Introduction
Breast cancer is one of the most common malignant
tumors in females. It has the highest incidence among this
population, and the age of onset is decreasing. The lifetime risk
of having breast cancer is 10% in women (1). Approximately 500,000 people succumb
to mortality each year from breast cancer, and the mortality rate
increases by 2–3% annually (2).
Early diagnosis and treatment have been particularly important in
breast cancer as the options for primary prevention are limited.
Identifying and studying the genes responsible for the growth and
malignancy of breast cancer is critical for disease therapy.
MicroRNAs (miRs) bind to target genes involved in
such pathologic processes as cell proliferation, differentiation
and apoptosis, and affect their expression in the tissues and cells
(3). miR-125a-5p has been reported
to be abnormally expressed in prostate carcinoma (4). Various studies have shown miR-125a-5p
to be closely related to tumor cell growth, differentiation and
metastasis (5–7). Nevertheless, its effects on the
proliferation, migration, and apoptosis of breast cancer cells and
the underlying mechanisms remain unknown.
Phosphatase and tensin homolog (PTEN) is a
well-characterized tumor suppressor, and it plays a key role in
cell proliferation, migration and cell renewal by regulating the
mitogen-activated protein kinase kinase (MEK)1/2/ERK1/2 pathway
(8). The MEK1/2/ERK1/2 signal
pathway and caspase-3 activation have been shown to be involved in
breast cancer cell proliferation, migration and apoptosis processes
(9). Studies revealed that
miR-125a-5p could regulate the MEK1/2/ERK1/2 signaling pathway in
multiple cell types (10,11). However, whether these proteins and
pathways are involved in the mechanisms underlying miR-125a-5p
regulation in breast cancer cells are unknown.
Thus, we aimed to evaluate the effects of
miR-125a-5p on the biological behaviors of MCF-7 breast cancer
cells and to investigate the underlying regulatory mechanisms.
Materials and methods
Cell culture and grouping
Human MCF-7 breast cancer cells were purchased from
Shanghai BioLeaf Biotech. The MCF-7 breast cancer cell line was
cultured in RPMI-1640 medium containing 10% fetal bovine serum
(Hyclone; GE Healthcare Life Sciences), 100 U/ml penicillin, and
100 µg/ml streptomycin in an incubator with 5% CO2 and
95% humidity at 37°C. Trypsin (0.25%) was used to digest the cells
when 80% of the cells adhered to the culture dish and for the
subculture, which was performed every 2–3 days.
Lentiviral-mediated transfection to
establish stable MCF-7 cell lines with miR-125a-5p overexpression
or reduced miR-125a-5p expression
MCF-7 cells were infected with green fluorescent
protein (GFP)-expressing lentivirus (1×107 multiplicity
of infection) that also contained miR-125a-5p overexpression vector
(LV-miR-125a-5p), miR-125a-5p silencing small interfering (si)RNA
(siRNA-LV-miR-125a-5p), and a scrambled sequence (LV-NC). The
medium was replaced 24 h after transfection, and the GFP-labeled
cells were counted 72 h after transfection. The positive cells were
screened with puromycin (final concentration of 1 mg/l) to
eventually produce MCF-7 cells with miR-125a-5p overexpression
(MCF-7miR-125a-5p cells), MCF-7 cells with miR-125a-5p
knockdown (MCF-7SimiR-125a-5p cells), and stable
LV-scramble infected MCF-7 cells (MCF-7NC), which served
as the control.
Detection of miR-125a-5p expression in
MCF-7 cells
The MCF-7miR-125a-5p,
MCF-7SimiR-125a-5p, and MCF-7NC cells were
seeded in 6-well plates. Once the MCF-7miR-125a-5p,
MCF-7SimiR-125a-5p and MCF-7NC cells reached
90% confluence, total RNAs of the harvested cells were extracted
using TRIzol® reagent (Thermo Fisher Scientific, Inc.) a
hairpin-it™ miRs reverse transcription-quantitative PCR (RT-qPCR)
kit was then used to synthesize miR-125a-5p cDNA. The RT primer
sequences were human miR-125a-5p-RT
(5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACCTGCAG-3′); U6-RT
(5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3′),
and the reverse transcription conditions were 25°C for 30 min, 42°C
for 30 min, and subsequently 85°C for 5 min. The products of RT
were used as templates for PCR. The primer sequences of qPCR were
5′-ACACTCCAGCTGGGTCCCTGAGACCCTTAA-3′ (miR-125a-5p forward primer),
5′-CTCAACTGGTGTGGAGT-3′ (miR-125a-5p reverse primer),
5′-CTCGCTTCGGCAGCACA-3′ (internal reference gene U6 forward
primer), and 5′-AACGCTTCACGAATTTGCGT-3′ (internal reference gene U6
reverse primer). The PCR conditions were 95°C denaturation for 3
min, 40 cycles of 95°C denaturation for 12 sec and 60°C annealing
and elongation for 40 sec. The RT-qPCR procedures were performed in
strict accordance with the manufacturer's instructions. Six
replicate wells were performed in each group, and each experiment
was performed six times. The relative expression levels of target
genes were calculated using the 2−∆∆Cq method (12).
Assessment of MCF-7 cell
proliferation
The harvested MCF-7miR-125a-5p,
MCF-7SimiR-125a-5p and MCF-7NC cells were
independently inoculated in six wells in 96-well plates (each well
contained 100 µl complete cell culture medium with 2×103
cells and incubated at 37°C for 5 days. Cells were incubated in
RPMI-164 + 10% FBS (HyClone; GE Healthcare Life Sciences). The
viability of cells in different groups was measured daily via an
MTT assay for 5 days. For the MTT assay, 20 µl MTT solution was
added to each well of cells for incubation at 37°C for 4 h,
followed by removal of the solution and addition of 150 µl dimethyl
sulfoxide, followed by incubation at 37°C for 20 min; the
absorbance was then measured at 490 nm using a microplate reader.
