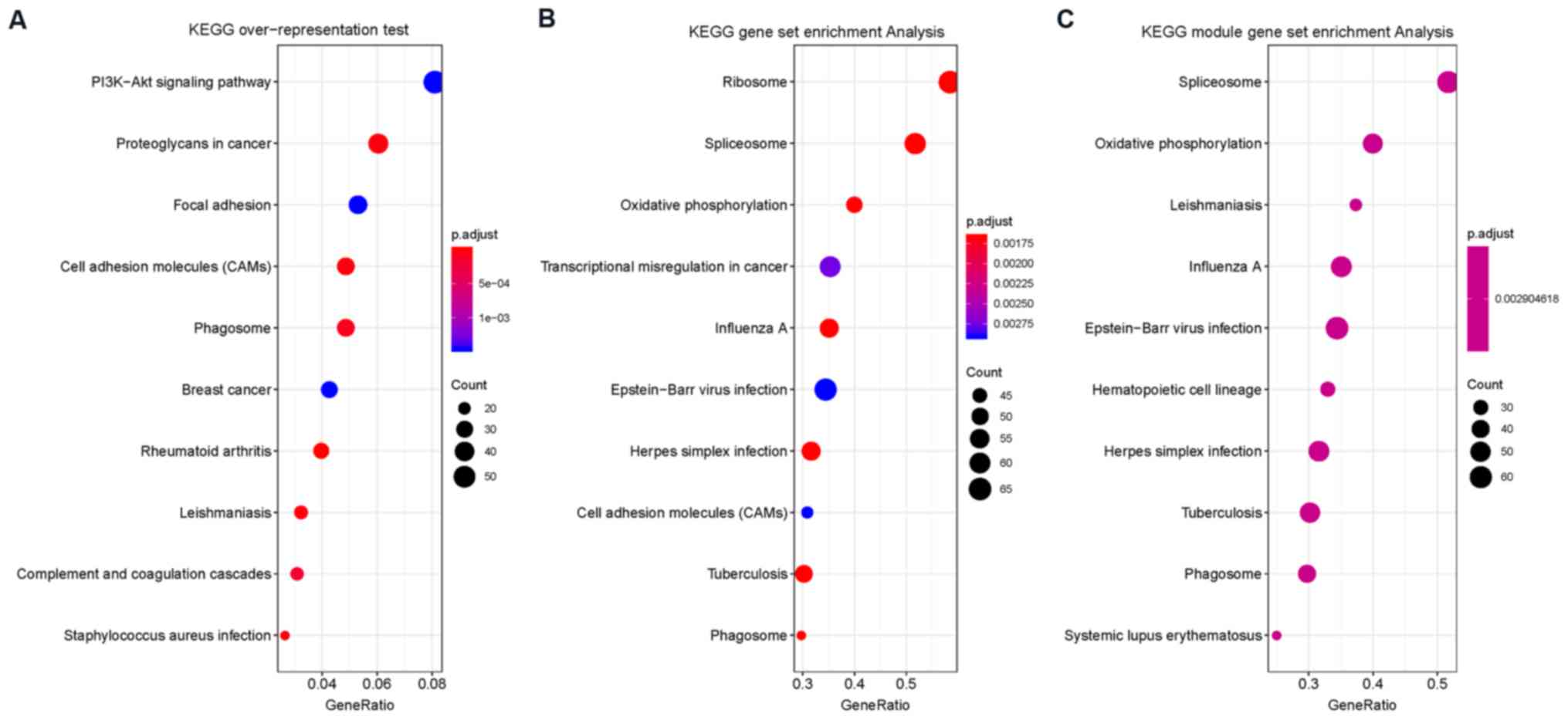
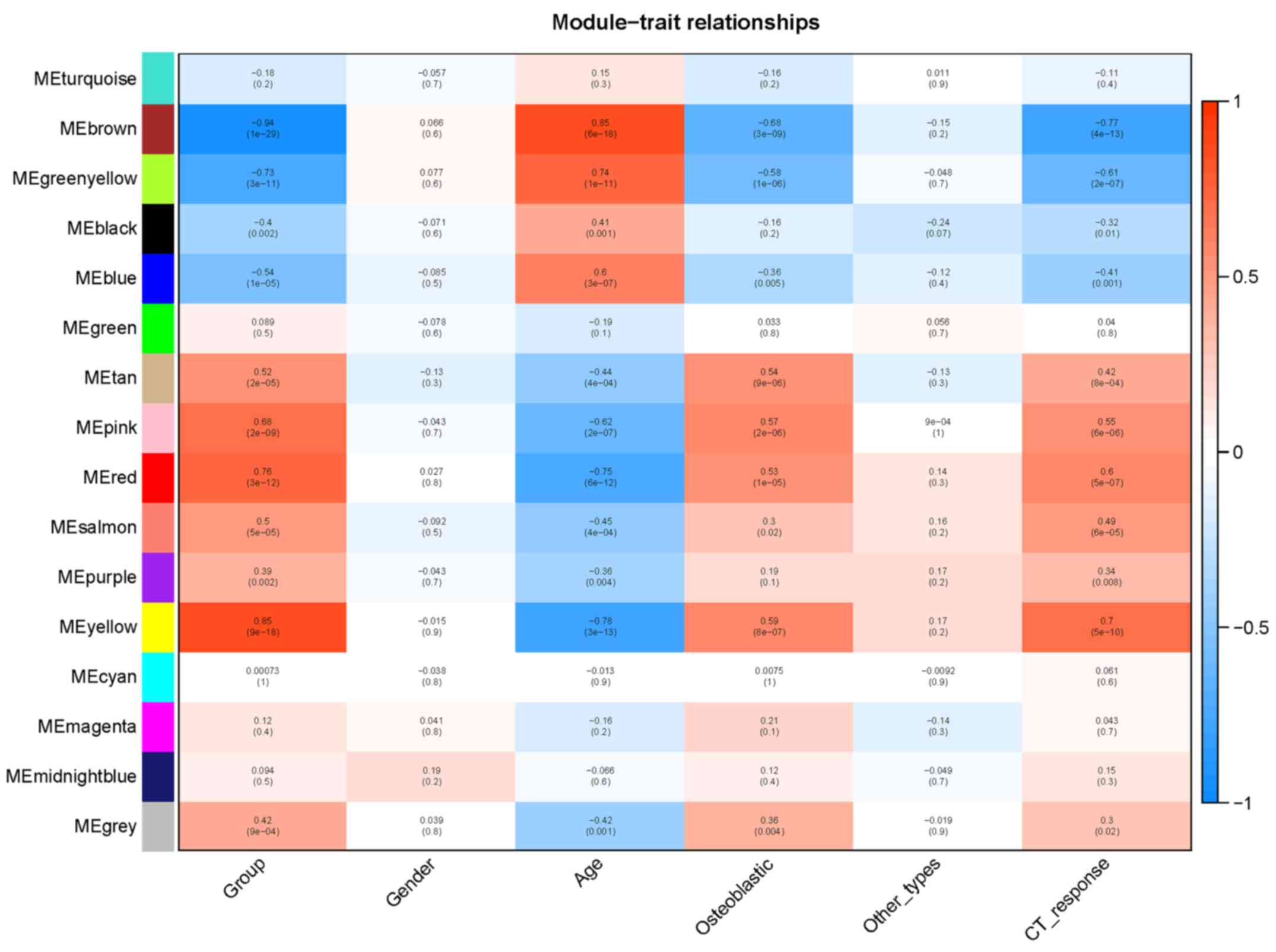
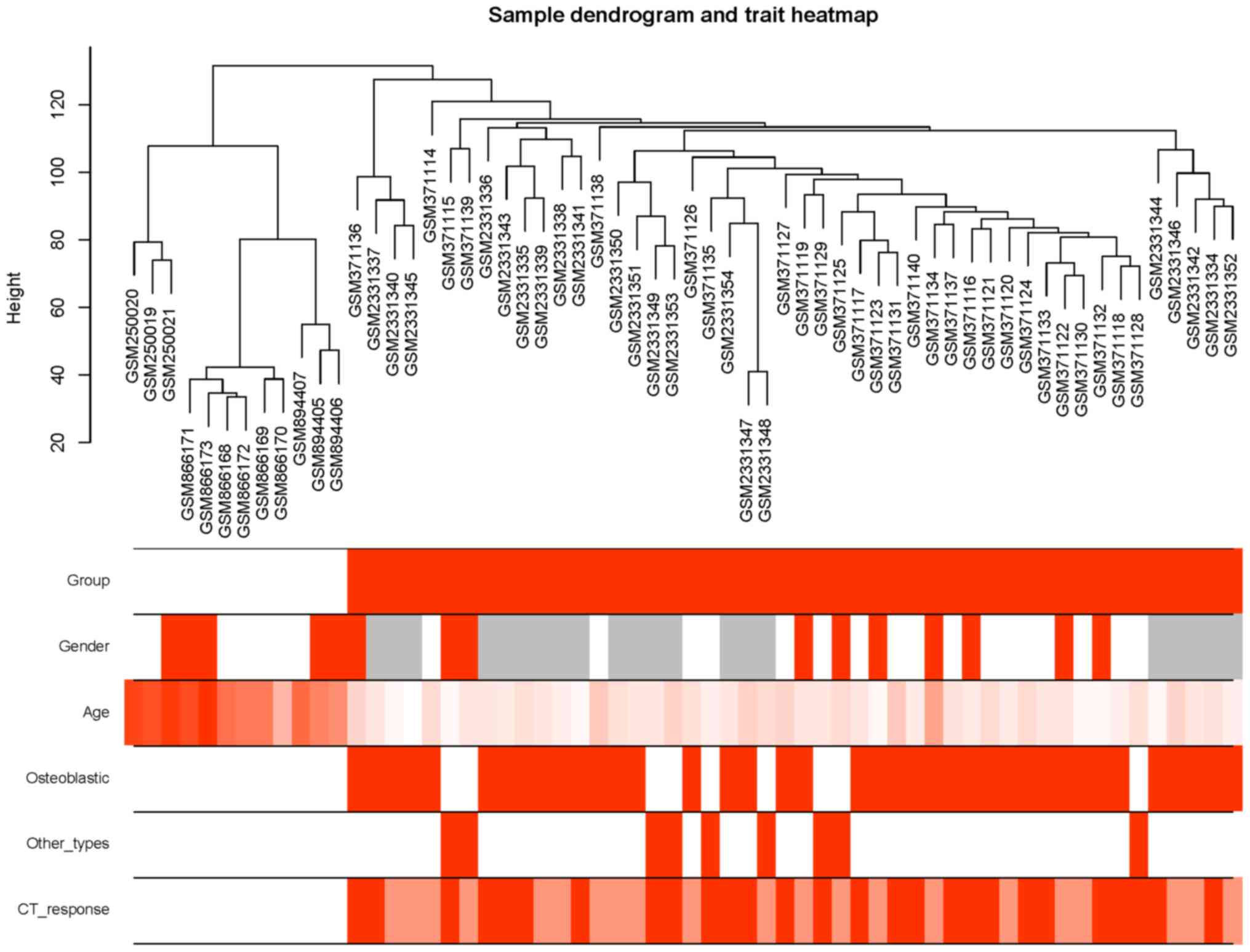
|
1
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Pediatric and adolescent osteosarcoma, Springer.
3–13. 2009. View Article : Google Scholar
|
|
3
|
Grimer RJ: Surgical options for children
with osteosarcoma. Lancet Oncol. 6:85–92. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Akiyama T, Dass CR and Choong PF: Novel
therapeutic strategy for osteosarcoma targeting osteoclast
differentiation, bone-resorbing activity and apoptosis pathway. Mol
Cancer Ther. 7:3461–3469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the Surveillance, Epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kempf-Bielack B, Bielack SS, Jürgens H,
Branscheid D, Berdel WE, Exner GU, Göbel U, Helmke K, Jundt G,
Kabisch H, et al: Osteosarcoma relapse after combined modality
therapy: An analysis of unselected patients in the Cooperative
Osteosarcoma Study Group (COSS). J Clin Oncol. 23:559–568. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kruzelock RP, Murphy EC, Strong LC, Naylor
SL and Hansen MF: Localization of a novel tumor suppressor locus on
human chromosome 3q important in osteosarcoma tumorigenesis. Cancer
Res. 57:106–159. 1997.PubMed/NCBI
|
|
8
|
Morrow JJ and Khanna C: Osteosarcoma
genetics and epigenetics: Emerging biology and candidate therapies.
