Introduction
Lung cancer, also known as primary bronchogenic
carcinoma of the lung, is a common malignancy, the incidence of
which has increased rapidly in previous years in China (1). Data published by the National Central
Cancer Registry of China in 2014 showed 605,900 new cases of lung
cancer in China, the highest of all malignant tumors; ~66% of the
patients were diagnosed too late for radical surgery (2,3). Of
those diagnosed too late, the 5-year survival rate of patients with
small cell lung cancer (SCLC), accounting for 15–17% of all lung
cancers, was only 6%; however, the median 5-year survival rate for
limited-stage SCLC was 28–60% (4).
Thus, early diagnosis is clearly essential for reducing SCLC
mortality. Many patients undergo a delayed examination and further
treatment because they exhibit no specific symptoms. Currently,
traditional medical diagnosis primarily relies on imaging
examinations (5); however, the
number of potentially transformed cell clones in patients with
early-stage lung cancer is lower than the minimum threshold of
current imaging and measurement technology. Therefore, searching
for sensitive and specific diagnostic methods applicable even
before imaging is necessary to increase the rate of early lung
cancer diagnosis.
Cancer proteomics is the study of cancer from the
perspective of protein and peptide fragments, and has great
potential for early cancer diagnosis. In recent years, mass
spectrometry (MS) of proteins and peptides has been increasingly
applied to exploratory studies on the early diagnosis of breast,
stomach and colon cancers, and other malignancies (6–8). The
present study employed matrix-assisted laser desorption/ionization
time-of-flight (MALDI-TOF)-MS and nanomagnetic beads to extract
serum peptides in combination with the ClinProt system to conduct
an exploratory analysis of the sera and urine of patients with SCLC
and healthy individuals, and screen for differentially expressed
peptides for establishing a foundation for the SCLC serum and urine
mass spectrometric diagnostic method. To the best of our knowledge,
this is the first exploratory study to identify small cell lung
cancer by blood and urine using MALDI-TOF-MS.
Materials and methods
Sample source and collection
In total, 155 patients with SCLC at the Department
of Lung Cancer of The Fifth Medical Centre, Chinese PLA General
Hospital, between October 2014 and April 2016, but only 72 patients
with SCLC were enrolled into the study according to the admission
criteria, with a median age of 58 (range, 36–83) years and a
male:female ratio of 1:1.54. Enrolled patients fulfilled the
following criteria: i) Histopathologically or cytologically
confirmed as a patient with small cell lung carcinoma [Extensive
Disease (ED) or Limited Disease (LD) stage] (9); ii) aged 18 years or older; iii)
Eastern Cooperative Oncology Group Performance Status score
(9) <2; and, iv) had not
previously undergone chemotherapy, radiotherapy, targeted therapy
or other cancer treatments. Over the corresponding period, 72 serum
and urine samples were collected from healthy individuals who
underwent a physical examination at the hospital and were age- and
sex-matched with the patients. The median age was 56 (range, 31–72)
years and the male:female ratio 1:1.32. These healthy individuals
were selected after fulfilling the following criteria: Older than
18 years, and without pulmonary nodules, pneumonia, tuberculosis,
or other abnormalities. This study was approved by the Ethics
Committee of The Fifth Medical Centre, Chinese PLA General Hospital
(no. 2012-11-171). All patients provided written informed consent
before undergoing any treatment, or serum or urine sampling. The
study overview is shown in Fig.
S1 and basic information regarding the characteristics of
patients and healthy subjects is presented in Table I.
 | Table I.Basic information on the
characteristics of patients with SCLC and healthy subjects. |
Table I.
Basic information on the
characteristics of patients with SCLC and healthy subjects.
| A, Training group
(n=108) |
|---|
|
|---|
| Characteristics | SCLC (n=54) | Healthy individual
(n=54) | P-value |
|---|
| Age (years) |
|
| 0.894 |
|
Median | 59 | 56 |
|
|
Range | 36–83 | 31–72 |
|
| Sex |
|
| 0.774 |
| Male | 33 | 32 |
|
|
Female | 21 | 22 |
|
| Ever smoked |
|
| 0.482 |
| Yes | 28 | 24 |
|
| No | 26 | 30 |
|
| Staging |
|
| – |
| LD | 5 | – |
|
| ED | 49 | – |
|
|
| B, Testing group
(n=36) |
|
|
Characteristics | SCLC
(n=18) | Healthy individual
(n=18) | P-value |
|
| Age (years) |
|
| 0.673 |
|
Median | 57 | 55 |
|
|
Range | 42–74 | 35–68 |
|
| Sex |
|
| 0.916 |
|
Male | 10 | 9 |
|
|
Female | 8 | 9 |
|
| Ever smoked |
|
| 0.910 |
|
Yes | 10 | 8 |
|
| No | 8 | 10 |
|
| Staging |
|
| – |
| LD | 0 | – |
|
| ED | 18 | – |
|
Serum (urine) sampling procedure
During fasting, 3 ml of peripheral venous whole
blood (15 ml of midstream urine) was collected at 8:00 am in a test
tube containing EDTA anticoagulant (urine collection tube). Samples
were maintained at 21°C for 2 h, then centrifuged at 2,500 × g for
10 min at 4°C. The supernatant (serum or urine) was collected,
divided into 150 µl aliquots, and stored in a freezer at −80°C.
Reagents and instrumentation
The following reagents and equipment were used in
the present study: Trifluoroacetic acid (TFA, Sigma-Aldrich; Merck
KGaA); acetonitrile (ACN; Thermo Fisher Scientific, Inc.);
α-Cyano-4-hydroxycinnamic acid (HCCA) and peptide mixture (Bruker
Corporation); copper ion-chelating nanomagnetic beads and buffer
system (National Center of Biomedical Analysis, Beijing, China);
MALDI-TOF-MS instrument (Ultraflex; Bruker Corporation); magnetic
bead separator (Bruker Corporation); 3K15 refrigerated benchtop
centrifuge (Sigma-Aldrich; Merck KGaA); MTP Anchorchip targets
(Var/384; Bruker Corporation); and ClinProTools version 3.0
analytical software (CPT; Bruker Corporation).