This experiment was performed six times.
In vitro scratch assay to detect MCF-7
cell migration
The harvested MCF-7miR-125a-5p,
MCF-7SimiR-125a-5p and MCF-7NC cells were
independently inoculated and incubated in six-well plates (each
well contained 2 ml complete cell culture medium with
5×104 cells). Each group of cells was transferred into
serum-free medium once they reached 90% confluence. Then, a 200-µl
pipette tip was used to scratch a straight line in the middle of
the well. The scratch width in each well was measured under an
inverted microscope and the measurement points were recorded. The
scratch width at the same position was measured again after
incubated at 37°C for 16 h. The difference between the two
measurements in each well was expressed in terms of cell migration
distance after 16 h. This experiment was performed six times.
Quantitative analysis of migration was calculated using the
following equation: (Cell-free area at 0 h-cell-free area at 16
h)/cell area at 0 h ×100%.
MCF-7 apoptosis analysis
Cell apoptosis was analyzed by Hoechst 33258
staining and an Annexin V-phycoerythrin (PE)/ 7-amino-actinomycin
(7-AAD) apoptosis detection kit (BD Biosciences). The harvested
MCF-7miR-125a-5p, MCF-7SimiR-125a-5p and
MCF-7NC cells were independently inoculated and
incubated in six-well plates (each well contained 2 ml complete
cell culture medium with 5×104 cells). Once the cells
reached 90% confluence, the cells were transferred to serum-free
medium for 24 h incubation at 37°C to stimulate apoptosis, which
was assessed using an apoptotic cell Hoechst 33258 dye kit
according to the manufacturer's protocol (Beyotime Institute of
Biotechnology) and observed under a fluorescence microscope (Leica
Microsystems GmbH; magnification, ×400). The apoptotic cells per
well were counted in five different fields of vision. The
percentage of apoptotic cells per field of vision divided by the
total number of cells per field of vision was obtained as the
apoptotic rate.
For Annexin V-PE/7-AAD apoptosis detection, MCF-7
cells after different treatments were collected by using 0.25%
trypsin and centrifuged at 200 × g at room temperature for 3 min.
Cells were then washed with PBS and resuspended in 100 µl 1X
annexin-binding buffer, which was included in the kit.
Subsequently, 5 µl PE-conjugated Annexin V and 5 µl 7-AAD were
added to the cell suspension, and incubated at room temperature for
15 min. The rate of apoptosis in MCF-7 cells was detected by flow
cytometry. The cells stained only with Annexin V were considered as
MCF-7 cells in early apoptosis, and those stained for both Annexin
V and 7-AAD were considered as late apoptotic cells. In this study,
we defined the apoptotic cells as both early and late apoptotic
cells.
Western blotting analysis of protein
expression
Protein extraction
After collecting the MCF-7 cells by centrifugation
(200 × g and 4°C for 3 min), the MCF-7 cells were lysed with RIPA
buffer (Applygen Technologies, Inc.) containing protease inhibitor
and subsequently centrifuged at 13,000 × g and 4°C for 10 min to
collect the supernatant (total protein) for later use.
Western blotting
The extracted proteins were separated using 12%
SDS-PAGE and subsequently transferred to polyvinylidene difluoride
(PVDF) membranes. The membranes were blocked at room temperature
with Tris-buffered saline (TBS) containing 5% fat-free dry milk
(i.e., 50 mmol/l Tris, 150 mmol/l NaCl, pH 7.6, and 5% fat-free dry
milk) for 1 h. After a few washes in TBS containing 0.5% Tween-20
(TBST), the protein blots were incubated with β-actin (1:1,000;
cat. no. E02102001; EarthOx), PTEN (1:1,000; cat. no. 9188S; Cell
Signaling Technology, Inc.), MEK1/2 (1:500; cat. no. 4694S; Cell
Signaling Technology, Inc.), ERK1/2 (1:1,000; cat. no. 4695S; Cell
Signaling Technology, Inc.), B-cell lymphoma-2 (Bcl-2, 1:500, cat.
no. 3498S; Cell Signaling Technology, Inc.), and caspase-3 (1:400;
cat. no. 9664S; Cell Signaling Technology, Inc.) primary antibodies
at 4°C overnight. After a few washes in TBST, the protein blots
were incubated with 1:50,000 horseradish peroxidase
(HRP)-conjugated goat-anti-mouse secondary antibodies (cat. no.
7076S; Cell Signaling Technology, Inc.) or HRP-conjugated
goat-anti-rabbit secondary antibodies (cat. no. 7074S; Cell
Signaling Technology, Inc.) at room temperature for 1 h. After
three washes in TBST, the protein blots were developed in enhanced
chemiluminescence (Amersham) substrate, and then results were
analyzed under Quantity one 4.6 software (Bio-Rad Laboratories,
Inc.). β-actin was used as an internal reference protein. Relative
protein expression was measured based on the ratio of the target
protein absorbance to the β-actin protein absorbance. This
experiment was performed six times.
Statistical analysis
Data were presented as the mean ± standard
deviation. Statistical analyses were performed using a t-test,
one-way ANOVA followed by a least significant distance post-hoc
test. GraphPad Prism 5 software (GraphPad Software, Inc.) was used
for analyzing the data. P<0.05 was considered to indicate a
statistically significant difference.
