Introduction
Atherosclerosis is a disease that involves several
cell types and a different set of inflammatory mediators (1). The first step is endothelial
activation that expresses certain adhesion molecules, including
vascular cell adhesion molecule (VCAM-1), intercellular adhesion
molecule (ICAM-1) and E-selectin.
A previous study confirmed that dendritic cells
(DCs) exist in healthy arteries and participate in diverse
pathogenic and protective mechanisms during atherosclerosis
(2). It has been demonstrated that
DCs participate in atherosclerosis through producing cytokines,
including tumor necrosis factor (TNF), interleukin (IL)-6 and IL-12
(3). DCs can be found in the
arterial intima of healthy individuals (4) and the number increased in
atherosclerotic lesions (5). In
mice, DCs are frequently found in the aortic intima which are
predisposed to atherosclerosis. Moreover, it has been demonstrated
that DCs accumulate in atherosclerotic areas (6–8).
Exosomes are a type of extracellular vesicle which
are released by almost all kinds of cells. They have attracted a
huge amount of attention because of the ability to transfer
information between cells. It has been demonstrated that exosomes
contain numerous biological components, including proteins, lipids,
RNA and DNA (9). Previous studies
showed that DC-derived exosomes (DC-exos) can exert biological
effects through membrane proteins or shuttled microRNAs
(miRNAs/miRs) (10–12). The authors have reported that
mDC-exos increase endothelial inflammation and atherosclerosis
through membrane TNF-α mediated nuclear factor (NF)-κB pathway
(13). This process was quickly
initiated by direct contact between exosomal membrane proteins and
human umbilical vein endothelial cell (HUVEC) ligands. However,
exosomes also contain a huge number of miRNAs and how these miRNAs
influence HUVECs remains unclear. The present study aimed to
investigate the role of mDC-exo miR-146a in inflammation of HUVECs.
To the best of our knowledge, this study is the first to
investigate the successive effect of exosomes on recipient cells in
different phases.
The present study first demonstrated that mDC-exos
increase endothelial expression of adhesion molecules. Then it was
found that HUVECs became resistant to a second stimulation after
the first stimulation by mDC-exo. The change of a set of miRNAs in
HUVECs was measured and a significant elevation of miR-146a was
observed. Finally, DC-exo was confirmed to shuttle miR-146a into
HUVECs and the shuttled miR-146a contributed to protect HUVECs from
a second stimulation through inhibiting interleukin-1
receptor-associated kinase (IRAK). The present study demonstrated
for the first time that exosomes make up a negative feedback loop
to regulate endothelial inflammation.
Materials and methods
Cell culture and cell
transfection
Male 6–8-week-old C57BL/6 mice (20–25 g) were
purchased from JSJ-lab (http://www.jsj-lab.com/) and manual cervical
dislocation was used to sacrifice the animals. Death was
ascertained by observing the cessation of either breathing or the
heartbeat. The mice were housed at a constant temperature (22±1°C)
in a 12-h light/dark cycle, with ad libitum access to a
standard diet and water. A total of four mice were used to obtain
DCs for every single experiment and a total of ~40 mice were used
in the whole study. As described previously, bone marrow DCs
(BMDCs) were from C57BL/6 mice (14). To eliminate the interference of
exosomes from fetal bovine serum, X–VIVO 15 was used (Lonza Group,
Ltd.) that contains no serum to culture BMDCs (15). Briefly, bone marrow progenitors
were washed out and 10×106 cells were placed into every
10 cm dish (generally 4–6 dishes of cells could be obtained in
every single experiment). The culture medium was X–VIVO 15 and
contains 10 ng/ml granulocyte-macrophage colony-stimulating factor,
1 ng/ml IL-4 (PeproTech, Inc.) and 1% antibiotics
(penicillin-streptomycin solution). Non-adherent cells were gently
washed out at 48 h since they were not DCs. The remaining clusters
were then continually cultured and the medium was changed every
other day (half of the original medium was kept to maintain the
micro-environment). On day 7, the cultured cells were
differentiated into immature DCs and they were ready for subsequent
studies. The present study was approved by the Institutional Review
Board of the Zhongshan Hospital, Fudan University and Shanghai
Institutes for Biological Sciences and were conducted in conformity
with the Public Health Service Policy on Humane Care and Use of
Laboratory Animals.
For the purpose of exosome isolation, DCs were
treated with lipopolysaccharide (LPS; 5 µg/ml) or PBS for 24 h to
generate mature or immature DCs. Then they were washed and replaced
with new medium. After another 48 h continuous culturing, the
culture medium was collected for exosome isolation. For
transfection studies, DCs were transfected with miRNA mimic or
inhibitor for 24 h and then treated with LPS or PBS for another
12–24 htogenerate mature or immature DCs. Then the cells were
washed and replaced with new medium. After another 36–48 h
culturing, the culture medium was collected for exosome isolation.
For transfection experiments, miR-146 mimics/inhibitors and
transfection reagent (riboFECT™ CP) were obtained from RiboBio Co.,
Ltd. The transfection concentration of miR-146 mimics and
inhibitors was 50 and 100 nM, respectively.
HUVECs were obtained from AllCells, LLC and cultured
in ECM (Scien Cell Research Laboratories, Inc.). The 8–10th
generations of HUVECs were used in this study and they were
harvested for mRNA or protein detection after treatment with
exosomes for 6 or 24 h, respectively.
Exosomes isolation, analysis, labeling
and neutralization
Exosomes were isolated using an exosome
precipitation solution (Exo-Quick; System Bioscience) following the
manufacturer's protocol with certain modifications (16). Briefly, the medium was harvested by
centrifugation at 3,000 × g for 15 min at 4°C and then 10,000 × g
for 30 min (at 4°C) to eliminate cell debris. Then the supernatant
was filtered with a 0.22-µm filter to further eliminate cell debris
and large particles. Finally, Exo-Quick was added to the medium at
a ratio of 1: 5 and the mixed solution was placed at 4°C overnight.
The solution was then centrifuged at 1,500 × g for 30 min at 4°C.