Crit Rev Oncog. 20:173–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Belchis DA, Meece CA, Benko FA, Rogan PK,
Williams RA and Gocke CD: Loss of heterozygosity and microsatellite
instability at the retinoblastoma locus in osteosarcomas. Diagn Mol
Pathol. 5:214–219. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Benassi MS, Molendini L, Gamberi G,
Ragazzini P, Sollazzo MR, Merli M, Asp J, Magagnoli G, Balladelli
A, Bertoni F and Picci P: Alteration of pRb/p16/cdk4 regulation in
human osteosarcoma. Int J Cancer. 84:489–493. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Durfee RA, Mohammed M and Luu HH: Review
of osteosarcoma and current management. Rheumatol Ther. 3:221–243.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lage K, Hansen NT, Karlberg EO, Eklund AC,
Roque FS, Donahoe PK, Szallasi Z, Jensen TS and Brunak S: A
large-scale analysis of tissue-specific pathology and gene
expression of human disease genes and complexes. Proc Natl Acad Sci
USA. 105:20870–20875. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang S and Cao J: A close examination of
double filtering with fold change and t test in microarray
analysis. BMC Bioinformatics. 10:4022009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reiner A, Yekutieli D and Benjamini Y:
Identifying differentially expressed genes using false discovery
rate controlling procedures. Bioinformatics. 19:368–375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:172005. View Article : Google Scholar
|
|
16
|
Whirl-Carrillo M, McDonagh EM, Hebert JM,
Gong L, Sangkuhl K, Thorn CF, Altman RB and Klein TE:
Pharmacogenomics knowledge for personalized medicine. Clin
Pharmacol Ther. 92:414–417. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chakravarty D, Gao J, Phillips SM, Kundra
R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et
al: OncoKB: A precision oncology knowledge base. JCO Precis Oncol.
May 16–2017.(Epub ahead of print). doi: 10.1200/PO.17.00011.
View Article : Google Scholar
|
|
18
|
Griffith M, Spies NC, Krysiak K, McMichael
JF, Coffman AC, Danos AM, Ainscough BJ, Ramirez CA, Rieke DT, Kujan
L, et al: CIViC is a community knowledgebase for expert
crowdsourcing the clinical interpretation of variants in cancer.
Nat Genet. 49:170–174. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hamidouche Z, Fromigué O, Ringe J, Häupl
T, Vaudin P, Pagès JC, Srouji S, Livne E and Marie PJ: Priming
integrin alpha5 promotes human mesenchymal stromal cell osteoblast
differentiation and osteogenesis. Proc Natl Acad Sci USA.
106:18587–18591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kobayashi E, Masuda M, Nakayama R,
Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A,
Tochigi N, et al: Reduced argininosuccinate synthetase is a
predictive biomarker for the development of pulmonary metastasis in
patients with osteosarcoma. Mol Cancer Ther. 9:535–544. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guilloton F, Caron G, Ménard C, Pangault
C, Amé-Thomas P, Dulong J, De Vos J, Rossille D, Henry C, Lamy T,
et al: Mesenchymal stromal cells orchestrate follicular lymphoma
cell niche through the CCL2-dependent recruitment and polarization
of monocytes. Blood. 119:2556–2567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
André T, Meuleman N, Stamatopoulos B, De
Bruyn C, Pieters K, Bron D and Lagneaux L: Evidences of early
senescence in multiple myeloma bone marrow mesenchymal stromal
cells. PLoS One. 8:e597562013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vella S, Tavanti E, Hattinger CM, Fanelli
M, Versteeg R, Koster J, Picci P and Serra M: Targeting CDKs with
roscovitine increases sensitivity to DNA damaging drugs of human
osteosarcoma cells. PLoS One. 11:e01662332016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Carlson M, Falcon S, Pages H and Li N:
hgu133plus2. db: Affymetrix human genome U133 Plus 2.0 Array
annotation data (chip hgu133plus2). R package version 2. 2012.
|
|
29
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu G, Wang LG, Yan GR and He QY: DOSE: An
R/Bioconductor package for disease ontology semantic and enrichment
analysis. Bioinformatics. 31:608–609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Metsalu T and Vilo J: ClustVis: A web tool
for visualizing clustering of multivariate data using Principal
Component Analysis and heatmap. Nucleic Acids Res. 43:W566–W570.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen B, Khodadoust MS, Liu CL, Newman AM
and Alizadeh AA: Profiling tumor infiltrating immune cells with
CIBERSORT. Methods Mol Biol. 1711:243–259. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pickup MW, Mouw JK and Weaver VM: The
extracellular matrix modulates the hallmarks of cancer. EMBO Rep.