Sample preparation and mass analysis
(peptide profiling)
Serum and urine samples from 72 patients with SCLC
and 72 healthy individuals were randomly divided into two groups in
a 3:1 ratio as follows: A training set comprising samples from 54
patients with SCLC (lung cancer serum group I), 54 patients with
SCLC (lung cancer urine group I), 54 healthy individuals (healthy
serum group I), and 54 healthy individuals (healthy urine group I)
was used to construct the classification model, and a testing set
comprising samples from 18 patients with SCLC (lung cancer serum
group II), 18 patients with SCLC (lung cancer urine group II), 18
healthy individuals (healthy serum group II), and 18 healthy
individuals (healthy urine group II) was used to validate the
model. No statistically significant difference in age, sex, smoking
status, histological type, or clinical stage was observed between
the patients in the training and testing groups. The clinical and
pathological characteristics of all patients are listed in Table I. Bead suspension was retrieved
from a 4°C refrigerator, and inverted repeatedly to evenly
resuspend the solution. To the 200-µl sample tubes, 7 µl of
solid-phase extraction (SPE)-CM magnetic bead suspension, 10 µl of
serum, and 95 µl of SPE-CB (Beijing Bioyong Technology Co., Ltd.)
were added, and triturated several times to thoroughly mix the
magnetic beads, SPE-CB and serum. The mixture was maintained at
21°C for 5 min. Samples were placed in the magnetic bead separator
for 1 min to separate the magnetic beads from the solution. After
the liquid became clear, the remaining liquid was removed. Samples
were removed from the magnetic bead separator and 100 µl of SPE-CW
(Beijing Bioyong Technology Co., Ltd.) was added to the sample, and
triturated several times to mix the magnetic beads and SPE-CW
thoroughly. The mixture was held at 21°C for 2 min. Samples were
placed in the magnetic bead separator for 1 min to separate the
magnetic beads from the solution. When the liquid became clear, the
remaining liquid was removed. The abovementioned step was repeated
twice. Then, 10 µl of SPE-CE (Bioyong Tech) was added to each
sample tube, and triturated 10 times to mix the magnetic beads and
SPE-CE thoroughly. The mixture was held at 21°C for 5 min. Samples
were placed in the magnetic bead separator for 1 min to separate
the magnetic beads from the solution. When the liquid became clear,
it was transferred to a dry sample tube, and stored in a freezer at
−20°C for MS analysis.
Targeting
Saturated HCCA, 0.1% TFA and 50% ACN were mixed in
an Eppendorf tube to form the matrix solution. The standard peptide
mixture (polypeptide mixture) was mixed thoroughly in a 2:1 ratio.
For external instrument calibration, 1 µl of this sample was used.
Next, 1 µl of each sample after magnetic bead processing and 1 µl
of the prepared matrix solution were mixed thoroughly in a 1:1
ratio. Then, 1 µl of this sample was placed on an MTP Anchorchip
target, and allowed to air-dry at 21°C.
MS analysis
After extracting serum peptides from the SPE-CM
magnetic beads, linear mode detection via MALDI-TOF-MS was
performed. After targeting, the MTP Anchorchip target was placed in
a MALDI-TOF mass spectrometer for analysis. To reduce operational
error and systematic error, one standard target (standardized
peptide mixture) was used for each sample group, before analysis,
as an external sample for external instrument calibration. The best
scanning mass-to-charge ratio (m/z) range for MALDI-TOF-MS is
0.8–10 kDa. Each scan accumulated 500 mass spectrum signals, and
each sample accumulated 3,000 signals. The accumulated spectra were
saved, thereby generating an accurate serum peptide mass
fingerprint map comprising peptide peaks with different m/z
ratios.
Data collection and preservation
The datasets generated and/or analyzed during the
current study are available in a repository hosted by Mendeley at
the following link: http://dx.doi.org/10.17632/btg4dknchy.1, which
includes the ‘final upload data for this experiment’, and whether
the data uploaded onto Mendeley was used to establish the
prediction models or validate the classification model, as well as
its stability and reliability (10).
Statistical analysis
CPT software was used for the statistical analysis
of all mass spectral data obtained. Before complete analysis using
CPT, the original mass spectra of the samples were processed,
including baseline correction, alignment and normalization using
CPT. Next, the statistical analysis of the serum peptide spectra of
the 54 patients with SCLC and 54 healthy individuals composing the
training group was conducted using the statistical algorithms
included in CPT. The peptides differentially expressed between the
two groups were obtained; three algorithms [genetic algorithm (GA),
supervised neural networks and quick classifier], which components
of the CPT, were used to establish the prediction models.
Similarly, the statistical analysis of urine
peptides revealed the urine peptides differentially expressed
between the 54 patients with SCLC and 54 healthy individuals in the
training group, and a urine classification model for SCLC diagnosis
was constructed from a number of peptides selected by CPT. The
categorical variables were analyzed using the χ2 test;
GA was used to identify differentially expressed peptides between
the 54 SCLC patients and 54 healthy controls; Cross Tabulations
analysis was used for sensitivity and specificity; at the same
time, the plasma and urine classification models were combined to
verify the specificity and sensitivity of their combinations. κ
test was used to perform consistency analysis. All statistical
tests were two-tailed, and a P-value of <0.0005 was considered
statistically significant. All statistical tests were performed
using SPSS software (version 19; IBM Corp.).