The exosome pellet was resuspended with PBS and stored at −80°C for
subsequent studies. The ultrastructure was obtained by transmission
electron microscopy. For transmission electron microscopy, 10 µl
exosome was deposited on TEM grids for 1 min at room temperature
and carefully blotted with soft paper. Then 10 µl 2% uranyl acetate
was added to the grid for 1 min at room temperature and carefully
blotted with soft paper before being left a dry environment for
5–10 min. Finally the grid was observed under transmission electron
microscopy at 80 kV. Protein markers, TSG101 (Abcam) and Alix
(Santa Cruz Biotechnology, Inc.), were determined by
immunoblotting. The miRNA content was measured using quantitative
PCR (Qiagen, Inc.).
Purified exosomes were labeled with a PKH67 (green)
kit (Sigma-Aldrich; Merck KGaA) according to protocols previously
reported (13,17). Briefly, the exosomes were added to
0.5 ml Diluent C. In parallel, 4 µl PKH67 dye was added to another
0.5 ml Diluent C and incubated with the exosome solution for 4 min
at room temperature. Then 2 ml 0.5% BSA/PBS (Sangon Biotech Co.,
Ltd.) was added to bind excess dye. To ensure the exosomes were
re-isolated as pure as possible, the labeled exosomes were
harvested by centrifugation at 100,000 × g for 70 min (at 4°C) and
then the exosome pellet was suspended with PBS for the next study.
Where necessary, labeled exosomes were pre-incubated with 2 µg/ml
annexin V (BD Pharmingen; Becton, Dickinson and Company) at 37°C
for 1.5 h (18). After the HUVECs
were fixed with 4% paraformaldehyde (Sangon Biotech Co., Ltd.) at
room temperature for 15 min, they were visualized with
immunofluorescent microscopy (magnification, ×400). For
immunofluorescence, the HUVEC cytoskeleton was stained with
Phalloidin from Sigma-Aldrich (Merck KGaA) according to
manufacturer's protocol at room temperature for 30 min. Cell nuclei
were then stained with DAPI at room temperature for 10 min.
Reverse transcription-PCR
Total RNA was extracted from cells by TRIzol reagent
(Sangon Biotech Co., Ltd.). A ReverTra Ace qPCR RT kit was used
(Toyobo Life Science) to generate cDNA from mRNA at 37°C for 30 min
and SYBR Premix Ex Taq (Takara Bio, Inc.) was used for qPCR with
the ABI 7500 Real-time PCR system. Primers for human VCAM-1,
ICAM-1, E-selectin are as follows: VCAM-1 forward
5′-GCTGCTCAGATTGGAGACTCA-3′, VCAM-1 reverse
5′-CGCTCAGAGGGCTGTCTATC-3′, ICAM-1 forward
5′-TCTGTGTCCCCCTCAAAAGTC-3′, ICAM-1 reverse
5′-GGGGTCTCTATGCCCAACAA-3′, E-selectin forward
5′-AATCCAGCCAATGGGTTCG-3′, E-selectin reverse
5′-GCTCCCATTAGTTCAAATCCTTCT-3′, GAPDH forward
5′-ATGGGGAAGGTGAAGGTCG-3′, and GAPDH reverse
5′-GGGGTCATTGATGGCAACAATA-3′. To amplify mature miRNA sequences
from cells and exosomes, total RNA was extracted using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
manufacturer's protocol and microRNA RT-qPCR syb kit (TaqMan) was
used for generating cDNA (at 42°C for 60 min) from microRNA and
qPCR detection. Primers for interested microRNA were purchased from
Invitrogen; Thermo Fisher Scientific, Inc., and primer sequences
are shown in Table I. The qPCR
conditions were as follows: 95°C for 2 min, 40 cycles of 95°C for
15 sec and 60°C for 35 sec. The relative expression levels of the
miRNAs were normalized to that of U6 by using the 2−ΔΔCq
cycle threshold method (19).
 | Table I.Forward and reverse primer sequences
for the miRs and U6. |
Table I.
Forward and reverse primer sequences
for the miRs and U6.
| miRNAs | Forward (5′-3′) | Reverse (5′-3′) |
|---|
| miR-146a |
GTGAGAACTGAATTCCAT | AACTGGTGTCGTGGAG |
| miR-222 |
GCTACATCTGGCTACTGG |
TCAACTGGTGTCGTGG |
| miR-17 |
AAAGTGCTTACAGTGCAGG |
AACTGGTGTCGTGGAG |
| miR-10a |
TACCCTGTAGATCCGAAT |
AACTGGTGTCGTGGAG |
| miR-31 |
CAAGATGCTGGCATAG |
CTCAACTGGTGTCGTG |
| miR-let-7g |
GGGTGAGGTAGTAGTTTGT |
CTCAACTGGTGTCGTG |
| miR-92a |
CCTATTGCACTTGTCCC |
TCAACTGGTGTCGTGG |
| miR-221 |
GCTACATTGTCTGCTGG |
CTGGTGTCGTGGAGTC |
| miR-21a |
CCTAGCTTATCAGACTGATG |
CTCAACTGGTGTCGTG |
| miR-217 |
CTGCATCAGGAACTGATT |
TCAACTGGTGTCGTGG |
| miR-181b |
ACGACAACATTCATTGC |
AACTCCACACCAGCAC |
| miR-126 |
CGTACCGTGAGTAATAATG |
CTCAACTGGTGTCGTG |
| miR-663 |
GTTTTAGGCGGGGCG |
TGGTGTCGTGGAGTCG |
Western blot analysis
The cultured cells were harvested and lysed by RIPA
supplemented with complete protease inhibitor cocktail tablets
(Roche Diagnostics). Cell debris was removed by centrifugation at
10,000 × g for 20 min at 4°C. Then the lysates (20 µg) were
separated by SDS-PAGE gels (10–15%, 150 V, 70 min) and transferred
them to PVDF membranes (Bio-Rad Laboratories, Inc.). After blocking
with 5% BSA for 1 h at room temperature, the PVDF membranes were
incubated with primary antibodies overnight at 4°C. Then, the
membranes were incubated with HRP-conjugated secondary antibodies
(Beyotime Institute of Biotechnology, cat. no. A0208, goat
anti-rabbit IgG, 1:1,000) at room temperature for 1 h. After
washing, the band intensity was analyzed using DAB Horseradish
Peroxidase Color Development kit (Beyotime Institute of
Biotechnology) and LAS-3000 Imaging System (Fuji Corporation).
