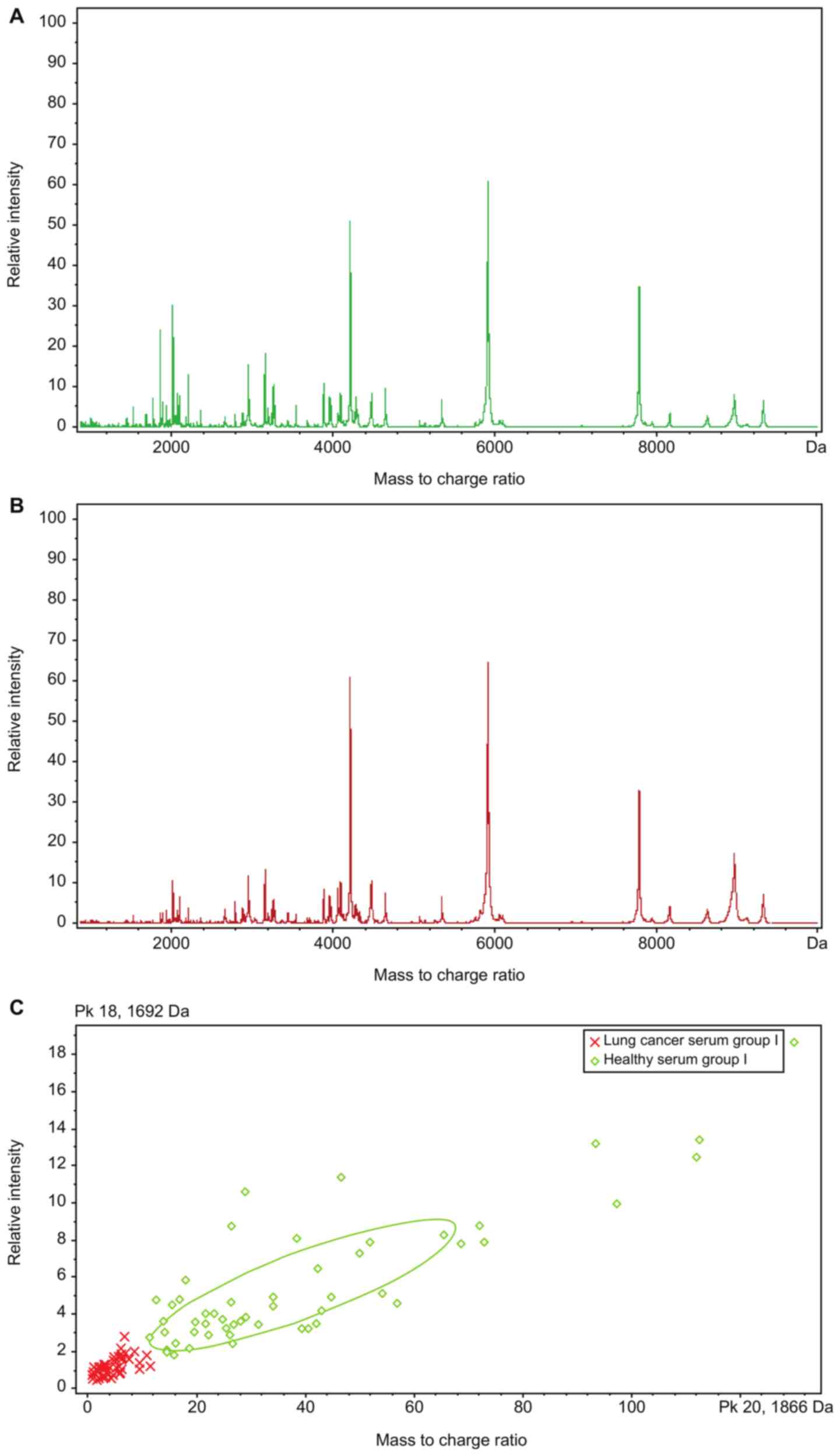
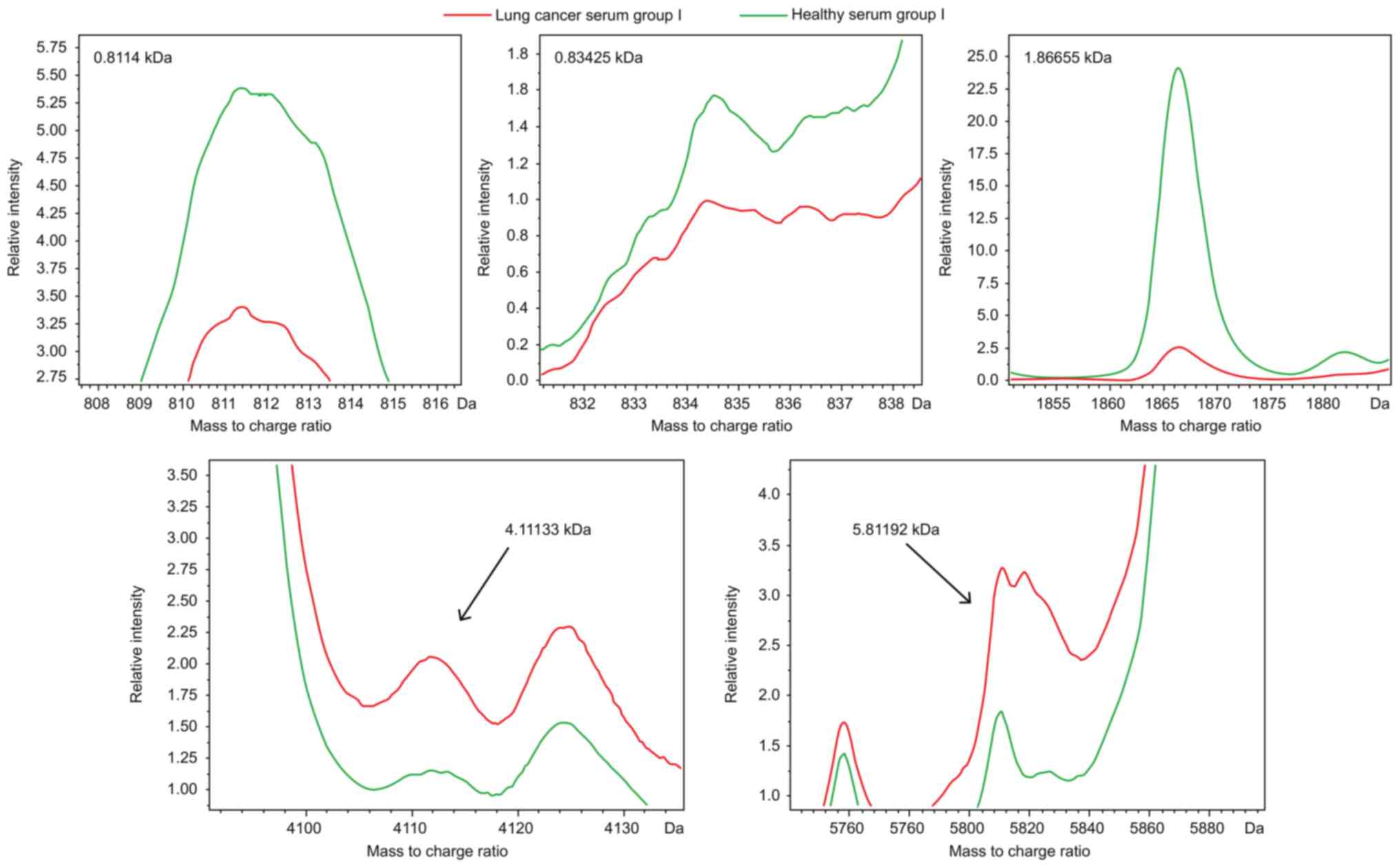
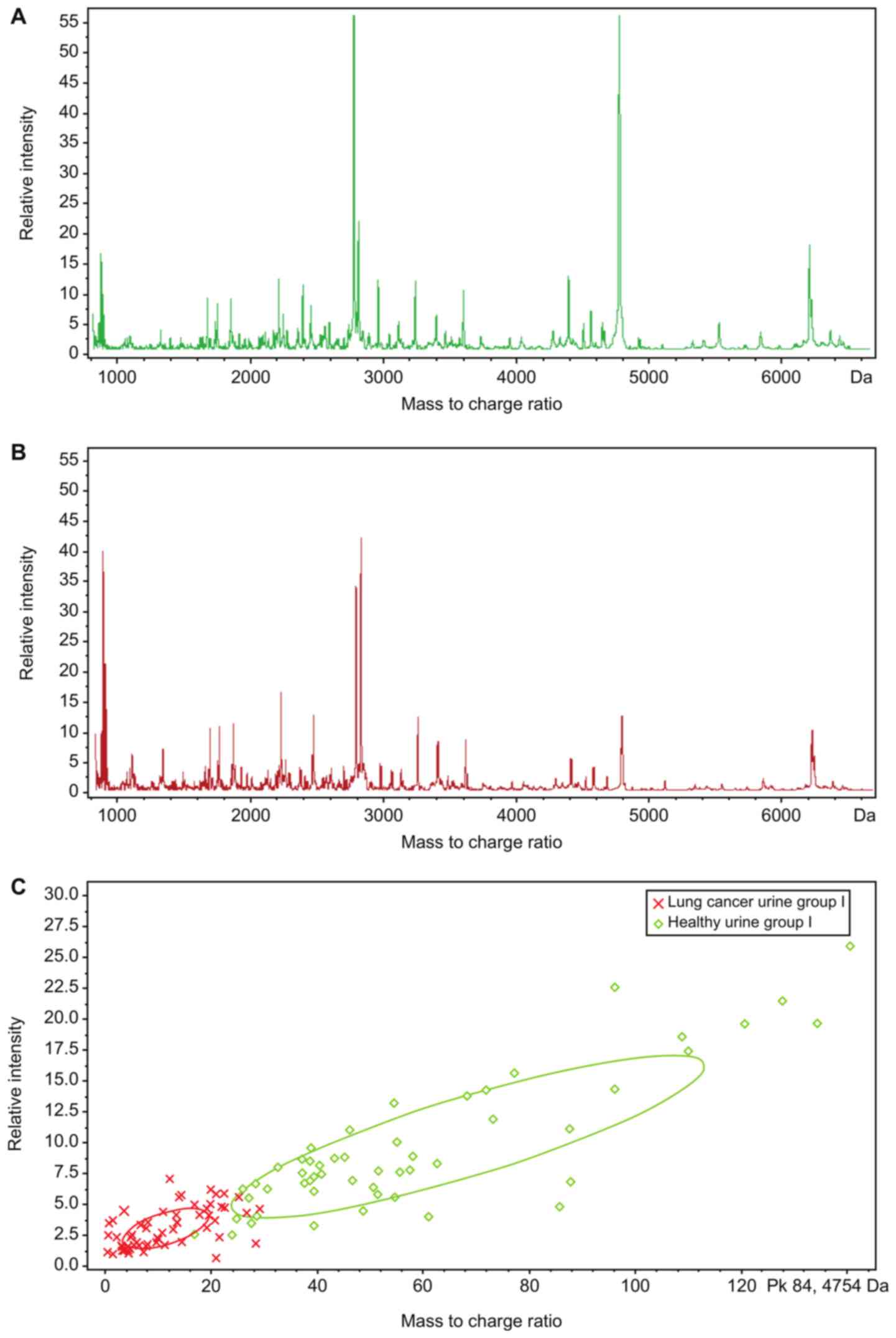
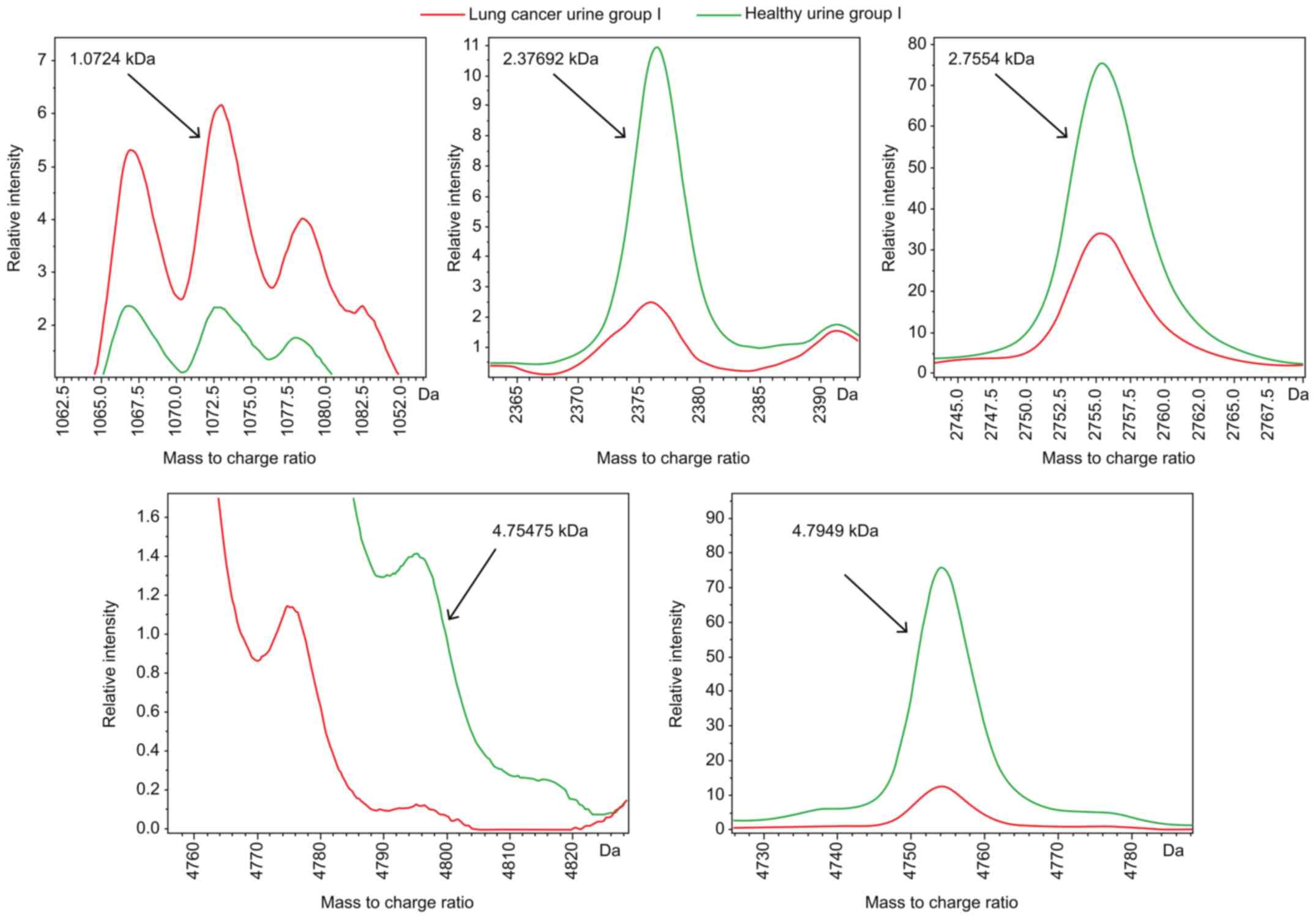
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer incidence and mortality
worldwide: IARC cancer base no. 11 [Internet]. International Agency
for Research on Cancer; Lyon: 2014
|
|
3
|
Kahnert K, Kauffmann-Guerrerom D and Huber
RM: SCLC-state of the art and what does the future have in store?
Clin Lung Cancer. 17:325–333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
National Central Cancer Registry of China,