Results
Serum peptide mass fingerprinting map
for training set
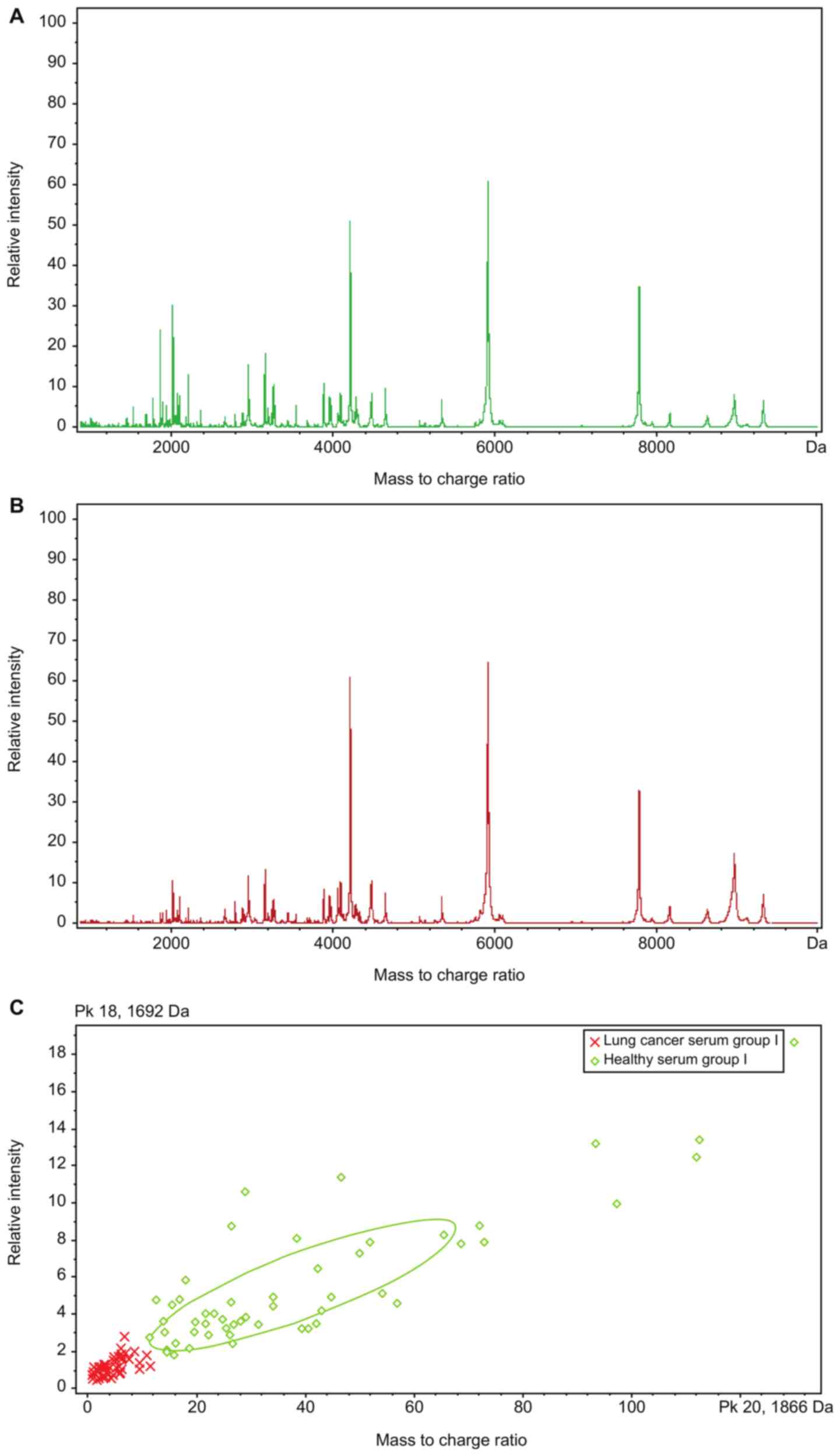
Fig. 1 shows the
serum peptide mass fingerprinting maps of lung cancer serum group I
and healthy serum group I obtained by performing MALDI-TOF-MS to
detect serum peptides in the training set extracted using the
magnetic beads for the 54 patients with SCLC and 54 healthy
individuals, respectively, in the training group.
Analysis of differentially expressed
peptides in serum samples of training set
The serum peptide mass fingerprinting maps of lung
cancer serum group I and healthy serum group I were analyzed using
CPT, which identified 126 peptide peaks in total; 19 peaks were
significantly different (P<0.0005; AUC≥0.8) between the 2 groups
(Table II).
 | Table II.Differential peaks in serum from the
patients with SCLC compared with healthy individuals in the
training group. |
Table II.
Differential peaks in serum from the
patients with SCLC compared with healthy individuals in the
training group.
| m/z (Da) | Peak areas of the
healthy group I (X±S) | Peak areas of the
SCLC group I (X±S) | Peak areas in
patients with SCLC compared with the healthy individuals | P-value |
|---|
| 1,866.37 | 24.8±17.7 | 2.85±1.61 | ↓ | <0.0005 |
| 1,779.22 | 7.7±5.4 | 1.19±0.59 | ↓ | <0.0005 |
| 1,692.13 | 3.47±2.21 | 0.77±0.33 | ↓ | <0.0005 |
| 3,541.28 | 5.56±3.18 | 2.61±1.46 | ↓ | <0.0005 |
| 5,251.47 | 0.5±0.18 | 0.87±0.32 | ↑ | <0.0005 |
| 2,210.62 | 13.3±9.48 | 4.03±4.15 | ↓ | <0.0005 |
| 2,366.84 | 4.26±2.86 | 1.45±1 | ↓ | <0.0005 |
| 1,968.41 | 1.94±0.69 | 1.28±0.34 | ↓ | <0.0005 |
| 8,952.22 | 8.37±6.19 | 16.59±9.9 | ↑ | <0.0005 |
| 4,055.64 | 3.66±2.07 | 9.41±6.81 | ↑ | <0.0005 |