Quantity One software (Bio-Rad Laboratories, Inc., version 4.6.2)
was used to calculate the expression of protein. An antibody
against Alix (1:500; cat. no. sc-53540) was purchased from Santa
Cruz Biotechnology, Inc. VCAM-1 (1:1,000; cat. no. ab134047),
ICAM-1 (1:1,000; cat. no. ab2213), E-Selectin (1:1,000; cat. no.
ab18981), IRAK (1:1,000; cat. no. ab238), TSG101 (1:1,000; cat. no.
ab125011) and GAPDH (1:10,000; cat. no. ab8245) were purchased from
Abcam. Phosphorylated-NF-κB p105 (1:1,000; cat. no. cst4806) and
p105 (1:1,000; cat. no. cst4717) were purchased from Cell Signaling
Technology, Inc.
Statistical analysis
All values are presented as the mean ± standard
deviation. A Student's t test (2 groups) or one-way analysis of
variance (3 or more groups with a least significant difference post
hoc test) was used to determine statistical significance between
the groups. All statistical analyses were performed by SPSS 17.0
(SSPS Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Successful isolation of exosomes from
DC culture medium
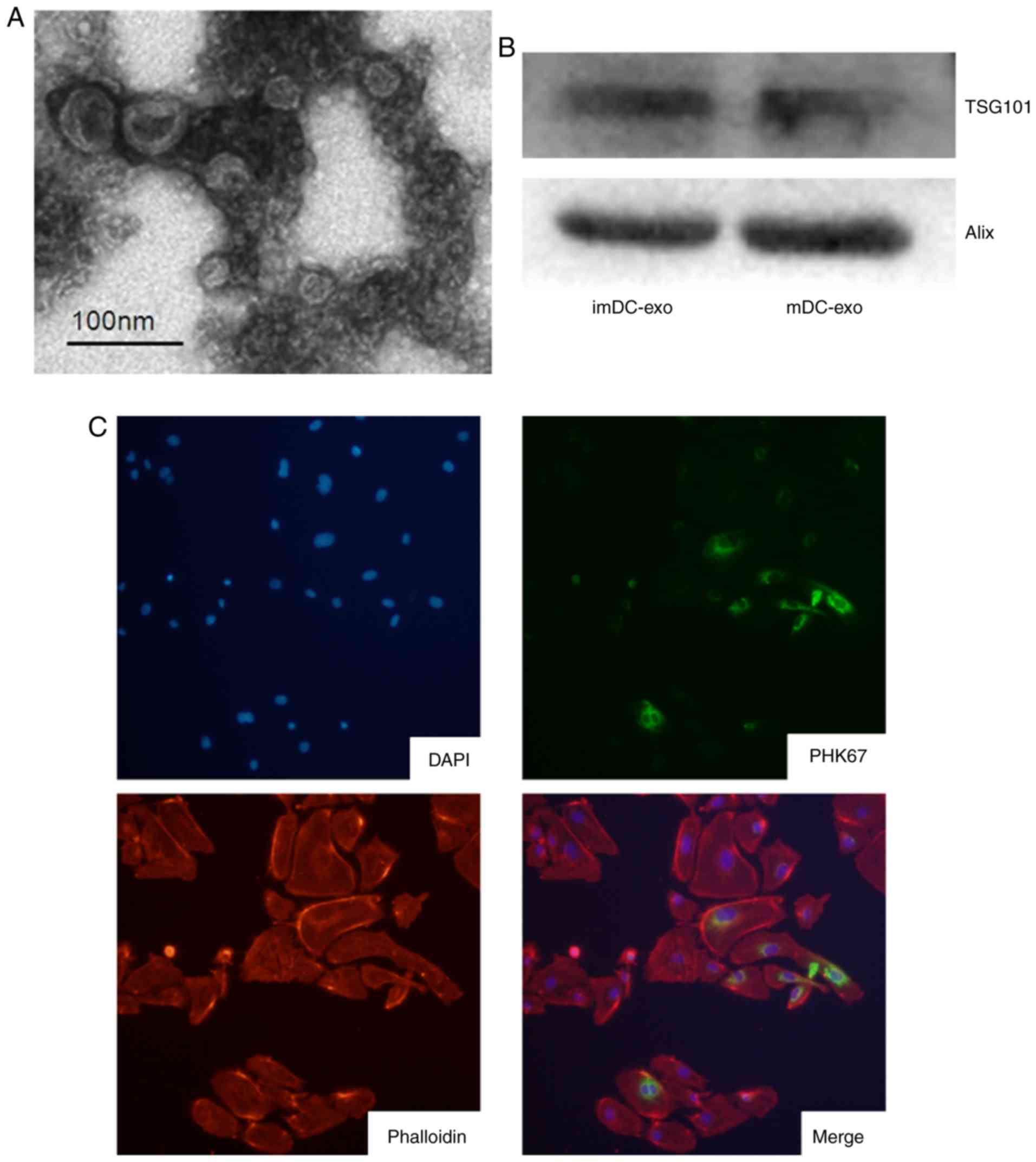
The ultrastructure of exosomes was observed using
transmission electron microscopy. Exosomes at a typical size of
30–100 nm in diameter were found by electron microscopic analysis
(Fig. 1A). Then the expression of
exosomes markers, TSG101 and Alix, was confirmed by immunoblotting
(Fig. 1B). These results suggest
successful isolation of exosomes from the culture medium. To study
whether other cells can uptake exosomes, DC-exo was labeled with
PKH67 and incubated with HUVECs. The results confirmed that HUVECs
could uptake exosomes (Fig.