. China Cancer Registration Annual Report. 2014.
|
|
5
|
Evans WK, Flanagan WM, Miller AB, Goffin
JR, Memon S, Fitzgerald N and Wolfson MC: Implementing low-dose
computed tomography screening for lung cancer in Canada:
Implications of alternative at-risk populations, screening
frequency, and duration. Curr Oncol. 23:e179–e187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeh CY, Adusumilli R, Kullolli M, Mallick
P, John EM and Pitteri SJ: Assessing biological and technological
variability in protein levels measured in pre-diagnostic plasma
samples of women with breast cancer. Biomark Res. 17:302017.
View Article : Google Scholar
|
|
7
|
Xu W, Li X, Lin N, Zhang X, Huang X, Wu T,
Tai Y, Chen S, Wu CH, Huang M and Wu S: Pharmacokinetics and tissue
distribution of five major triterpenoids after oral administration
of Rhizoma Alismatis extract to rats using ultra high-performance
liquid chromatography-tandem mass spectrometry. J Pharm Biomed
Anal. 146:314–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Popp R, Li H, Le Blanc A, Mohammed Y,
Aguilar-Mahecha A, Chambers AG, Lan C, Poetz O, Basik M, Batist G
and Borchers CH: Immuno-matrix-assisted laser desorption/ionization
assays for quantifying AKT1 and AKT2 in breast and colorectal
cancer cell lines and tumors. Anal Chem. 89:10592–10600. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
National Comprehensive Cancer Network:
NCCN Clinical Practice Guidelines in Oncology-Non-Small Cell Lung
Cancer, version 2.2016. http://www.nccn.org/professionals/physician_gls/f_guidelines.AspDecember
04–2015
|
|
10
|
Panpan IV: Exploratory study on
application of MALDI-TOF-MS to detect serum and urine peptides
related to small cell lung carcinoma. Mendeley Data, v1, 2018.
