Introduction
Prostate cancer (PCa) is the second most common type
of cancer among men globally, and constitutes ~15% of all cancer
diagnoses worldwide (1). Digital
rectal examination, measurement of the serum level of prostate
specific antigen (PSA) and biopsy from a prostate transrectal
ultrasonography are the most common diagnostic tools for PCa
(2). Additionally, with advances
in genetic analysis, alterations have been identified in a number
of gene regions in patients with PCa, including prostate antigen 3,
androgen-dependent transmembrane serine 2 and S-transferase P1
(3–5). However, genetic analysis exhibits a
low specificity and can increase the number of unnecessary biopsies
performed without reducing patient mortality (6). Previous studies have associated the
tribbles pseudokinase 1 gene with the development of a number of
tumors, including colorectal leukemia and hepatocellular cancers
(7–9). It has been shown that transmembrane
protease, serine 2:ETS-related gene (TMPRSS2:ERG) fusion is
associated with diagnosing PCa in urine samples and DNA-based
molecular templates (10).
However, due to the lack of effective diagnostic methods during the
early stages of the disease, the mortality rate of PCa remains high
(10). Therefore, it is crucial to
understand the molecular mechanisms associated with PCa
carcinogenesis, proliferation and recurrence.
Microarray technology and bioinformatics analysis
led to the identification of 273 differentially expressed genes
(DEGs) and functional pathways in the carcinogenesis and
progression of PCa. In the current study, two mRNA microarray
datasets from Gene Expression Omnibus (GEO) were analyzed to
identify DEGs between PCa tissues and non-cancerous tissues.
Subsequently, the molecular mechanisms of PCa carcinogenesis and
progression were investigated using Gene Ontology (GO), Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis and protein-protein interaction (PPI) network analyses. In
conclusion, a total of 273 DEGs and 8 hub genes were identified in
the current study, and these genes may be candidate biomarkers for
PCa.
Materials and methods
Database
GEO (http://www.ncbi.nlm.nih.gov/geo) (11) is a public functional genomics
database. GSE3325 (12) and
GSE6919 (13) were downloaded from
GEO (Affymetrix Human Genome U133 Plus 2.0 Array). The GSE3325
dataset contained 12 PCa tissue samples and 12 non-cancerous
samples. GSE6919 contained 8 PCa samples and 8 non-cancerous
samples.
Identification of DEGs
The Affy package (version 1.52.0) (14) was used to preprocess the raw
expression data in the R statistical software (R ×64 3.5.3;
http://cran.r-project.org). DEGs were
subsequently identified between PCa and normal samples using the
limma (version 3.34.7) package of the R statistical software
(https://bioconductor.org/packages/release/bioc/html/limma.html).
DEGs with log2FC >1 and P<0.01 were selected in the
microarray data.
Enrichment analysis of DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; version 6.7; http://david.ncifcrf.gov) (15), which provides functional annotation
information of genes and proteins, was used to perform DEG analysis
of KEGG pathway enrichment (16)
and GO annotation (17). P<0.05
was set as the threshold value.
Module analysis and construction of
the PPI network
The Search Tool for the Retrieval of Interacting
Genes (version 10.0; http://string-db.org) (18), which offers comprehensive
information on PPIs, was used to create the PPI network. The
molecular interaction networks were visualized using Cytoscape
(version 3.4.0; http://cytoscape.org/) (19). The Molecular Complex Detection
(MCODE) (version 1.4.2) of Cytoscape was used to identify densely
connected regions (20). The PPI
networks were visualized using Cytoscape software. The hub genes
and the most significant module in the PPI networks were identified
using MCODE.
Hub genes selection and analysis
The hub genes were selected and their co-expression
genes were analyzed using cBioPortal (http://www.cbioportal.org) (21). Hierarchical clustering of hub genes
was constructed using the University of California, Santa Cruz
Cancer Browser (http://genome-cancer.ucsc.edu) (22). The overall survival and
disease-free survival analyses of hub genes (the cutoff was the
median expression value) were performed using Kaplan-Meier analysis
in cBioPortal. The hazard ratio (HR) with 95% confidence intervals
and log-rank P-value was also computed. The expression of PDZ
binding kinase (PBK) and Krüppel-like factor 4 (KLF4) in cancer
tissues were analyzed and presented using the online database
Serial Analysis of Gene Expression (SAGE; http://www.ncbi.nlm.nih.gov/SAGE) (23). The relationship between expression
patterns and tumor-node-metastasis (TNM) stage, Gleason grade and
recurrence status were analyzed using the online database Oncomine
(http://www.oncomine.com) (24).