1C).
mDC-exos increase endothelial
expression of adhesion molecules through quick activation of the
NF-κB pathway
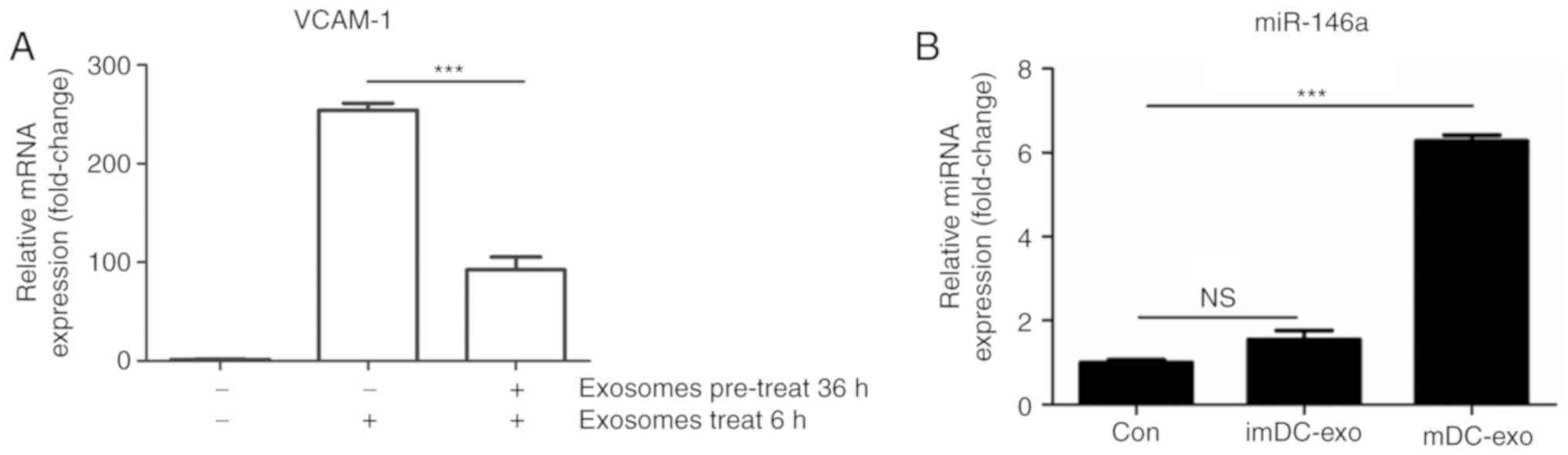
mDC-exo were co-cultured with HUVECs and it was
found that the protein expression of VCAM-1, ICAM-1 and E-Selectin
significantly increased (P<0.05; Fig. 2A and B). To explore the mechanism
of endothelial inflammation, p-p105 of the NF-κB signaling pathway
was measured and a quick activation of this pathway (within 0.5 h)
was demonstrated (Fig. 2C). The
authors' previously published study demonstrated that mDC-exo can
quickly activate the NF-κB signaling pathway by direct contact
between exosomal membrane proteins and HUVEC ligands (13). The present study aimed to
investigate the effect of exosomal miRNAs after the influence of
exosomal proteins. The mRNA expression of adhesion molecules at
different time (as long as 36 h) was determined and it was
confirmed that the first stage effect of mDC-exo on HUVECs
gradually vanished 36 h after incubation (Fig. 2D-F).
 | Figure 2.mDC-exo increase endothelial
expression of adhesion molecules through quick activation of NF-κB
pathway. (A) Western blotting and (B) analysis of exosomes from
mDCs were incubated with HUVECs for 24 h and the expression of
VCAM-1, ICAM-1 and E-Selectin was detected by immunoblotting. (C)
mDC-exo were incubated with HUVECs for 0.5 h and p-p105 of NF-κB
pathway was detected by immunoblotting. mDC-exos were incubated
with HUVECs for indicated time and the mRNA expression of (D)
VCAM-1, (E) ICAM-1 and (F) E-Selectin was detected by quantitative
PCR. *P<0.05, **P<0.01 and ***P<0.001. ICAM, intercellular
adhesion molecule; VCAM, vascular cellular adhesion molecule;
HUVECs, human umbilical vein endothelial cell; DC, dendritic cell;
exo, exosome; p, phosphorylated; NF, nuclear factor; NS, not
significant; m, mature. |
HUVECs were less vulnerable to a
second stimulation and miR-146a might play a role
To investigate the effect of exosomal miRNAs on
HUEVCs, the experiments were conducted as follows: HUVECs were
pre-treated with mDC-exo for 12 h and then they were allowed a
24-hrecovery period, after which the HUVECs were treated with
mDC-exo for another 6 h. According to the findings above, the mRNA
expression of adhesion molecules would reduce to a very low level
at 36 h. In addition, exosomal miRNAs would have been released into
HUVECs and impose an effect on the cells. The second stimulation of
mDC-exos could reflect the effect of DC-exosomal miRNAs on HUVECs,
whether they are protective or injurious. The results showed that
VCAM-1 expression was significantly reduced (P<0.001; Fig. 3A), indicating a protective role of
DC-exosomal miRNAs on HUVECs.
Next, which miRNA was involved in the protection of
HUVECs was investigated. To this end, a recent review was
retrieved, which comprehensively reviewed current evidence of
miRNAs in endothelial inflammation and atherosclerosis (20). In that review, the authors
concluded that 13 miRNAs are involved in adhesion molecule
expression. They are miR-146a/b, miR-181b, miR-126, miR-663,
miR-21, miR-217, miR-10a, miR-31, miR-221, miR-222, miR-17,
miR-let-7g and miR-92a. their expression in HUVECs were measured
after DC-exo stimulation and it was found that miR-146a expression
increased 6-fold (Fig. 3B). It was
noticed that the expression of miR-222 and miR-17 also slightly
increased (Table II). miR-146a,
miR-222 and miR-17 were demonstrated to be protective miRNAs
(21–24) and miR-146a was chosen as the target
of further research since it increased the most significantly.
 | Table II.Expression of 13 miRNAs in immature
and mature dendritic cell exosome. |
Table II.