15:1243–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schlegel J, Sambade MJ, Sather S, Moschos
SJ, Tan AC, Winges A, DeRyckere D, Carson CC, Trembath DG, Tentler
JJ, et al: MERTK receptor tyrosine kinase is a therapeutic target
in melanoma. J Clin Invest. 123:2257–2267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu Y, Gaillard S, Phillip JM, Huang TC,
Pinto SM, Tessarollo NG, Zhang Z, Pandey A, Wirtz D, Ayhan A, et
al: Inhibition of spleen tyrosine kinase potentiates
paclitaxel-induced cytotoxicity in ovarian cancer cells by
stabilizing microtubules. Cancer Cell. 28:82–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matsusaka S, Cao S, Hanna DL, Sunakawa Y,
Ueno M, Mizunuma N, Zhang W, Yang D, Ning Y, Stintzing S, et al:
CXCR4 polymorphism predicts progression-free survival in metastatic
colorectal cancer patients treated with first-line
bevacizumab-based chemotherapy. Pharmacogenomics J. 17:543–550.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Musolino A, Naldi N, Bortesi B, Pezzuolo
D, Capelletti M, Missale G, Laccabue D, Zerbini A, Camisa R,
Bisagni G, et al: Immunoglobulin G fragment C receptor
polymorphisms and clinical efficacy of trastuzumab-based therapy in
patients with HER-2/neu-positive metastatic breast cancer. J Clin
Oncol. 26:1789–1796. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tamura K, Shimizu C, Hojo T, Akashi-Tanaka
S, Kinoshita T, Yonemori K, Kouno T, Katsumata N, Ando M, Aogi K,
et al: FcγR2A and 3A polymorphisms predict clinical outcome of
trastuzumab in both neoadjuvant and metastatic settings in patients
with HER2-positive breast cancer. Ann Oncol. 22:1302–1307. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pander J, van Huis-Tanja L, Böhringer S,
van der Straaten T, Gelderblom H, Punt C and Guchelaar HJ: Genome
wide association study for predictors of progression free survival
in patients on capecitabine, oxaliplatin, bevacizumab and cetuximab
in first-line therapy of metastatic colorectal cancer. PLoS One.
10:e01310912015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hartmaier RJ, Richter AS, Gillihan RM,
Sallit JZ, McGuire SE, Wang J, Lee AV, Osborne CK, O'Malley BW,
Brown PH, et al: A SNP in steroid receptor coactivator-1 disrupts a
GSK3β phosphorylation site and is associated with altered tamoxifen
response in bone. Mol Endocrinol. 26:220–177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pascual T, Apellániz-Ruiz M, Pernaut C,
Cueto-Felgueroso C, Villalba P, Álvarez C, Manso L, Inglada-Pérez
L, Robledo M, Rodríguez-Antona C and Ciruelos E: Polymorphisms
associated with everolimus pharmacokinetics, toxicity and survival
in metastatic breast cancer. PLoS One. 12:e01801922017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Volz N, Stintzing S, Zhang W, Yang D, Ning
Y, Wakatsuki T, El-Khoueiry RE, Li JE, Kardosh A, Loupakis F, et
al: Genes involved in pericyte-driven tumor maturation predict
treatment benefit of first-line FOLFIRI plus bevacizumab in
patients with metastatic colorectal cancer. Pharmacogenomics J.
15:69–76. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rokavec M, Schroth W, Amaral SM, Fritz P,
Antoniadou L, Glavac D, Simon W, Schwab M, Eichelbaum M and Brauch
H: A polymorphism in the TC21 promoter associates with an
unfavorable tamoxifen treatment outcome in breast cancer. Cancer
Res. 68:9799–9808. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Glynn SA, Boersma BJ, Howe TM, Edvardsen
H, Geisler SB, Goodman JE, Ridnour LA, Lønning PE, Børresen-Dale
AL, Naume B, et al: A mitochondrial target sequence polymorphism in
manganese superoxide dismutase predicts inferior survival in breast
cancer patients treated with cyclophosphamide. Clin Cancer Res.