Results
Identification of DEGs in PCa
After standardization of the microarray results, a
total of 1,024 DEGs in GSE6919 and 2,371 DEGs in GSE3325 were
identified. The overlap between the 2 datasets contained 273 genes,
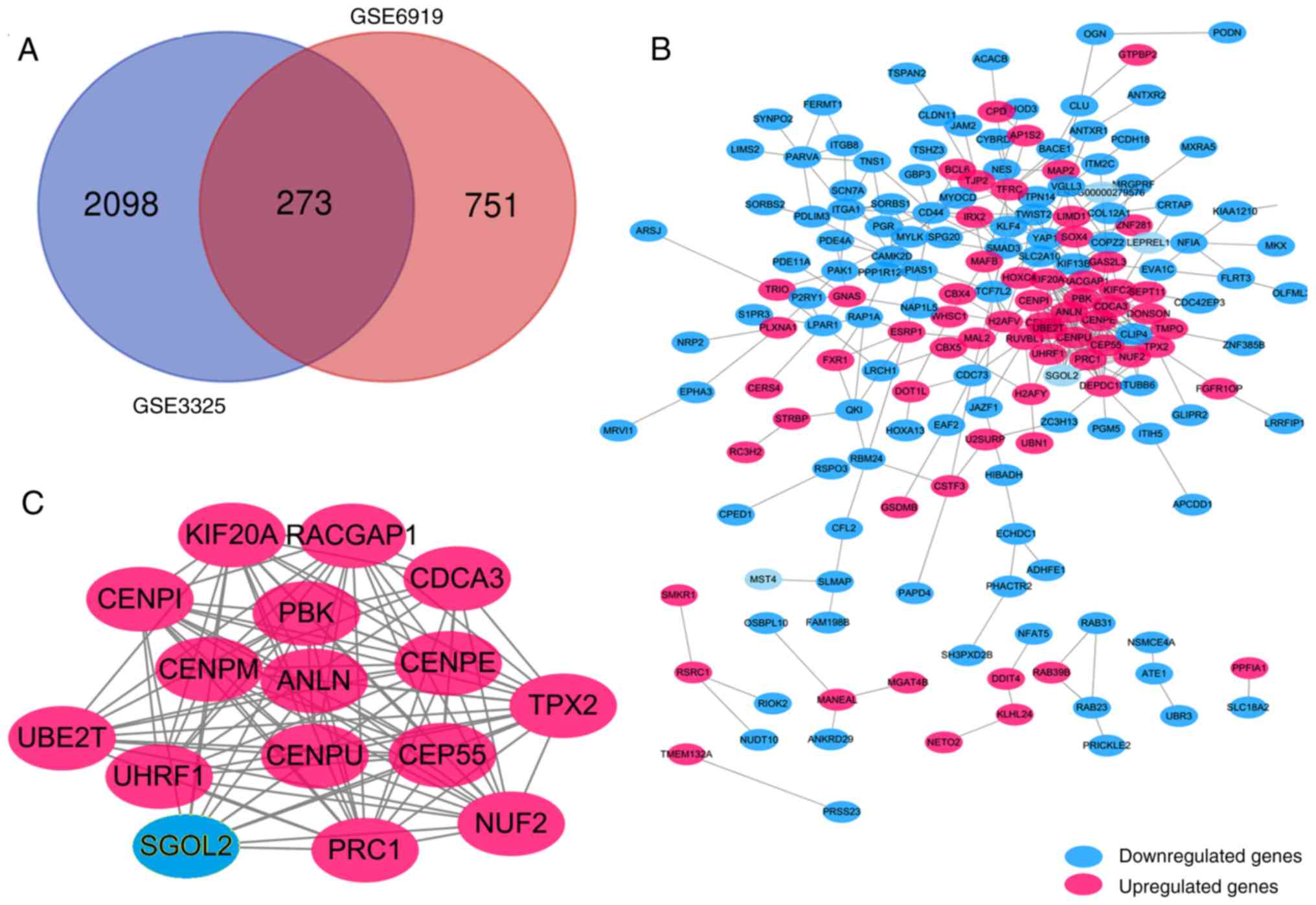
as presented in Fig. 1A,
consisting of 173 downregulated genes and 100 upregulated genes in
PCa tissues.