Expression of 13 miRNAs in immature
and mature dendritic cell exosome.
| miRNAs | Change | Fold-change |
|---|
| miR-146a | Up | 6.28 |
| miR-222 | Up | 2.15 |
| miR-17 | Up | 2.06 |
| miR-10a | Up | 1.63 |
| miR-31 | Up | 1.59 |
| miR-let-7g | Up | 1.55 |
| miR-92a | Up | 1.54 |
| miR-221 | Up | 1.44 |
| miR-21a | Up | 1.27 |
| miR-217 | Down | 0.99 |
| miR-181b | Down | 0.87 |
| miR-126 | Down | 0.69 |
| miR-663 | Down | 0.60 |
Elevated miR-146a in HUVECs is partly
due to the miRNA cargo in exosomes
Next the present study wanted to investigate whether
the miRNAs contained in mDC-exo contribute to the elevation of
miR-146a in HUVECs. First, the expression of miR-146a in exosomes
was determined and it was found that expression was increased in
mature DC-exo compared with in immature DC-exo (Fig. 4A). This result was consistent with
a previous study, which performed a microarray to investigate the
differential expression of miRNAs in immature and mature DC-exos
(12). Since miR-146a can be
induced in endothelial cells upon exposure to proinflammatory
cytokines (21), the direct
contact of exosomes and HUVECs induce the NF-κB signaling pathway
and it surely will increase miR-146a expression. Therefore, the
present study needs to confirm that the elevated miR-146a in HUVECs
is partly due to the shuttled cargo in exosomes. Annexin V (AV), a
well-known inhibitor of exosome uptake was used to investigate
whether the contained miR-146a in mDC-exo would influence its level
in HUVECs. The effect of AV was confirmed by immunofluorescence
(Fig. 4B). Then the present study
found that whether AV was present or not, the HUVECs expression of
VCAM-1 induced by mDC-exo remained unchanged (Fig. 4C). This result showed that,
although the uptake of exosomes was inhibited, activation of the
NF-κB pathway by direct contact of exosomes and HUVECs was not
influenced. As a result, the intrinsic expression of miR-146a in
HUVECs induced by the NF-κB signaling pathway should be similar.
Additionally, it was found that the expression of miR-146a in
HUVECs was significantly decreased in mDC-exo-AV group compared
with in the mDC-exo group (P<0.05), suggesting the inhibition of
exosome uptake decreased the shuttling of miR-146a from exosomes to
HUVECs (Fig. 4D).
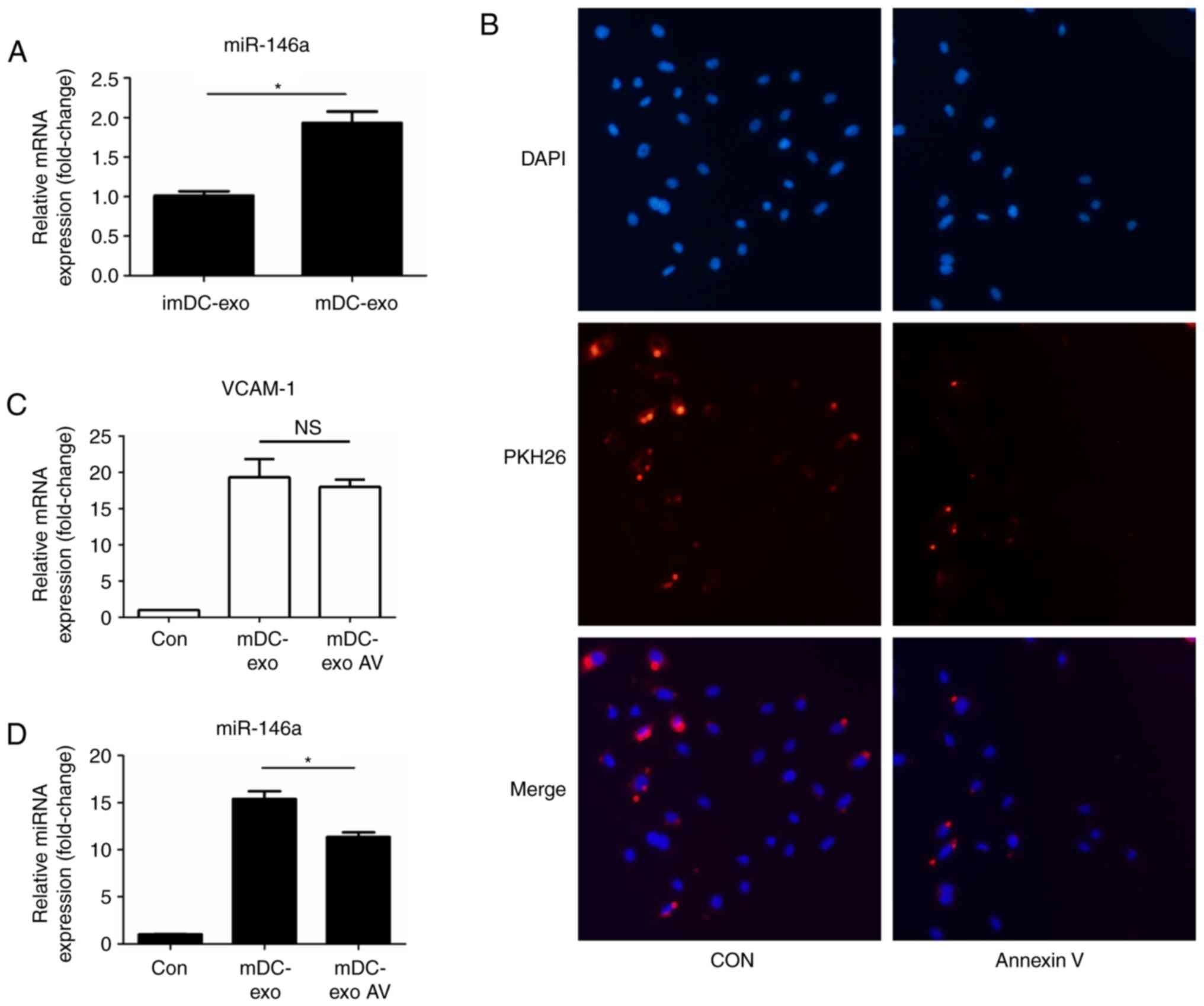 | Figure 4.Elevated miR-146a in HUVECs partly due
to the miRNAs cargo in exosomes. (A) Expression of miR-146a in
imDC-exo and mDC-exo. (B) Fluorescence confocal of HUVECs treated
with PKH26-labled exosomes (red) which were preincubated for 1.5 h
with annexin V (2 µg/ml). HUVECs were stained with nuclei with DAPI
(blue). (C) Expression of VCAM-1 in HUVECs treated with mDC-exo
which were pre-incubated with annexin V or not. (D) Expression of
miR-146a in HUVECs treated with mDC-exo which were pre-incubated
with annexin V. *P<0.05. Magnification, ×400. HUVECs, human
umbilical vein endothelial cells; DC, dendritic cell; exo, exosome;