| 2,082.49 | 8.75±6.06 | 3.56±2.34 | ↓ | <0.0005 |
| 3,444.06 | 1.8±0.84 | 2.96±1.44 | ↑ | <0.0005 |
| 839.2 | 4.38±1.87 | 2.47±1.4 | ↓ | <0.0005 |
| 1,450.69 | 2.61±1.8 | 1.1±0.55 | ↓ | <0.0005 |
| 1,980.53 | 2.46±0.96 | 1.47±0.55 | ↓ | <0.0005 |
| 1,011.84 | 2.53±1.7 | 0.98±0.67 | ↓ | <0.0005 |
| 1,887.67 | 2.68±1.47 | 1.44±1.1 | ↓ | <0.0005 |
| 1,882.02 | 2.56±1.81 | 1.04±0.89 | ↓ | <0.0005 |
| 4,111.85 | 1.45±0.53 | 2.33±1.09 | ↑ | <0.0005 |
The clustering analytical method of CPT (Fig. 1C) was used for comparison analysis
of the serum peptide mass fingerprinting maps of lung cancer serum
group I and healthy serum group I, and GAs were obtained. The
result of this algorithm showed that the optimal template comprised
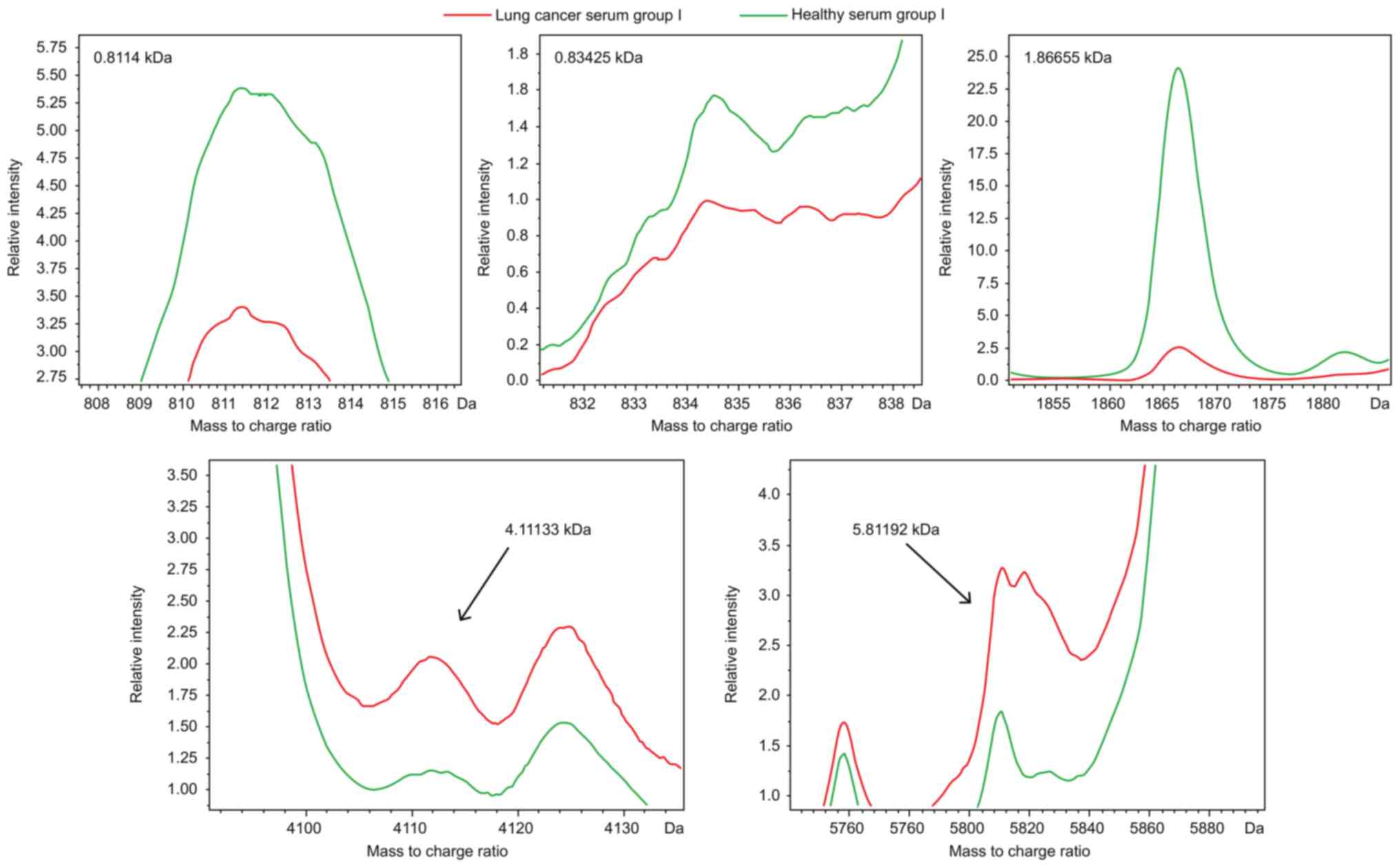
5 peptides whose m/z peptide peaks were at 0.8114, 0.83425,
1.86655, 4.11133 and 5.81192 kDa (Fig.
2). The resulting model had a lung cancer detection rate of
99.04% and cross-validation rate of 91.47%.
For the sera of the 18 patients with lung cancer and
18 healthy individuals in the testing set, the serum peptide mass
fingerprinting map was similarly obtained using CPT, and the model
constructed using GA was validated. The 18 patients with lung
cancer and 18 healthy individuals were accurately identified (1
false negative among the 18 patients with lung cancer, 2 false
positives among the 18 healthy individuals; the others were
correctly identified). Blinded validation showed that the model had
a specificity of 90% and sensitivity of 95%.
Analysis of differentially expressed
peptides in urine samples of training set
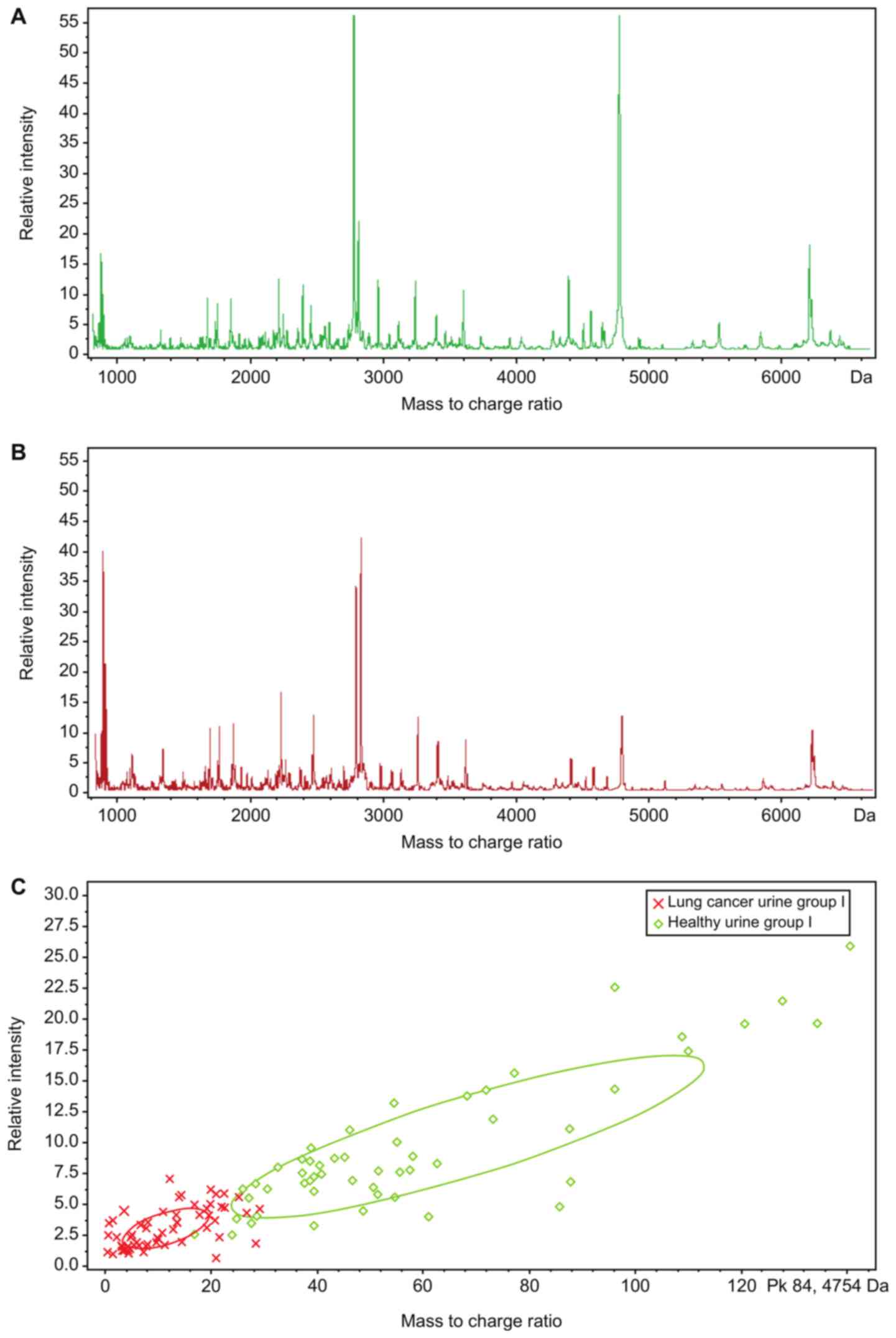
Fig. 3 shows the
urine peptide mass fingerprinting maps of lung cancer urine group I