PPI network and module analysis
The PPI network of DEGs (Fig. 1B) and the most significant module
were identified using Cytoscape (Fig.
1C). The functional analyses of DEGs demonstrated that genes in
this module were mainly enriched in nucleotide binding, small
molecule binding, focal adhesion and the regulation of the actin
cytoskeleton (Table I).
 | Table I.GO and KEGG pathway enrichment
analysis of DEGs in the most significant module. |
Table I.
GO and KEGG pathway enrichment
analysis of DEGs in the most significant module.
| Pathway ID | Pathway
description | Count in gene
set | FDR |
|---|
| GO:0000166 | Nucleotide
binding | 6 | 0.003 |
| GO:1901265 | Nucleoside
phosphate binding | 6 | 0.003 |
| GO:0036094 | Small molecule
binding | 6 | 0.004 |
| GO:0008092 | Cytoskeletal
protein binding | 5 | <0.001 |
| hsa04510 | Focal adhesion | 8 | 0.007 |
| hsa04810 | Regulation of actin
cytoskeleton | 8 | 0.008 |
Functional enrichment analyses of
DEGs
Functional and pathway enrichment analyses of DEGs
were performed using DAVID. GO analysis revealed that the
biological processes of DEGs were significantly enriched in cell
adhesion, negative regulation of cell proliferation, cell division
and extracellular matrix organization (Table II). Molecular functions of DEGs
were enriched in protein binding, GTP binding, mannose-binding,
chromatin binding and chromatin binding (Table II). Cell components enriched with
DEGs included the nucleus, cytoplasm, perinuclear region of the
cytoplasm and focal adhesion (Table
II). KEGG pathway analysis revealed that DEGs were mainly
enriched in focal adhesion, regulation of the actin cytoskeleton,
tight junction, coagulation cascades and gap junctions.
 | Table II.KEGG and GO enrichment analyses of