Results
Overexpression and knockdown of
miR-125a-5p in MCF-7 cells
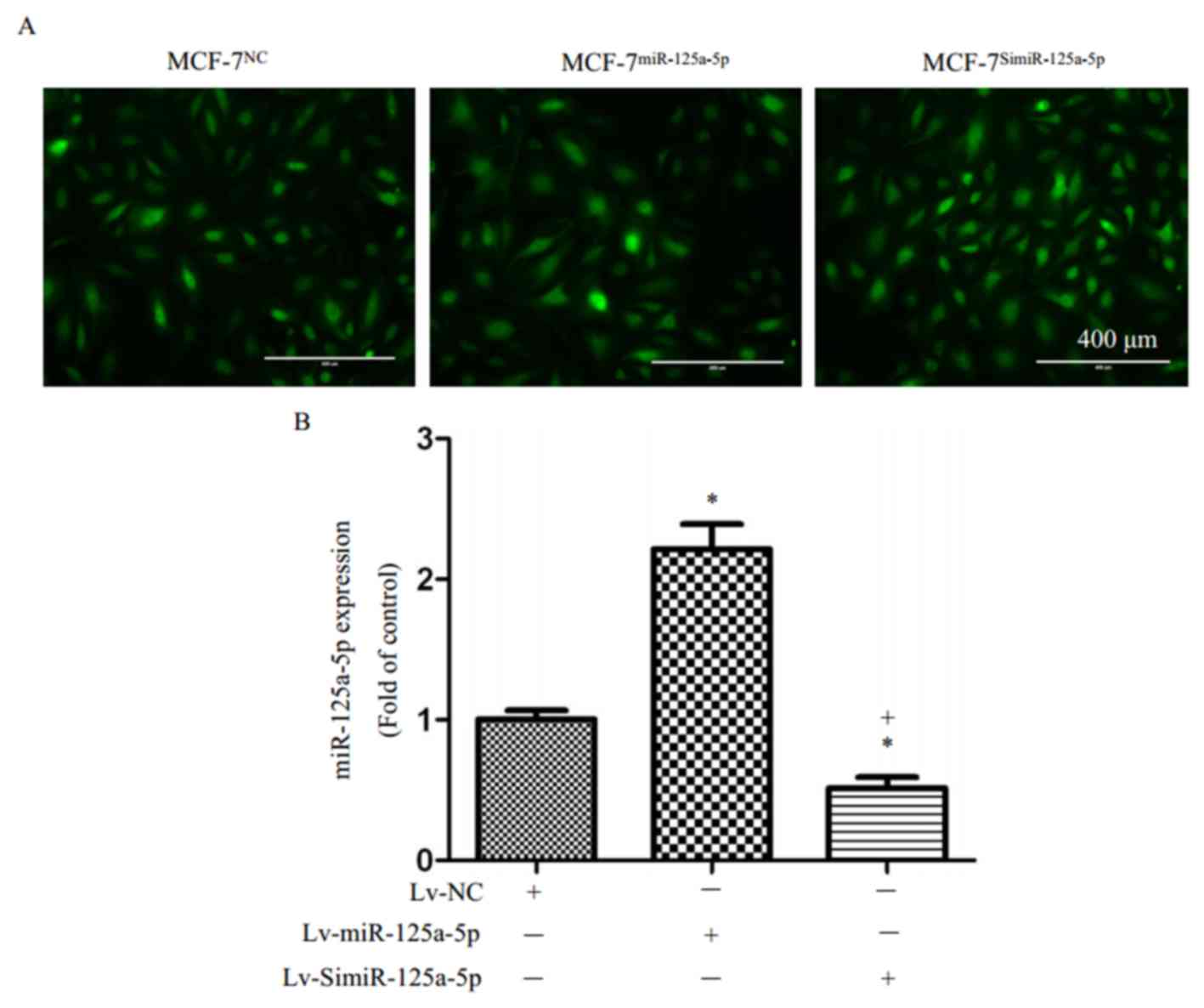
MCF-7 cells were labeled with green fluorescence
after transfection with LV-scramble, siRNA-LV-miR-125a-5p, or
LV-miR-125a-5p and selection of stable clones (Fig. 1A). The results of RT-qPCR showed
that miR-125a-5p expression in the MCF-7miR-125a-5p
cells was significantly higher than in the MCF-7NC cells
(P<0.05; Fig. 1B); in addition,
there miR-125a-5p expression was significantly reduced in the
MCF-7SimiR-125a-5p cells than in MCF-7NC
cells (P<0.05; Fig. 1B).