VCAM, vascular cellular adhesion molecule; microRNA, miR; NS, not
significant; im, immature; m, mature; Con, control. |
Exosomes can shuttle miR-146a to
HUVECs and protect the recipient cells through inhibiting IRAK
To further confirm whether mDC-exo could shuttle
miRNAs to recipient cells, imaging experiments were conducted to
visualize the transfer of labeled miRNAs between cells. First, DCs
were transfected with a Cy3-labeled miRNA control. After 24 h of
transfection, DCs were replaced with fresh culture medium. After
another 36 h, the culture supernatant was collected for exosome
isolation. Then, HUVECs were incubated with the isolated exosomes
for 24 h. It was found that DC-exos, which contained the
Cy3-labeled miRNA control, were endocytosed by HUVECs, as
visualized by fluorescence microscopy (Fig. 5A). To confirm this result, a
miR-146a mimic/inhibitor was transfected into BMDCs and co-culture
their exosomes with HUVECs. An increase of miR-146a upon mimic
transfection and a decrease upon inhibitor transfection was
observed (Fig. 5B). To test
whether the shuttled miRNA can functionally affect the recipient
cells, HUVECs were pre-treated with exosomes derived from DCs
transfected with miR-146a mimic or inhibitor for 36 h. Then the
HUVECs were once again stimulated with mDC-exo. It was found that
exosomes transfected with the miR-146a mimic could attenuate the
elevated expression of adhesion molecules and exosomes transfected
with miR-146a inhibitor could increase the expression of adhesion
molecules (Fig. 5C). A previous
study has shown that miR-146a can regulate cell response through
targeting IRAK (25). It was
confirmed that IRAK-1 expression was significantly inhibited in the
miR-146a mimic group and rescued in the miR-146a inhibitor group
(P<0.001; Fig. 5D). These
results indicate that exosomes derived from BMDCs can shuttle miRNA
to the recipient cells and the shuttled miR-146a can protect HUVECs
from further injury by mDC-exo.
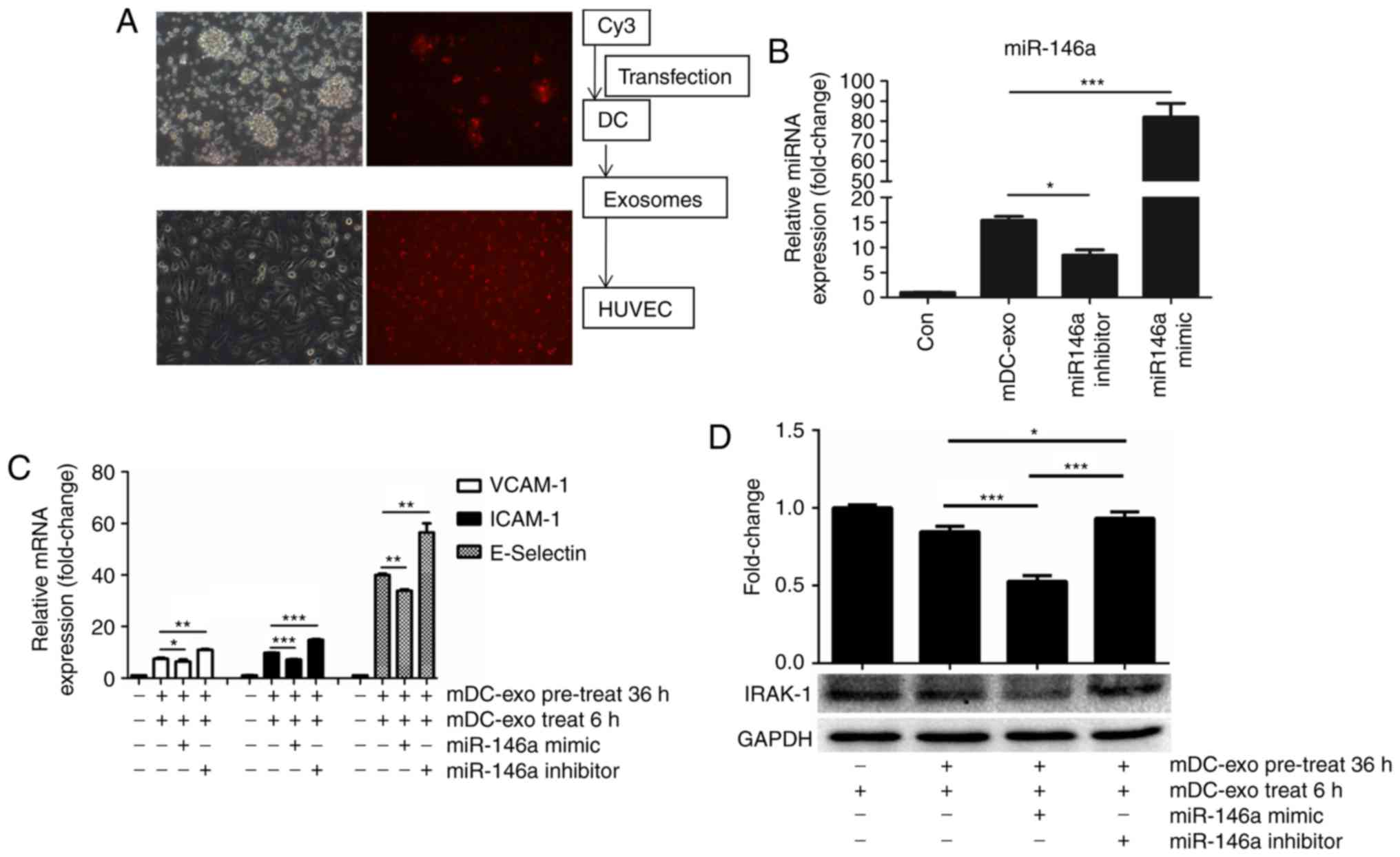 | Figure 5.Exosomes can shuttle miR-146a to
HUVECs and protect the recipient cells through inhibiting IRAK. (A)
Upper two images show that Cy3 was transfected into DCs clusters
(the left one was plain image and the right was fluorescence
image). In the lower two images, exosomes from Cy3 transfected DCs
were co-cultured with HUVECs and uptake by HUVECs are shown. (B)
Expression of miR-146a in HUVECs treated with exosomes from DCs
transfected with miR-146a mimic or inhibitor. (C) HUVECs were
pre-treated with exosomes from DCs transfected with miR-146a mimic
or inhibitor for 12 h. After a 24-h recovery period, the HUVECs
were once again treated with DCs-exos for another 6 h. Expression
of adhesion molecules, VCAM-1, ICAM-1 and E-Selectin, were detected
by qPCR. (D) Based on the design of Fig. 5C, expression of IRAK was detected
by immunoblotting. *P<0.05, **P<0.01 and ***P<0.001.