and healthy urine group I, obtained using MALDI-TOF-MS to detect
urine peptides in the training set extracted from the magnetic
beads for the 54 patients with SCLC and 54 healthy individuals in
the training group.
The urine peptide mass fingerprinting maps of lung
cancer urine group I and healthy urine group I were analyzed using
CPT, which identified 132 peptide peaks in total; 8 peaks were
significantly different (P<0.0005; AUC≥0.8) between the two
groups (Table III).
 | Table III.Differential peaks in urine from the
patients with SCLC compared with healthy individuals in the
training group. |
Table III.
Differential peaks in urine from the
patients with SCLC compared with healthy individuals in the
training group.
| m/z (Da) | Peak areas of the
healthy group I (X±S) | Peak areas of the
SCLC group I (X±S) | Peak areas in SCLC
patients compared with the healthy individuals | P-value |
|---|
| 4,754.75 | 0.5±0.18 | 0.87±0.32 | ↓ | <0.0005 |
| 2,376.92 | 4.26±2.86 | 1.45±1.00 | ↓ | <0.0005 |
| 4,794.90 | 0.61±0.19 | 0.85±0.26 | ↓ | <0.0005 |
| 4,712.28 | 1.8±0.84 | 2.96±1.44 | ↓ | <0.0005 |
| 4,625.74 | 9.98±3.28 | 7.45±2.76 | ↓ | <0.0005 |
| 4,900.42 | 2±0.92 | 4.47±4.54 | ↓ | <0.0005 |
| 6,341.19 | 5.95±2.33 | 3.6±1.8 | ↑ | <0.0005 |
| 2,755.40 | 2.93±1.25 | 1.9±0.95 | ↓ | <0.0005 |
The clustering analytical method of CPT (Fig. 3C) was used for comparison analysis
of the urine peptide mass fingerprinting maps of lung cancer urine
group I and healthy urine group I, and GAs were obtained. The
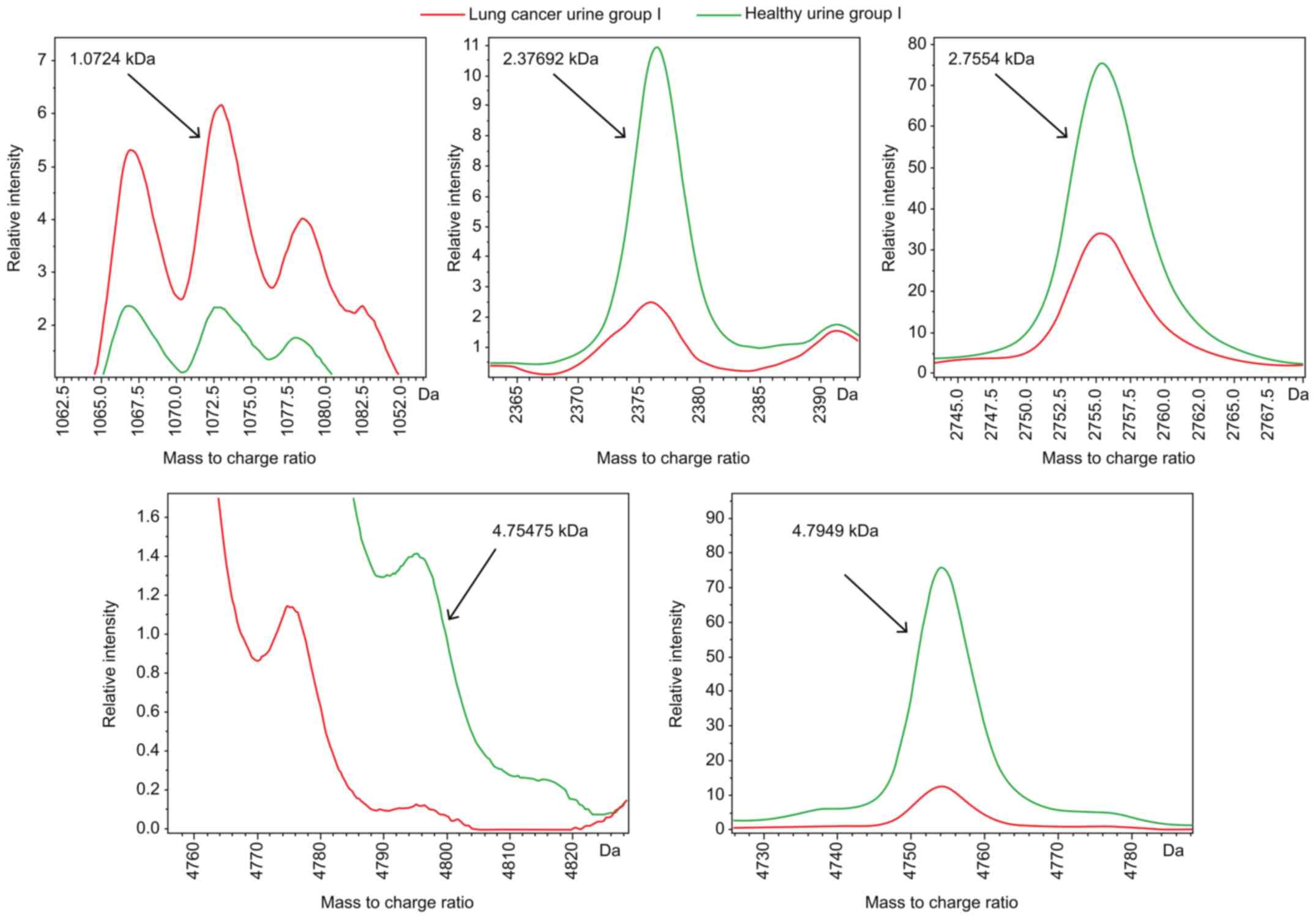
result of this algorithm showed that the optimal template comprised
5 peptides whose m/z peptide peaks were at 1.0724, 2.37692, 2.7554,
4.75475 and 4.7949 kDa (Fig. 4).