miR-125a-5p is involved in MCF-7 cell
proliferation and migration
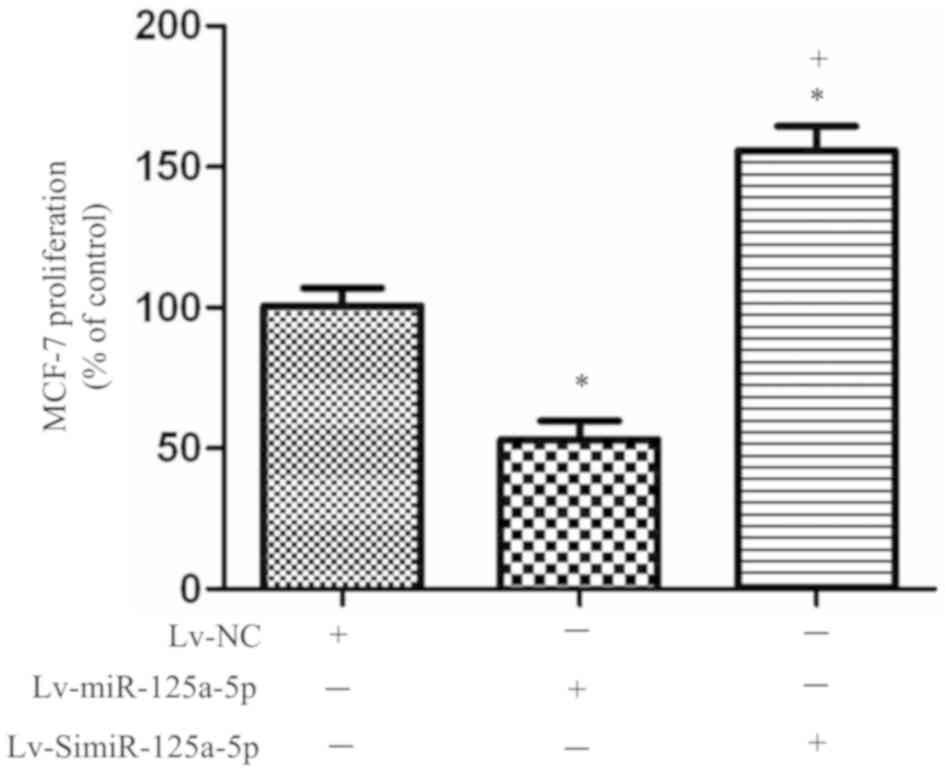
The results of the MTT assay showed the viability of
MCF-7 cells in different groups. Starting from the 3rd day of
experiment, the proliferation rate of the
MCF-7miR-125a-5p cells was significantly lower than that
of MCF-7NC cells (P<0.05; Fig. 2), while the proliferation rate of
the MCF-7simiR-125a-5p cells was significantly higher
than that of MCF-7NC cells (P<0.05; Fig. 2). This suggested that high
miR-125a-5p expression inhibited the proliferation of MCF-7 cells,
and low miR-125a-5p expression promoted this ability within MCF-7
cells.
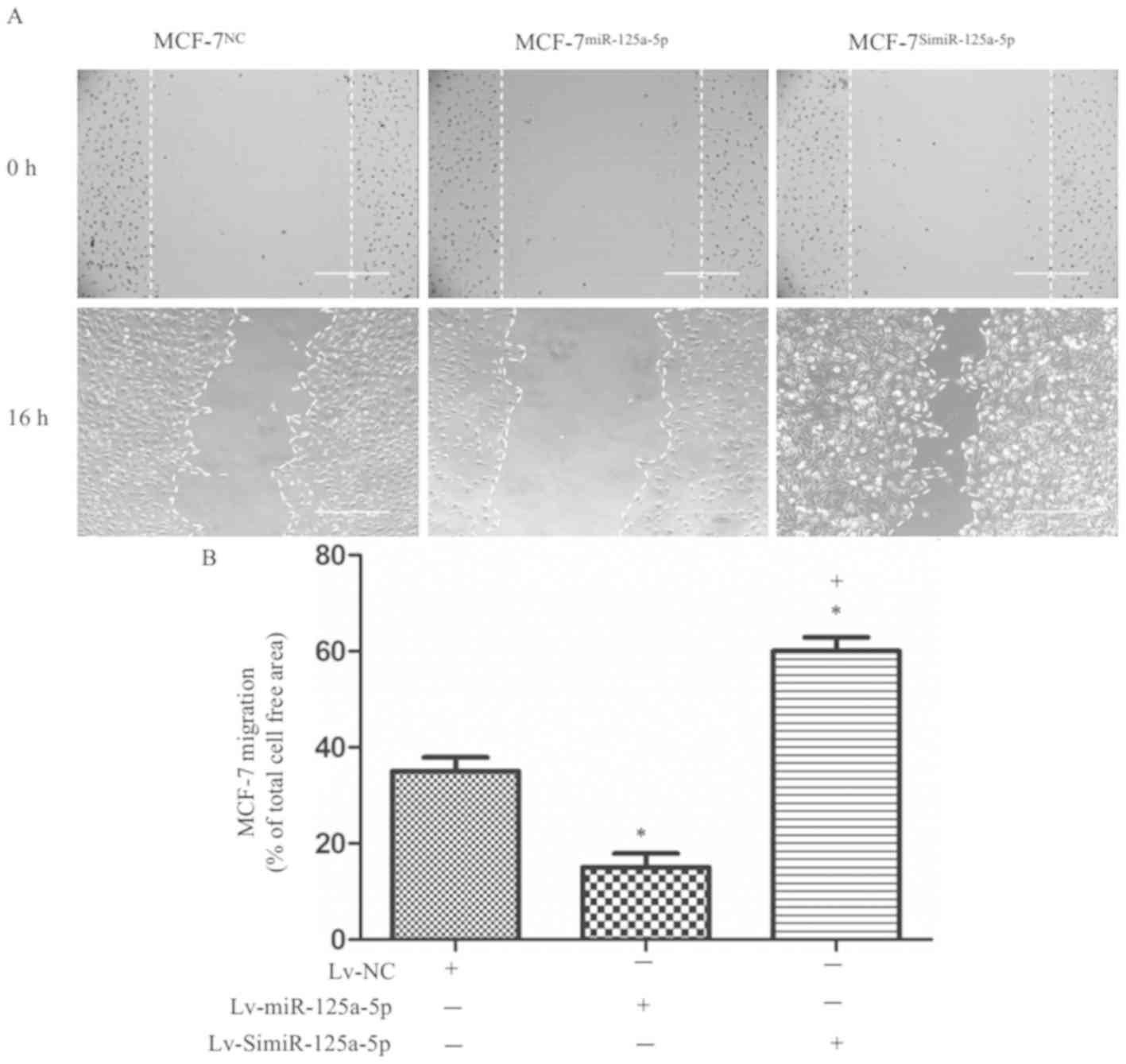
The results of in vitro scratch assay
revealed that migration ability of MCF-7miR-125a-5p
cells was significantly lower than the MCF-7NC cells
(P<0.05; Fig. 3A and B), while
that of the MCF-7SimiR-125a-5p cells was significantly
higher than the MCF-7NC cells (P<0.05; Fig. 3A and B). These results indicated
that high miR-125a-5p expression inhibited the migration of MCF-7
cells. Conversely, low miR-125a-5p expression in MCF-7 cells
resulted in enhanced migration of the cells.