Magnification, ×40. HUVECs, human umbilical vein endothelial cells;
DC, dendritic cell; exo, exosome; miR, microRNA; VCAM, vascular
adhesion molecule; ICAM, intercellular adhesion molecule; m,
mature; IRAK, interleukin-1 receptor-associated kinase. |
Discussion
In this present study, mDC-exos were shown to
increase endothelial expression of adhesion molecules. Then it was
found that mDC-exo could shuttle miR-146a into HUVECs and the
shuttled miR-146a contributed to protecting HUVECs from a second
stimulation. The present study demonstrated for the first time that
DCs-derived exosomes make up a negative feedback loop to control
inflammation.
It has been demonstrated that DCs play an important
role in both pro-atherogenic (3,26,27)
and regulatory (7,28) responses within the artery wall.
Vascular DCs seem to be involved in endothelial inflammation in
three roles as follows: The uptake and storage of lipids; the
clearance of lipids, apoptotic cells, and debris from the artery
wall, and the maintenance of vascular Treg responses (29). In this study, DCs were shown to
regulate endothelial inflammation through released exosomes.
Exosomes can be released by numerous cell types.
Classically in cell biology, cells communicate with each other by
direct interaction or secreted soluble factors. These factors can
act on the cell itself (autocrine signaling) or on both neighboring
(paracrine signaling) and distant cells (endocrine signaling).
During the past decade, exosomes have been found to be potent
vehicles of intercellular communication. In this study, it was
shown that DCs can release exosomes and these exosomes can activate
endothelial cells.
A major group of exosomal cargos is RNAs (9). Since the beginning of the discovery
that exosomes contain RNA, miRNAs shuttled by exosomes have
attracted huge attention in all research fields. Previous studies
have confirmed that exosomes and exosomal miRNAs play an important
role in cardiac repair after myocardial infarction (30–32).
However, they are less investigated in inflammation and
atherosclerosis. Up to now, ~12 miRNAs have been identified in the
process of endothelial injury (20,33).
In this study, these miRNAs were detected in HUVECs stimulated by
DC-exo and it was found that miR-146a was changed the most.
miR-146a has been shown to play a role in numerous diseases, such
as atherosclerosis, aging and neurodegenerative disease (1,34,35).
In the present study, it was then confirmed that DC-exo could
shuttle miR-146a into HUVECs and the shuttled miR-146a contribute
to protecting HUVECs from a second stimulation.
This is interesting because it is a new kind of
negative feedback regulatory loop. A regulatory feedback loop,
whether negative or positive, commonly exists in numerous life
processes, including the response to injury of endothelial cells.
It has been reported that miRNAs provide negative feedback control
of inflammation in HUVECs (21,23).
With the authors' previous (13)
and present findings, the knowledge of this feedback system is
extended, indicating that exosomes can exert an influence on
recipient cells in a timely manner as follows: Quick and direct
effect by direct contact of proteins, slow and indirect effect by
shuttled miRNAs.
In conclusion, the present study showed that mDC-exo
increased endothelial expression of adhesion molecules. Then after
being up-taken by HUVECs, mDC-exo could shuttle miR-146a into
recipient cells and the shuttled miR-146a contributes to protecting
HUVECs from a second stimulation. These data suggest a negative
feedback loop of inflammation regulation by mDC-exos.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Grants of
Shanghai Sailing Program (no. 16YF1401600), the National Key
R&D Program of China (grant no. 2016YFC1301200) and the Natural
Science Foundation of China (grant no. 81770350).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ and WG designed the study, performed the research
and wrote the manuscript. RW performed the molecular biology
experiment and immunofluorescence assay. HL helped in the molecular
biology experiment and was involved in the immunofluoresence assay.
JG designed and supervised the study.