The resulting model had a lung cancer detection rate of 87.81% and
cross-validation rate of 79.49%.
For the urine samples of the 18 patients with lung
cancer and 18 healthy individuals in the testing set, the urine
peptide mass fingerprinting map was similarly obtained using CPT,
and the model constructed using GA was validated. The 18 patients
with lung cancer and 18 healthy individuals were accurately
identified (2 false negatives among the 18 patients with lung
cancer, 4 false positives among the 18 healthy individuals; the
others were correctly identified). Blinded validation showed that
the model had a specificity of 80% and a sensitivity of 90%.
Peptide peak identification
To further study the specific peptide peaks for
serum and urine, 3 peptide peaks with simultaneously upregulated
expression in the sera and urine of the patients with SCLC were
examined. Their m/z ratios were 2.3764, 0.8778 and 0.8616 kDa. A
search of the liquid chromatography/MS secondary mass spectrum
database of our laboratory determined the 2.3764-kDa peptide peak
to correspond to fibrinogen α, having the sequence SYK MAD EAG SEA
DHE GTH STK RGH AKS RPV, the 0.8778-kDa peptide peak to correspond
to glucose-6-phosphate isomerase, having the sequence RNG FKS HAL
QLN NRQ I, and the 0.8616-kDa peptide peak to correspond to
cyclin-dependent kinase-1, having the sequence SSK ITH RIH WES ASL
LR.
Consistency analysis of serum model
and urine model blind sample verification results
The results of the blind test results of the serum
model and the urine model were further analyzed. It was found that
in 18 patients with lung cancer and 18 healthy individuals, in the
serum model, 1 patient with lung cancer was false negative, and
this patient gave the same result in the urine model. Of the 2
patients who showed false positive results, 1 was still a false
positive in the urine model, and the other was diagnosed correctly
to be a healthy individual. The remaining patients showing false
positive or false negative results in the urine model were
diagnosed correctly in the serum model. It can be seen that the
sensitivity and specificity of the serum model are higher than
those of the urine model, and the serum model is in good agreement
with the urine model (κ=0.720; P<0.001). The results of the
blind sample verification results are shown in Table IV.
 | Table IV.Consistency analysis of serum model
and urine model blind sample verification results. |
Table IV.
Consistency analysis of serum model
and urine model blind sample verification results.
|
| Serum model |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Urine model | SCLC | Healthy | Total | κ | P-value |
|---|
| SCLC | 16 | 4 | 20 | 0.720 | <0.001 |
| Healthy | 3 | 13 | 16 |
|
|
| Total | 19 | 17 | 36 |
|
|
Discussion
Mass spectrometry analysis is an important detection
method in cancer proteomics. Currently, the mass spectrometric
techniques common in biology include MALDI-TOF-MS, electrospray
ionization (ESI)-MS, and surface-enhanced laser
desorption/ionization (SELDI)-TOF-MS. High tolerance to salt and
impurities and high sensitivity render MALDI-TOF-MS superior to
other MS techniques (11). After
reflective and delayed extraction techniques were introduced for
the method, the resolution of the method increased greatly,
reaching ≤10 ppm for detecting small molecular peptides of ~2 kDa
(12,13).
Considering the high sensitivity, high throughput
and other advantages of MS for protein identification, numerous
studies have attempted to use MS detection for early-stage cancer
diagnosis, primarily via MS analysis of samples from patients with
cancer and healthy individuals, thereby discovering differentially
expressed cancer-specific proteins or peptides. For example, Monari
et al (14) used IMAC30-Cu
and H50 SELDI-TOF-MS chips to study 44 patients with lung cancer
and 19 healthy individuals; the study observed 28 significantly
different peaks and constructed a model. The sensitivity and
specificity of the IMAC30-Cu chip were 70.45 and 68.42%,
respectively, and the sensitivity and specificity of the H50 chip
were 72.73 and 73.68%, respectively. Han et al (15) used metal affinity protein chips and
SELDI-TOF technology to study serum samples and used 3 proteins to
construct a classification tree model. Blinded validation revealed
a sensitivity of 89%, specificity of 91%, and accuracy of 90%.
Hocker et al (16) used
ESI-MS to analyze serum samples of 43 patients with early-stage
lung cancer and 21 healthy individuals, and the method using the
differentially expressed proteins identified for differentiating
between the cancer patients and healthy individuals had an overall
efficacy of 80% and sensitivity of 84%. Results from these studies
demonstrate that MS detection methods can recognize cancer-related
molecules contained in the sera of patients with lung cancer, and
can potentially be used for early lung cancer diagnosis.