miR-125a-5p increases
PTEN/MEK1/2/ERK1/2 pathway protein expression in MCF-7 cells
PTEN protein expression in
MCF-7miR-125a-5p cells was significantly higher than in
MCF-7NC cells (P<0.05; Fig. 4A and B). In contrast, significantly
reduced PTEN protein expression was exhibited by
MCF-7SimiR-125a-5p cells compared with
MCF-7NC and MCF-7miR-125a-5p cells
(P<0.05; Fig. 4A and B). In
addition, we found that overexpression of miR-125a-5p significantly
decreased the phosphorylation levels of MEK1/2 and ERK1/2 in MCF-7
cells compared with the MCF-7NC group (P<0.05;
Fig. 4A,C and D). Additionally,
silencing miR-125a-5p significantly increased the phosphorylation
levels of MEK1/2 and ERK1/2 in MCF-7 cells compared with the
MCF-7miR-125a-5p group (P<0.05, Fig. 4A,C and D). These data suggested
that the PTEN/MEK1/2/ERK1/2 signaling pathway may have contributed
to the effects of miR-125a-5p on inhibiting MCF-7 cell
proliferation and migration.
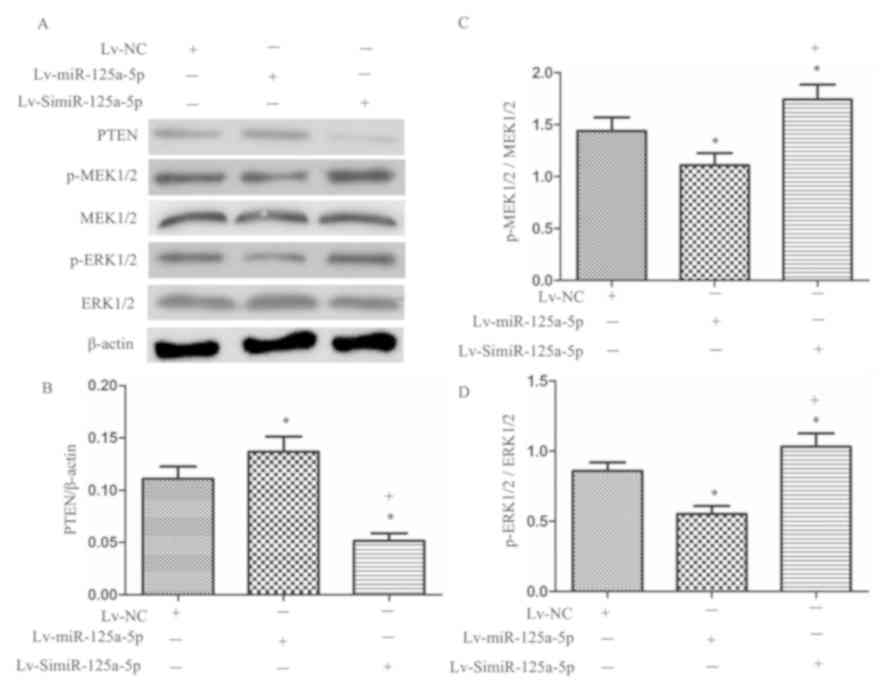 | Figure 4.Effects of miR-125a-5p on PTEN,
p-MEK1/2/MEK1/2, and p-ERK1/2/ERK1/2 expression in MCF-7 cells. (A)
Expression of PTEN, p-MEK1/2/MEK1/2, and p-ERK1/2/ERK1/2 in MCF-7
cells. (B) Summarized data of the PTEN expression. (C) Summarized
data of the p-MEK1/2/MEK1/2 level. (D) Summarized data of the
p-ERK1/2/ERK1/2 level. *P<0.05, vs. MCF-7NC;
+P<0.05, vs. MCF-7miR-125a-5p; n=6/group.