Ethics approval and consent to
participate
This research was approved by the Institutional
Review Board of The Zhongshan Hospital, Fudan University and
Shanghai Institutes for Biological Sciences.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hopkins PN: Molecular biology of
atherosclerosis. Physiol Rev. 93:1317–1542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zernecke A: Dendritic cells in
atherosclerosis: Evidence in mice and humans. Arterioscler Thromb
Vasc Biol. 35:763–770. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koltsova EK and Ley K: How dendritic cells
shape atherosclerosis. Trends Immunol. 32:540–547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Millonig G, Niederegger H, Rabl W,
Hochleitner BW, Hoefer D, Romani N and Wick G: Network of
vascular-associated dendritic cells in intima of healthy young
individuals. Arterioscler Thromb Vasc Biol. 21:503–508. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yilmaz A, Lochno M, Traeg F, Cicha I,
Reiss C, Stumpf C, Raaz D, Anger T, Amann K, Probst T, et al:
Emergence of dendritic cells in rupture-prone regions of vulnerable
carotid plaques. Atherosclerosis. 176:101–110. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Busch M, Westhofen TC, Koch M, Lutz MB and
Zernecke A: Dendritic cell subset distributions in the aorta in
healthy and atherosclerotic mice. PLoS One. 9:e884522014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Choi JH, Cheong C, Dandamudi DB, Park CG,
Rodriguez A, Mehandru S, Velinzon K, Jung IH, Yoo JY, Oh GT and
Steinman RM and Steinman RM: Flt3 signaling-dependent dendritic
cells protect against atherosclerosis. Immunity. 35:819–831. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Galkina E, Kadl A, Sanders J, Varughese D,
Sarembock IJ and Ley K: Lymphocyte recruitment into the aortic wall
before and during development of atherosclerosis is partially
L-selectin dependent. J Exp Med. 203:1273–1282. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles. 4:270662015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alexander M, Hu R, Runtsch MC, Kagele DA,
Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM and
O'Connell RM: Exosome-delivered microRNAs modulate the inflammatory
response to endotoxin. Nat Commun. 6:73212015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Munich S, Sobo-Vujanovic A, Buchser WJ,
Beer-Stolz D and Vujanovic NL: Dendritic cell exosomes directly
kill tumor cells and activate natural killer cells via TNF
superfamily ligands. Oncoimmunology. 1:1074–1083. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Montecalvo A, Larregina AT, Shufesky WJ,
Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G,
Wang Z, et al: Mechanism of transfer of functional microRNAs
between mouse dendritic cells via exosomes. Blood. 119:756–766.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao W, Liu H, Yuan J, Wu C, Huang D, Ma Y,
Zhu J, Ma L, Guo J, Shi H, et al: Exosomes derived from mature
dendritic cells increase endothelial inflammation and
atherosclerosis via membrane TNF-α mediated NF-κB pathway. J Cell
Mol Med. 20:2318–2327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu C, Gong Y, Yuan J, Zhang W, Zhao G, Li
H, Sun A, KaiHu, Zou Y and Ge J: microRNA-181a represses
ox-LDL-stimulated inflammatory response in dendritic cell by
targeting c-Fos. J Lipid Res. 53:2355–2363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gatti E, Velleca MA, Biedermann BC, Ma W,
Unternaehrer J, Ebersold MW, Medzhitov R, Pober JS and Mellman I:
Large-scale culture and selective maturation of human Langerhans
cells from granulocyte colony-stimulating factor-mobilized CD34+
progenitors. J Immunol. 164:3600–3607. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fabbri M, Paone A, Calore F, Galli R,
Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al:
MicroRNAs bind to Toll-like receptors to induce prometastatic
inflammatory response. Proc Natl Acad Sci USA. 109:E2110–E2116.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bang C, Batkai S, Dangwal S, Gupta SK,
Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, et al:
Cardiac fibroblast-derived microRNA passenger strand-enriched
exosomes mediate cardiomyocyte hypertrophy. J Clin Invest.
124:2136–2146. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H,
Liu J, Pan T, Chen J, Wu M, et al: Exosomes mediate the
cell-to-cell transmission of IFN-α-induced antiviral activity. Nat
Immunol. 14:793–803. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Andreou I, Sun X, Stone PH, Edelman ER and
Feinberg MW: miRNAs in atherosclerotic plaque initiation,
progression, and rupture. Trends Mol Med. 21:307–318. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng HS, Sivachandran N, Lau A, Boudreau
E, Zhao JL, Baltimore D, Delgado-Olguin P, Cybulsky MI and Fish JE:
MicroRNA-146 represses endothelial activation by inhibiting
pro-inflammatory pathways. EMBO Mol Med. 5:1017–1034. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu N, Zhang D, Chen S, Liu X, Lin L,
Huang X, Guo Z, Liu J, Wang Y, Yuan W and Qin Y: Endothelial
enriched microRNAs regulate angiotensin II-induced endothelial
inflammation and migration. Atherosclerosis. 215:286–293. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suárez Y, Wang C, Manes TD and Pober JS:
Cutting edge: TNF-induced microRNAs regulate TNF-induced expression
of E-selectin and intercellular adhesion molecule-1 on human
endothelial cells: Feedback control of inflammation. J Immunol.
184:21–25. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park H, Huang X, Lu C, Cairo MS and Zhou
X: MicroRNA-146a and microRNA-146b regulate human dendritic cell
apoptosis and cytokine production by targeting TRAF6 and IRAK1
proteins. J Biol Chem. 290:2831–2841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Niessner A and Weyand CM: Dendritic cells
in atherosclerotic disease. Clin Immunol. 134:25–32. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Galkina E and Ley K: Immune and
inflammatory mechanisms of atherosclerosis (*). Annu Rev Immunol.
27:165–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Subramanian M, Thorp E, Hansson GK and
Tabas I: Treg-mediated suppression of atherosclerosis requires
MYD88 signaling in DCs. J Clin Invest. 123:179–188. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alberts-Grill N, Denning TL, Rezvan A and
Jo H: The role of the vascular dendritic cell network in
atherosclerosis. Am J Physiol Cell Physiol. 305:C1–C21. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y, Shen Z and Yu XY: Transport of
microRNAs via exosomes. Nat Rev Cardiol. 12:1982015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Boon RA and Dimmeler S: MicroRNAs in
myocardial infarction. Nat Rev Cardiol. 12:135–142. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sahoo S and Losordo DW: Exosomes and
cardiac repair after myocardial infarction. Circ Res. 114:333–344.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Economou EK, Oikonomou E, Siasos G,
Papageorgiou N, Tsalamandris S, Mourouzis K, Papaioanou S and
Tousoulis D: The role of microRNAs in coronary artery disease: From
pathophysiology to diagnosis and treatment. Atherosclerosis.
241:624–633. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kumar S, Vijayan M, Bhatti JS and Reddy
PH: MicroRNAs as peripheral biomarkers in aging and age-related
diseases. Prog Mol Biol Transl Sci. 146:47–94. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kumar S and Reddy PH: Are circulating
microRNAs peripheral biomarkers for Alzheimer's disease? Biochim
Biophys Acta. 1862:1617–1627. 2016. View Article : Google Scholar : PubMed/NCBI
|