However, the sensitivity and specificity of blinded
validation in the aforementioned studies were relatively low, with
no further validation being subsequently performed for the
following reasons: i) All traditional methods for extracting
proteins and peptides from samples, including pre-processing
techniques, such as 2-dimensional gel electrophoresis, protein
chip-based methods, or solid phase extraction, have operational
complexities and low extraction efficiencies, resulting in the loss
of usable information; and ii) ESI, SELDI-TOF-MS, and other mass
spectrometric detection methods have low sensitivity and
resolution, and reflect limited information. These limitations have
restricted their application in disease-specific marker discovery
and clinical diagnosis. Hence, the present study used the CPT
detection system developed by Bruker Daltonics (17). It includes a magnetic bead
separation system, a MALDI-TOF-MS system, analytical software and
automated body fluid sample processing system. Its advantages are
that it contains the latest Anchor-Chip patented sample targeting
technology, which can greatly increase the analyte concentration,
and increase sensitivity to 10–100 times that of conventional
chips, and the resolution, repeatability and reproducibility of
MALDI-TOF-MS are higher than those of SELDI-TOF and other mass
spectrometric techniques (18). At
various world-class hospitals and medical research institutes, this
technology has been widely used in studies on the early diagnosis
of ovarian cancer, lung adenocarcinoma and multiple myeloma
patients (19–21).
The present study used the CPT detection system for
the sera and urine of patients with SCLC and healthy individuals,
and discovered 19 differentially expressed peptides in serum and 8
differentially expressed peptides in urine. Next, the statistical
software screened 5 peptide peaks for constructing the serum and
urine SCLC diagnostic classification models. Blinded model
validation was performed, and satisfactory results were obtained,
demonstrating that the classification model of the present study
had a certain theoretical value for identifying patients with SCLC,
although it still requires further clinical validation. The
classification model of the present study was unlike traditional
diagnostic models based on a single index for detection; rather, it
used a group of peptide indicators as detection criteria. It used
biological mass spectrometry as the support platform, and
bioinformatics as the bridge for peptide research, and via changes
in the blood peptidome of patients with lung cancer, specific
cancer-related information could be obtained to establish cancer
diagnosis. When blood flows through the tumor microenvironment, it
may cause various slight changes in unknown components of serum
proteins/peptides, making the use of markers in combination more
effective than the use of a single marker. In the present study,
the statistically significantly differentially expressed proteins
observed in both serum and urine were fibrinogen α,
glucose-6-phosphate isomerase and cyclin-dependent kinase-1.
Fibrinogen is a protein with coagulant activity synthesized by the
liver; a number of studies have shown that fibrinogen is
upregulated in cancers of the urinary system, reproductive system
and gastrointestinal tract (22–24).
The present study identified high fibrinogen α
expression in the sera and urine of patients with SCLC, which may
be associated with the hypercoagulable state of patients with
cancer. Glucose-6-phosphate isomerase is an important enzyme in
humans. The main function is to catalyze the exchange between
D-glucose-6-phosphate and D-fructose-6-phosphate, which is an
important enzyme for glycolysis and gluconeogenesis. Although there
are no reports on its involvement in cancer development and
progression, to the best of the authors' knowledge.
Cyclin-dependent kinase 1 is a protein kinase family member, and
depends on binding with cell cycle proteins to serve key roles in
the systematic progress of the cell cycle. When cell cycle proteins
are lacking, or cyclin-dependent kinase inhibitors are present,
cell division is arrested and cell death may occur (25). Future studies will employ ELISA to
validate the expression of the three peptides identified in the
serum and urine samples of the patients with SCLC. The majority of
SCLC cases are already in LD stages when discovered (there were
only 5 ED patients in the present study). The model has been
established and validated in patients with LD SCLC, but whether
this model can be used for ED SCLC patient diagnosis remains to be
verified. The future aims of this study are to increase the number
of LD SCLC samples used, validate the reproducibility and
reliability of the diagnostic classification model, further
identify the peptides corresponding to the m/z peaks, and evaluate
protein/peptide expression, to identify novel tumor markers of lung
cancer, and study their associations with the molecular mechanisms
of lung cancer development.
The use of the specific serum and urine
peptide-spectrum model for SCLC was revealed to be accurate in a
small cohort of patients. In the future, a large-scale validation
with actual clinical operation should be conducted, and it is
expected to create the foundation for developing a serum- and
urine-based diagnosis that is rapid, non-invasive and requires a
small sample quantity.
The present study selected the sera and urine of
patients with SCLC and healthy individuals as the research
subjects, used MALDI-TOF-MS with the CPT system to detect serum and
urine peptides, performed small-scale validation of specific
SCLC-related peptides, and achieved high sensitivity and
specificity, as the samples originated primarily from LD-stage
patients. Samples from more ED-stage patients, as well as dynamic
monitoring of samples from high-risk populations, should be
included in future studies, to further validate the sensitivity and
specificity of this diagnostic model.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was funded by the Chinese National
Instrumentation Program (Beijing, China; grant no.
2011YQI70067).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in a repository hosted by Mendeley at
the following link: http://dx.doi.org/10.17632/btg4dknchy.1.
Authors' contributions
XLiu, ZL and PL conceived and designed this study.
KH and BX acquired the data. PL, CT, SY and HQ analyzed and
interpreted the data. The original draft was written, and the
subsequent review and editing for important intellectual content
was performed by HG, PL and XLi.
Ethics approval and consent to
participate
This study was conducted using protocols approved by
the ethics committee of The Fifth Medical Centre, Chinese PLA
General Hospital for drug clinical trials (no. 2012-11-171). All
patients provided written informed consent before undergoing any
treatment or serum or urine sampling. Patients with heart, liver,
kidney, or other major organ diseases were excluded.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AUC
|
area under curve
|
|
CPT
|
ClinProTools
|
|
ED
|
Extensive Disease
|
|
HCCA
|
hydroxycinnamic acid
|
|
LD
|
Limited Disease
|
|
MALDI-TOF-MS
|
matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry
|
|
SCLC
|
small cell lung cancer
|
|
SPE
|
solid-phase extraction
|
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer incidence and mortality
worldwide: IARC cancer base no. 11 [Internet]. International Agency
for Research on Cancer; Lyon: 2014
|
|
3
|
Kahnert K, Kauffmann-Guerrerom D and Huber
RM: SCLC-state of the art and what does the future have in store?