ERK, extracellular signal-regulated kinase; Lv, lentiviral; MEK,
mitogen-activated protein kinase kinase; miR, microRNA; NC,
negative control; p, phosphorylated; PTEN, phosphatase and tensin
homolog. |
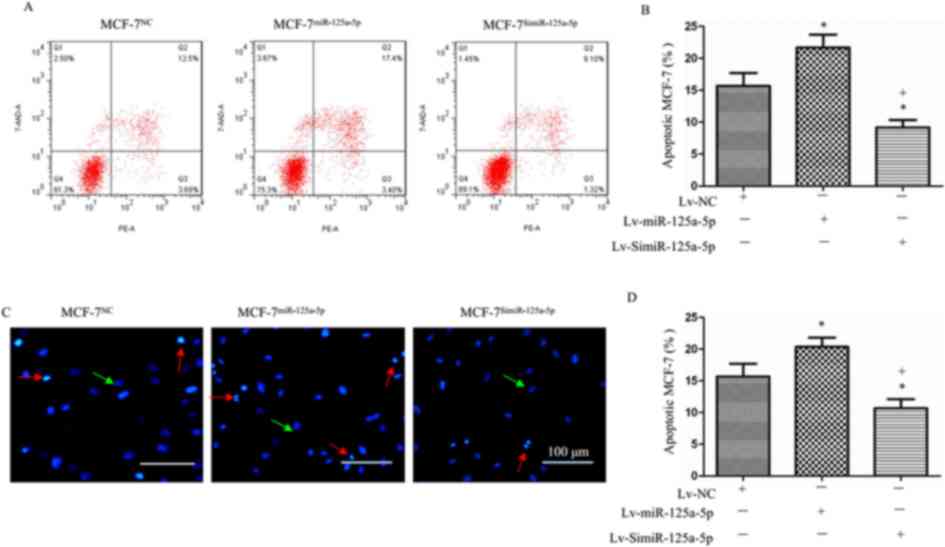
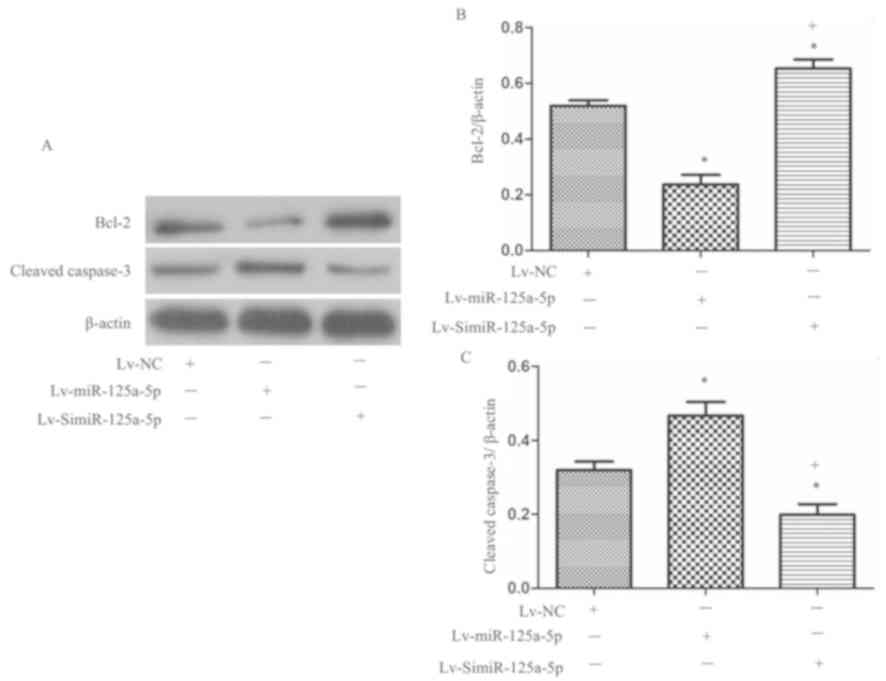
miR-125a-5p pomotes the apoptosis of
MCF-7 cells, which is accompanied with increased cleaved caspase-3
and decreased Bcl-2 levels
Hoechst 33258 staining and flow cytometry showed
that the rate of apoptosis among MCF-7miR-125a-5p cells
was significantly higher than that of the MCF-7NC cells
(P<0.05; Fig. 5A-D), while the
apoptotic rate of the MCF-7SimiR-125a-5p cells was
significantly lower than that of the MCF-7NC cells
(P<0.05, Fig. 5A-D). These
results indicated that high levels of miR-125a-5p promoted, while
low levels of miR-125a-5p reduced the apoptosis of MCF-7 cells.
In addition, via western blotting, we monitored the
levels of cleaved caspase-3 and Bcl-2, which are associated with
induction of apoptosis (13). The
results showed that overexpression of miR-125a-5p significantly
increased cleaved caspase-3 expression, while decreased Bcl-2
expression was observed compared with the control (P<0.05;
Fig. 6A-C). Silencing miR-125a-5p
in MCF-7 cells revealed significantly decreased cleaved caspase-3
protein expression and increased Bcl-2 expression compared with the
control and overexpression groups (P<0.05; Fig. 6A-C).
Discussion
In the present study, we found that miR-125a-5p
inhibited MCF-7 migration and proliferation, but promoted
apoptosis. The effects of miR-125a-5p on MCF-7 cell migration and
proliferation may be associated with an increase in PTEN, and
decrease in MEK1/2 and ERK1/2 protein expression, while the
apoptotic effects could be associated with upregulated cleaved
caspase-3 levels.
The survival time of breast cancer patients is
closely related to the pathological stage of breast cancer: The
earlier the pathological stage at the time of diagnosis, the higher
the recovery rate (14). For this
reason, and as there are currently no effective primary prevention
methods, early diagnosis and treatment are particularly important
in breast cancer (2,15). Mammography, ultrasonography,
computed tomography, magnetic resonance imaging and biopsies are
commonly used for the diagnosis of breast cancer (16). However, each diagnostic method has
its limitations. Breast cancer treatments mainly include surgery,
chemotherapy, radiotherapy, endocrine therapy, and biological
targeted therapy (14). Although
these treatments prolong patient survival, and the postoperative
recurrent and metastatic rates are still relatively high, it is
important to study the mechanism underlying the development and
progression of breast cancer. Thus, novel therapeutic targets for
breast cancer could be identified.
miRs are non-coding single-stranded small RNAs. They
are ~18–25 nucleotides in length with high evolutionarily
conservation (17). It binds to
its target mRNAs to inhibit translation and induce degradation of
the mRNA, altering the levels of expression of the target protein
(18). miR-125a expression is low
in breast cancer tissues, and was negatively correlated with tumor
size, lymph node metastasis and clinical stage of breast cancer
(19). Previous studies have
reported that miR-125a is closely related to tumor cell growth,
differentiation, and metastasis (20,21).