http://dx.doi.org/10.17632/btg4dknchy.1
|
|
11
|
Cho A and Normile D: Nobel prize in
chemistry: Mastering macromolecules. Science. 298:527–528. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dekker LJ, Boogerd W, Stockhammer G,
Dalebout JC, Siccama I, Zheng P, Bonfrer JM, Verschuuren JJ,
Jenster G, Verbeek MM, et al: MALDI-TOF mass spectrometry analysis
of cerebrospinal fluid tryptic peptide profiles to diagnose
Leptomeningeal metastases in patients with breast cancer. Mol Cell
Proteomics. 4:1341–1349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Villanueva J, Shaffer DR, Philip J,
Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H,
Brogi E, Boyd J, et al: Differential exoprotease activities confer
tumor-specific serum peptidome patterns. J Clin Invest.
116:271–284. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Monari E, Casali C, Cuoghi A, Nesci J,
Bellei E, Bergamini S, Fantoni LI, Natali P, Morandi U and Tomasi
A: Enriched sera protein profiling for detection of non-small cell
lung cancer biomarkers. Proteome Sci. 9:552011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han KQ, Huang G, Gao CF, Wang XL, Ma B,
Sun LQ and Wei ZJ: Identification of lung cancer patients by serum
protein profiling using surface-enhanced laser
desorption/ionization time-of-flight mass spectrometry. Am J Clin
Oncol. 31:133–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hocker JR, Peyton MD, Lerner MR, Mitchell
SL, Lightfoot SA, Lander TJ, Bates-Albers LM, Vu NT, Hanas RJ,
Kupiec TC, et al: Serum discrimination of early-stage lung cancer
patients using electrospray-ionization mass spectrometry. Lung
Cancer. 74:206–211. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ketterlinus R, Hsieh SY, Teng SH, Lee H
and Pusch W: Fishing for biomarkers: Analyzing mass spectrometry
data with the new ClinProTools software. Biotechniques. 38
(Suppl):S37–S40. 2009. View Article : Google Scholar
|
|
18
|
Telesmanich NR, Agafonova VV, Chemisova
OS, Chaĭka IA, Vodop'ianov AS, Goncharenko EV and Telicheva VO:
Proteome mass-spectrometric analysis and typing of Vibrio cholerae
strains isolated in the territory of Russian Federation in
2010–2012. Zh Mikrobiol Epidemiol Immunobiol. 97–101. 2014.(In
Russian). PubMed/NCBI
|
|
19
|
Qiu F, Liu HY, Dong ZN, Feng YJ, Zhang XJ
and Tian YP: Searching for potential ovarian cancer biomarkers with
matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry. Am J Biomed Sci. 1:80–90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Xu B, Tang C, Li X, Qin H, Wang W,
Wang H, Wang Z, Li L, Li Z, et al: The exploration of peptide
biomarkers in malignant pleural effusion of lung cancer using
Matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry. Dis Markers. 2017:31604262017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deulofeu M, Kolářová L, Salvadó V, María
Peña-Méndez E, Almáši M, Štork M, Pour L, Boadas-Vaello P,
Ševčíková S, Havel J and Vaňhara P: Rapid discrimination of
multiple myeloma patients by artifcial neural networks coupled with
mass spectrometry of peripheral blood plasma. Sci Rep. 9:79752019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coverley D, Higgins G, West D, Jackson OT,
Dowle A, Haslam A, Ainscough E, Chalkley R and White J: A
quantitative immunoassay for lung cancer biomarker CIZ1b in patient
plasma. Clin Biochem. 50:336–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim YS, Gu BH, Choi BC, Kim MS, Song S,
Yun JH, Chung MK, Choi CH and Baek KH: Apolipoprotein A-IV as a
novel gene associated with polycystic ovary syndrome. Int J Mol
Med. 31:707–716. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu C, Luo Z, Tang D, Liu L, Yao D, Zhu L
and Wang Z: Identification of carboxyl terminal peptide 410 of
fibrinogen as a potential serum biomarker for gastric cancer.
Tumour Biol. 37:6963–6970. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parrilla A, Cirillo L, Thomas Y, Gotta M,
Pintard L and Santamaria A: Mitotic entry: The interplay between
Cdk1, Plk1 and Bora. Cell Cycle. 15:3177–3182. 2016. View Article : Google Scholar : PubMed/NCBI
|