15:4165–4173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bosó V, Herrero MJ, Santaballa A, Palomar
L, Megias JE, de la Cueva H, Rojas L, Marqués MR, Poveda JL,
Montalar J and Aliño SF: SNPs and taxane toxicity in breast cancer
patients. Pharmacogenomics. 15:1845–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
DiLeo MV, Strahan GD, den Bakker M and
Hoekenga OA: Weighted correlation network analysis (WGCNA) applied
to the tomato fruit metabolome. PLoS One. 6:e266832011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zlotnik A, Burkhardt AM and Homey B:
Homeostatic chemokine receptors and organ-specific metastasis. Nat
Rev Immunol. 11:597–606. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Holm NT, Byrnes K, Li BD, Turnage RH,
Abreo F, Mathis JM and Chu QD: Elevated levels of chemokine
receptor CXCR4 in HER-2 negative breast cancer specimens predict
recurrence. J Surg Res. 141:53–59. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jiang YP, Wu XH, Shi B, Wu WX and Yin GR:
Expression of chemokine CXCL12 and its receptor CXCR4 in human
epithelial ovarian cancer: An independent prognostic factor for
tumor progression. Gynecol Oncol. 103:226–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Longo-Imedio MI, Longo N, Treviño I,
Lázaro P and Sánchez-Mateos P: Clinical significance of CXCR3 and
CXCR4 expression in primary melanoma. Int J Cancer. 117:861–865.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao H, Guo L, Zhao H, Zhao J, Weng H and
Zhao B: CXCR4 over-expression and survival in cancer: A system
review and meta-analysis. Oncotarget. 6:5022–5040. 2015.PubMed/NCBI
|
|
54
|
Akashi T, Koizumi K, Tsuneyama K, Saiki I,
Takano Y and Fuse H: Chemokine receptor CXCR4 expression and
prognosis in patients with metastatic prostate cancer. Cancer Sci.
99:539–542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xie L, Wei J, Qian X, Chen G, Yu L, Ding Y
and Liu B: CXCR4, a potential predictive marker for docetaxel
sensitivity in gastric cancer. Anticancer Res. 30:2209–2216.
2010.PubMed/NCBI
|
|
56
|
Yang Z, Chen Y, Fu Y, Yang Y, Zhang Y,
Chen Y and Li D: Meta-analysis of differentially expressed genes in
osteosarcoma based on gene expression data. BMC Med Genet.
15:802014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu Y, Mi Y, Mueller T, Kreibich S,
Williams EG, Van Drogen A, Borel C, Frank M, Germain PL, Bludau I,
et al: Multi-omic measurements of heterogeneity in HeLa cells
across laboratories. Nat Biotechnol. 37:314–322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Venning FA, Wullkopf L and Erler JT:
Targeting ECM disrupts cancer progression. Front Oncol. 5:2242015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ivanov V, Ivanova S, Roomi M, Kalinovsky
T, Niedzwiecki A and Rath M: Naturally produced extracellular
matrix inhibits growth rate and invasiveness of human osteosarcoma
cancer cells. Med Oncol. 24:209–217. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lu P, Weaver VM and Werb Z: The
extracellular matrix: A dynamic niche in cancer progression. J Cell
Biol. 196:395–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Provenzano PP, Eliceiri KW, Campbell JM,
Inman DR, White JG and Keely PJ: Collagen reorganization at the
tumor-stromal interface facilitates local invasion. BMC Med.
4:382006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kim BG, An HJ, Kang S, Choi YP, Gao MQ,
Park H and Cho NH: Laminin-332-rich tumor microenvironment for
tumor invasion in the interface zone of breast cancer. Am J Pathol.
178:373–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee S, Stewart S, Nagtegaal I, Luo J, Wu
Y, Colditz G, Medina D and Allred DC: Differentially expressed
genes regulating the progression of ductal carcinoma in situ to
invasive breast cancer. Cancer Res. 72:4574–4586. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Miyata Y, Kanda S, Nomata K, Hayashida Y
and Kanetake H: Expression of metalloproteinase-2,
metalloproteinase-9, and tissue inhibitor of metalloproteinase-1 in
transitional cell carcinoma of upper urinary tract: Correlation
with tumor stage and survival. Urology. 63:602–608. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Akira S, Uematsu S and Takeuchi O:
Pathogen recognition and innate immunity. Cell. 124:783–801. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fabriek BO, van Bruggen R, Deng DM,
Ligtenberg AJ, Nazmi K, Schornagel K, Vloet RP, Dijkstra CD and van
den Berg TK: The macrophage scavenger receptor CD163 functions as
an innate immune sensor for bacteria. Blood. 113:887–892. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Héninger E, Krueger TE and Lang JM:
Augmenting antitumor immune responses with epigenetic modifying
agents. Front Immunol. 6:292015.PubMed/NCBI
|
|
68
|
Schwefel D, Arasu BS, Marino SF, Lamprecht
B, Köchert K, Rosenbaum E, Eichhorst J, Wiesner B, Behlke J, Rocks
O, et al: Structural insights into the mechanism of GTPase
activation in the GIMAP family. Structure. 21:550–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fischer U, Michel A and Meese EU:
Expression of the gene for hematopoietic cell specific protein is
not restricted to cells of hematopoietic origin. Int J Mol Med.