A study by Scott et al (20) has shown that miR-125a downregulates
human epidermal growth factor receptor 2 (ERBB2) and ERBB3 protein
and mRNA expression by targeting ERBB2 and ERBB3; furthermore,
miR-125a inhibited the migration and invasion of SKBR3 breast
cancer cells. To the bet of our knowledge, our study, for the first
time found that miR-125a-5p inhibited the proliferation and
migration, and promoted the apoptosis of MCF-7 cells. Collectively,
these findings demonstrated that miR-125a-5p is a candidate target
for breast cancer therapy.
The MEK1/2/ERK1/2 pathway is extensively involved in
the proliferation and the migration-associated protein expression
of tumor cells (22), and serves
an important role in tumor development and progression. A study by
Hsieh et al (19) indicated
that human aldo-keto reductase family 1 member B10 promotes the
invasion and migration of MCF-7 cells by activating the
MEK1/2/ERK1/2 pathway. Another study reported that the activation
of ERK1/2/p38 MAPK signaling is involved in the regulation of
invasion and metastasis of MDA-MB-231 breast cancer cells (23). These studies suggest that abnormal
activation of the MEK1/2/ERK1/2 pathway may be an important
mechanism underlying the development of breast cancer. In this
study, MEK1/2/ERK1/2 signaling was inhibited in MCF-7 breast cancer
cells with high miR-125a-5p expression. In contrast, we observed
that knockdown of miR-125a-5p expression led to activation of
MEK1/2/ERK1/2 signaling. PTEN, located on human chromosome 10, is a
tumor suppressor gene that inhibits the growth and migration of
various tumors by negatively regulating the PI3K/Akt/mTOR pathway
(24,25). A recent study revealed that PTEN
also inhibits the migration of type 2 endometrial cancer cells by
negatively regulating the MEK1/2/ERK1/2 pathway (26). In this study, high levels of
miR-125a-5p expression promoted PTEN expression in MCF-7 cells,
while knockdown of miR-125a-5p expression significantly reduced
PTEN expression in MCF-7 cells. This suggested that miR-125a-5p
inhibited the protein expression of the MEK1/2/ERK1/2 pathway
proteins by upregulating the expression of PTEN, playing a tumor
suppressive role.
Caspase-3 is an important pro-apoptotic factor that
plays an important role in the process of apoptosis. Bcl-2 is an
important anti-apoptotic gene that serves a key role in the
regulation of early apoptosis in cells (13). One recent study has shown that
cyanidin-3-glucoside inhibits MDA-MB-231 breast cancer cells by
promoting caspase-3 expression and downregulating Bcl-2 expression
(27). In this study, we found
that high miR-125a-5p expression promoted the apoptosis of MCF-7
cells, while knockdown of miR-125a-5p expression significantly
lowered the rate of apoptosis among MCF-7 cells. Our study into the
underlying mechanism showed that high levels of miR-125a-5p
expression promoted the expression of pro-apoptotic protein,
caspase-3 and inhibited the expression of anti-apoptotic protein
Bcl-2. These results indicated that miR-125a-5p might be involved
in regulating apoptosis in MCF-7 breast cancer cells by regulating
the caspase/Bcl-2 pathway.
In summary, the present study demonstrated a
potential role of miR-125a-5p in the pathogenesis of breast cancer
via control of cancer cell proliferation, apoptosis and migration.
In addition, our data indicated that miR-125a-5p exerted these
functions in breast cancer cells by upregulating PTEN expression
and inhibiting the activity of the MEK1/2/ERK1/2 pathway. However,
further in-depth and in vivo investigations into the role,
and the precise mechanisms underlying miR-125a-5p regulation in
breast cancer should be conducted in the future.
Acknowledgements
Not applicable.
Funding
This work was supported by the Medical Scientific
Research Foundation of Guangdong Province (grant no. A2018490) and
the Science and Technology Innovation Fund of Guangdong Medical
University (grant nos. M2015012 and M2016049).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZL, JL and QP designed the experiments. ZL, QP, ZZ,
CH, ZY and YZ performed the experiments. ZL, JL, QP analyzed the
data. ZL and QP interpreted the data. ZL and QP drafted the
manuscript. JL and QP revised the manuscript. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Amin A, Shriver CD, Henry LR, Lenington S,
Peoples GE and Stojadinovic A: Breast cancer screening compliance
among young women in a free access healthcare system. J Surg Oncol.
97:20–24. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Ward EM, Johnson CJ, Cronin KA,
Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al:
Annual report to the Nation on the status of cancer, 1975–2014,
featuring survival. J Natl Cancer Inst. 109:2017. View Article : Google Scholar
|
|
3
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fu Y and Cao F: MicroRNA-125a-5p regulates
cancer cell proliferation and migration through NAIF1 in prostate
carcinoma. Onco Targets Ther. 8:3827–3835. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo X, Wu Y and Hartley RS: MicroRNA-125a
represses cell growth by targeting HuR in breast cancer. RNA Biol.