Clin Lung Cancer. 17:325–333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
National Central Cancer Registry of China,
. China Cancer Registration Annual Report. 2014.
|
|
5
|
Evans WK, Flanagan WM, Miller AB, Goffin
JR, Memon S, Fitzgerald N and Wolfson MC: Implementing low-dose
computed tomography screening for lung cancer in Canada:
Implications of alternative at-risk populations, screening
frequency, and duration. Curr Oncol. 23:e179–e187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeh CY, Adusumilli R, Kullolli M, Mallick
P, John EM and Pitteri SJ: Assessing biological and technological
variability in protein levels measured in pre-diagnostic plasma
samples of women with breast cancer. Biomark Res. 17:302017.
View Article : Google Scholar
|
|
7
|
Xu W, Li X, Lin N, Zhang X, Huang X, Wu T,
Tai Y, Chen S, Wu CH, Huang M and Wu S: Pharmacokinetics and tissue
distribution of five major triterpenoids after oral administration
of Rhizoma Alismatis extract to rats using ultra high-performance
liquid chromatography-tandem mass spectrometry. J Pharm Biomed
Anal. 146:314–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Popp R, Li H, Le Blanc A, Mohammed Y,
Aguilar-Mahecha A, Chambers AG, Lan C, Poetz O, Basik M, Batist G
and Borchers CH: Immuno-matrix-assisted laser desorption/ionization
assays for quantifying AKT1 and AKT2 in breast and colorectal
cancer cell lines and tumors. Anal Chem. 89:10592–10600. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
National Comprehensive Cancer Network:
NCCN Clinical Practice Guidelines in Oncology-Non-Small Cell Lung
Cancer, version 2.2016. http://www.nccn.org/professionals/physician_gls/f_guidelines.AspDecember
04–2015
|
|
10
|
Panpan IV: Exploratory study on
application of MALDI-TOF-MS to detect serum and urine peptides
related to small cell lung carcinoma. Mendeley Data, v1, 2018.
http://dx.doi.org/10.17632/btg4dknchy.1
|
|
11
|
Cho A and Normile D: Nobel prize in
chemistry: Mastering macromolecules. Science. 298:527–528. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dekker LJ, Boogerd W, Stockhammer G,
Dalebout JC, Siccama I, Zheng P, Bonfrer JM, Verschuuren JJ,
Jenster G, Verbeek MM, et al: MALDI-TOF mass spectrometry analysis
of cerebrospinal fluid tryptic peptide profiles to diagnose
Leptomeningeal metastases in patients with breast cancer. Mol Cell
Proteomics. 4:1341–1349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Villanueva J, Shaffer DR, Philip J,
Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H,
Brogi E, Boyd J, et al: Differential exoprotease activities confer
tumor-specific serum peptidome patterns. J Clin Invest.
116:271–284. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Monari E, Casali C, Cuoghi A, Nesci J,
Bellei E, Bergamini S, Fantoni LI, Natali P, Morandi U and Tomasi
A: Enriched sera protein profiling for detection of non-small cell
lung cancer biomarkers. Proteome Sci. 9:552011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han KQ, Huang G, Gao CF, Wang XL, Ma B,
Sun LQ and Wei ZJ: Identification of lung cancer patients by serum
protein profiling using surface-enhanced laser
desorption/ionization time-of-flight mass spectrometry. Am J Clin
Oncol. 31:133–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hocker JR, Peyton MD, Lerner MR, Mitchell
SL, Lightfoot SA, Lander TJ, Bates-Albers LM, Vu NT, Hanas RJ,
Kupiec TC, et al: Serum discrimination of early-stage lung cancer
patients using electrospray-ionization mass spectrometry. Lung
Cancer. 74:206–211. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ketterlinus R, Hsieh SY, Teng SH, Lee H
and Pusch W: Fishing for biomarkers: Analyzing mass spectrometry
data with the new ClinProTools software. Biotechniques. 38
(Suppl):S37–S40. 2009. View Article : Google Scholar
|
|
18
|
Telesmanich NR, Agafonova VV, Chemisova
OS, Chaĭka IA, Vodop'ianov AS, Goncharenko EV and Telicheva VO:
Proteome mass-spectrometric analysis and typing of Vibrio cholerae
strains isolated in the territory of Russian Federation in
2010–2012. Zh Mikrobiol Epidemiol Immunobiol. 97–101. 2014.(In
Russian). PubMed/NCBI
|
|
19
|
Qiu F, Liu HY, Dong ZN, Feng YJ, Zhang XJ
and Tian YP: Searching for potential ovarian cancer biomarkers with
matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry. Am J Biomed Sci. 1:80–90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Xu B, Tang C, Li X, Qin H, Wang W,
Wang H, Wang Z, Li L, Li Z, et al: The exploration of peptide
biomarkers in malignant pleural effusion of lung cancer using
Matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry. Dis Markers. 2017:31604262017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deulofeu M, Kolářová L, Salvadó V, María
Peña-Méndez E, Almáši M, Štork M, Pour L, Boadas-Vaello P,
Ševčíková S, Havel J and Vaňhara P: Rapid discrimination of
multiple myeloma patients by artifcial neural networks coupled with
mass spectrometry of peripheral blood plasma. Sci Rep. 9:79752019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coverley D, Higgins G, West D, Jackson OT,
Dowle A, Haslam A, Ainscough E, Chalkley R and White J: A
quantitative immunoassay for lung cancer biomarker CIZ1b in patient
plasma. Clin Biochem. 50:336–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim YS, Gu BH, Choi BC, Kim MS, Song S,
Yun JH, Chung MK, Choi CH and Baek KH: Apolipoprotein A-IV as a
novel gene associated with polycystic ovary syndrome. Int J Mol
Med. 31:707–716. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu C, Luo Z, Tang D, Liu L, Yao D, Zhu L
and Wang Z: Identification of carboxyl terminal peptide 410 of
fibrinogen as a potential serum biomarker for gastric cancer.
Tumour Biol. 37:6963–6970. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parrilla A, Cirillo L, Thomas Y, Gotta M,
Pintard L and Santamaria A: Mitotic entry: The interplay between
Cdk1, Plk1 and Bora. Cell Cycle. 15:3177–3182. 2016. View Article : Google Scholar : PubMed/NCBI
|