15:611–615. 2005.PubMed/NCBI
|
|
70
|
Morris P, Shaman J, Attaya M, Amaya M,
Goodman S, Bergman C, Monaco JJ and Mellins E: An essential role
for HLA-DM in antigen presentation by class II major
histocompatibility molecules. Nature. 368:551–554. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fling SP, Arp B and Pious D: HLA-DMA and
-DMB genes are both required for MHC class II/peptide complex
formation in antigen-presenting cells. Nature. 368:554–548. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kinseth MA, Jia Z, Rahmatpanah F, Sawyers
A, Sutton M, Wang-Rodriguez J, Mercola D and McGuire KL: Expression
differences between African American and Caucasian prostate cancer
tissue reveals that stroma is the site of aggressive changes. Int J
Cancer. 134:81–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wu Y, Tworkoski K, Michaud M and Madri JA:
Bone marrow monocyte PECAM-1 deficiency elicits increased
osteoclastogenesis resulting in trabecular bone loss. J Immunol.
182:2672–2679. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kaplan R, Morse B, Huebner K, Croce C,
Howk R, Ravera M, Jaye M and Schlessinger J: Cloning of three human
tyrosine phosphatases reveals a multigene family of receptor-linked
protein-tyrosine-phosphatases expressed in brain. Proc Natl Acad
Sci USA. 87:7000–7004. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zuccotti P, Cartelli D, Stroppi M, Pandini
V, Venturin M, Aliverti A, Battaglioli E, Cappelletti G and Riva P:
Centaurin-α2 interacts with β-tubulin and stabilizes microtubules.
PLoS One. 7:e528672012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Autieri MV, Carbone C and Mu A: Expression
of allograft inflammatory factor-1 is a marker of activated human
vascular smooth muscle cells and arterial injury. Arterioscler
Thromb Vasc Biol. 20:1737–1744. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Talmadge JE and Gabrilovich DI: History of
myeloid-derived suppressor cells. Nat Rev Cancer. 13:739–752. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kelleher FC and O'Sullivan H: Monocytes,
macrophages and osteoclasts in osteosarcoma. J Adolesc Young Adult
Oncol. 6:396–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Han Q, Shi H and Liu F: CD163(+) M2-type
tumor-associated macrophage support the suppression of
tumor-infiltrating T cells in osteosarcoma. Int Immunopharmacol.
34:101–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Syn NL, Teng MWL, Mok TSK and Soo RA:
De-novo and acquired resistance to immune checkpoint targeting.
Lancet Oncol. 18:e731–e741. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Lussier DM, O'Neill L, Nieves LM, McAfee
MS, Holechek SA, Collins AW, Dickman P, Jacobsen J, Hingorani P and
Blattman JN: Enhanced T-cell immunity to osteosarcoma through
antibody blockade of PD-1/PD-L1 interactions. J Immunother.
38:96–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yoshida A, Minowa MT, Takamatsu S, Hara T,
Oguri S, Ikenaga H and Takeuchi M: Tissue specific expression and
chromosomal mapping of a human UDP-N-acetylglucosamine:
Alpha1,3-d-mannoside beta1, 4-N-acetylglucosaminyltransferase.
Glycobiology. 9:303–310. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Buddingh EP, Kuijjer ML, Duim RA, Bürger
H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PC,
Lankester AC and Cleton-Jansen AM: Tumor-infiltrating macrophages
are associated with metastasis suppression in high-grade
osteosarcoma: A rationale for treatment with macrophage activating
agents. Clin Cancer Res. 17:2110–2119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
van Ravenswaay Claasen HH, Kluin PM and
Fleuren GJ: Tumor infiltrating cells in human cancer. On the
possible role of CD16+ macrophages in antitumor cytotoxicity. Lab
Invest. 67:166–174. 1992.PubMed/NCBI
|