6:575–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cai M, Chen Q, Shen J, Lv C and Cai L:
Epigenetic silenced miR-125a-5p could be self-activated through
targeting Suv39H1 in gastric cancer. J Cell Mol Med. 22:4721–4731.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang WK, Akçakaya P, Gangaev A, Lee L,
Zeljic K, Hajeri P, Berglund E, Ghaderi M, Åhlén J, Bränström R, et
al: miR-125a-5p regulation increases phosphorylation of FAK that
contributes to imatinib resistance in gastrointestinal stromal
tumors. Exp Cell Res. 371:287–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Jin T, Huang X, Liu X, Liu Z, Jia
Y and Hao J: Effects of the tumor suppressor PTEN on biological
behaviors of activated pancreatic stellate cells in pancreatic
fibrosis. Exp Cell Res. 373:132–144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong Q, Yang B, Han JG, Zhang MM, Liu W,
Zhang X, Yu HL, Liu ZG, Zhang SH, Li T, et al: A novel hydrogen
sulfide-releasing donor, HA-ADT, suppresses the growth of human
breast cancer cells through inhibiting the PI3K/AKT/mTOR and
Ras/Raf/MEK/ERK signaling pathways. Cancer Lett. 455:60–72. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang Y, Ye J, Jiao J, Zhang J, Lu Y,
Zhang L, Wan D, Duan L, Wu Y and Zhang B: Down-regulation of
miR-125a-5p is associated with salivary adenoid cystic carcinoma
progression via targeting p38/JNK/ERK signal pathway. Am J Transl
Res. 9:1101–1113. 2017.PubMed/NCBI
|
|
11
|
Pan Q, Liao X, Liu H, Wang Y, Chen Y, Zhao
B, Lazartigues E, Yang Y and Ma X: MicroRNA-125a-5p alleviates the
deleterious effects of ox-LDL on multiple functions of human brain
microvessel endothelial cells. Am J Physiol Cell Physiol.
312:C119–C130. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kluck RM, Bossy-Wetzel E, Green DR and
Newmeyer DD: The release of cytochrome c from mitochondria: A
primary site for Bcl-2 regulation of apoptosis. Science.
275:1132–1136. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de la Mare JA, Contu L, Hunter MC, Moyo B,
Sterrenberg JN, Dhanani KC, Mutsvunguma LZ and Edkins AL: Breast
cancer: Current developments in molecular approaches to diagnosis
and treatment. Recent Pat Anticancer Drug Discov. 9:153–175. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jafari SH, Saadatpour Z, Salmaninejad A,
Momeni F, Mokhtari M, Nahand JS, Rahmati M, Mirzaei H and Kianmehr
M: Breast cancer diagnosis: Imaging techniques and biochemical
markers. J Cell Physiol. 233:5200–5213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
M Braden A, V Stankowski R, M Engel J and
A Onitilo A: Breast cancer biomarkers: Risk assessment, diagnosis,
prognosis, prediction of treatment efficacy and toxicity, and
recurrence. Curr Pharm Des. 20:4879–4898. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai
CY, Hou MF, Lee JN, Wu DC, Wang SC and Tsai EM: miR-125a-5p is a
prognostic biomarker that targets HDAC4 to suppress breast
tumorigenesis. Oncotarget. 6:494–509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Du L and Feng R: c-Src regulates
cell cycle proteins expression through protein kinase B/glycogen
synthase kinase 3 beta and extracellular signal-regulated kinases
1/2 pathways in MCF-7 cells. Acta Biochim Biophys Sin (Shanghai).
45:586–592. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jafari SM, Joshaghani HR, Panjehpour M and
Aghaei M: A2B adenosine receptor agonist induces cell cycle arrest
and apoptosis in breast cancer stem cells via ERK1/2
phosphorylation. Cell Oncol (Dordr). 41:61–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Düzgün ŞA, Yerlikaya A, Zeren S, Bayhan Z,
Okur E and Boyacı İ: Differential effects of p38 MAP kinase
inhibitors SB203580 and SB202190 on growth and migration of human
MDA-MB-231 cancer cell line. Cytotechnology. 69:711–724. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mashayekhi S, Yousefi B, Tohidi E, Darband
SG, Mirza-Aghazadeh-Attari M, Sadighparvar S, Kaviani M,
Shafiei-Irannejad V, Kafil HS, Karimian A, et al: Overexpression of
tensin homolog deleted on chromosome ten (PTEN) by ciglitazone
sensitizes doxorubicin-resistance leukemia cancer cells to
treatment. J Cell Biochem. 120:15719–15729. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ocana A, Vera-Badillo F, Al-Mubarak M,
Templeton AJ, Corrales-Sanchez V, Diez-Gonzalez L, Cuenca-Lopez MD,
Seruga B, Pandiella A and Amir E: Activation of the PI3K/mTOR/AKT
pathway and survival in solid tumors: Systematic review and
meta-analysis. PLoS One. 9:e952192014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiong S, Cheng JC, Klausen C, Zhao J and
Leung PC: TGF-β1 stimulates migration of type II endometrial cancer
cells by down-regulating PTEN via activation of SMAD and ERK1/2
signaling pathways. Oncotarget. 7:61262–61272. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cho E, Chung EY, Jang HY, Hong OY, Chae
HS, Jeong YJ, Kim SY, Kim BS, Yoo DJ, Kim JS and Park KH:
Anti-cancer effect of cyanidin-3-glucoside from mulberry via
caspase-3 cleavage and DNA fragmentation in vitro and in vivo.
Anticancer Agents Med Chem. 17:1519–1525. 2017. View Article : Google Scholar : PubMed/NCBI
|