DEGs. |
Table II.
KEGG and GO enrichment analyses of
DEGs.
| Term | Description | Count in gene
set | P-value |
|---|
| GO:0007155 | Cell adhesion | 12 | 0.049 |
| GO:0008285 | Negative regulation
of cell proliferation | 11 | 0.044 |
| GO:0051301 | Cell division | 10 | 0.050 |
| GO:0030308 | Negative regulation
of cell growth | 7 | 0.006 |
| GO:0030198 | Extracellular
matrix organization | 7 | 0.051 |
| GO:0005634 | Nucleus | 99 | <0.001 |
| GO:0005737 | Cytoplasm | 93 | <0.001 |
| GO:0048471 | Perinuclear region
of cytoplasm | 17 | 0.010 |
| GO:0005925 | Focal adhesion | 16 | <0.001 |
| GO:0009986 | Cell surface | 14 | 0.033 |
| GO:0005515 | Protein
binding | 147 | <0.001 |
| GO:0005525 | GTP binding | 13 | 0.007 |
| GO:0003682 | Chromatin
binding | 12 | 0.019 |
| GO:0019901 | Protein kinase
binding | 11 | 0.035 |
| hsa04510 | Focal adhesion | 11 | 0.007 |
| hsa04810 | Regulation of actin
cytoskeleton | 9 | 0.008 |
| hsa04530 | Tight junction | 6 | 0.003 |
| hsa04540 | Gap junction | 6 | 0.005 |
Analyses of the 8 hub genes
In the present study, a total of 8 hub genes were
identified and these hub genes were presented in Table III. The criteria for selection
were as follows: MCODE scores >5, degree cut-off=2, node score
cut-off=0.2, Max depth=100 and k-score=2. Among the 8 genes, PBK,
RAP1A, GNAS and RAB39B were upregulated, while COPZ2, KLF4, BACE1
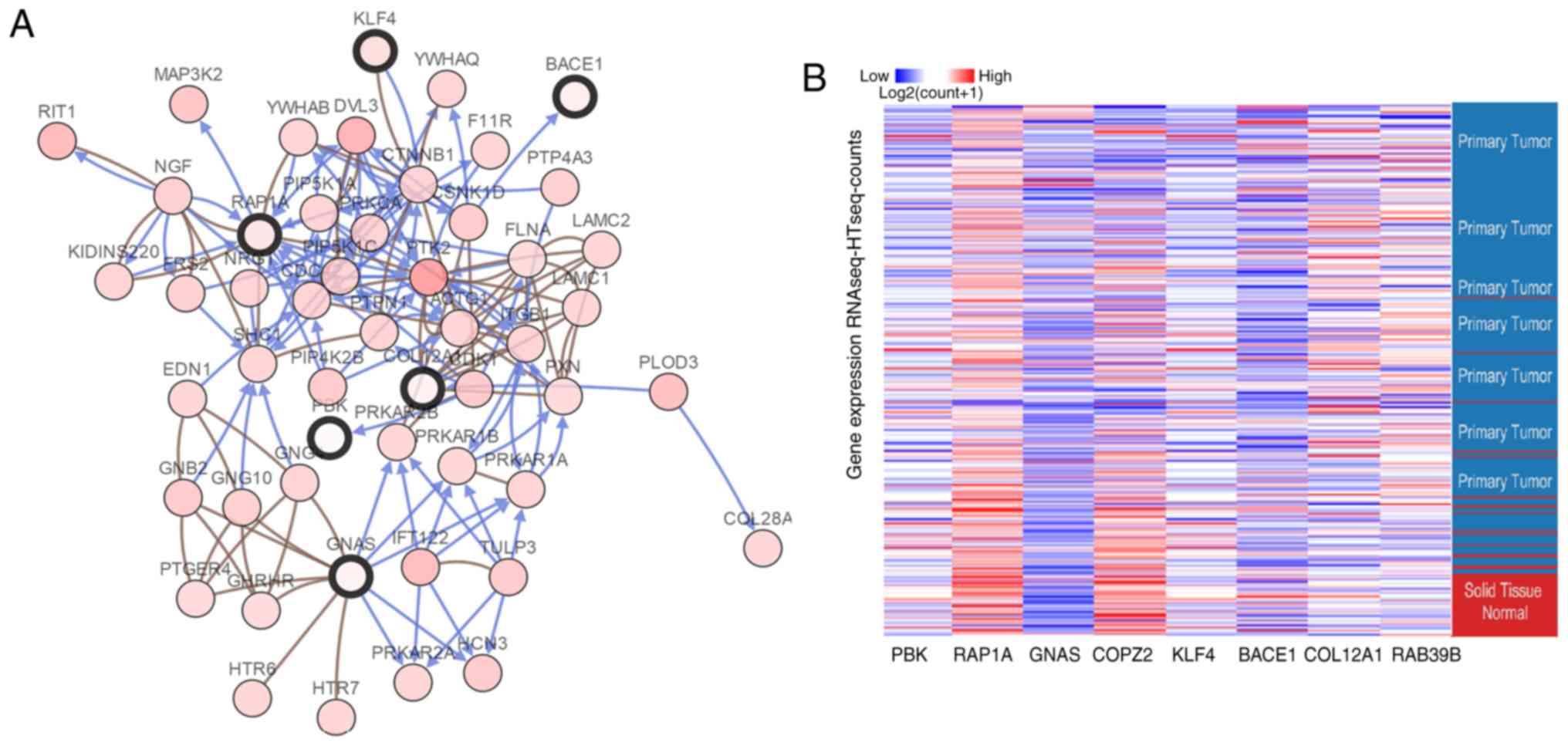
and COL12A1 were downregulated A network of the hub genes and their
co-expression genes were analyzed using the cBioPortal online
platform (Fig. 2A). Hierarchical
clustering demonstrated that the hub genes could differentiate PCa
samples from noncancerous samples (Fig. 2B). Subsequently, the overall
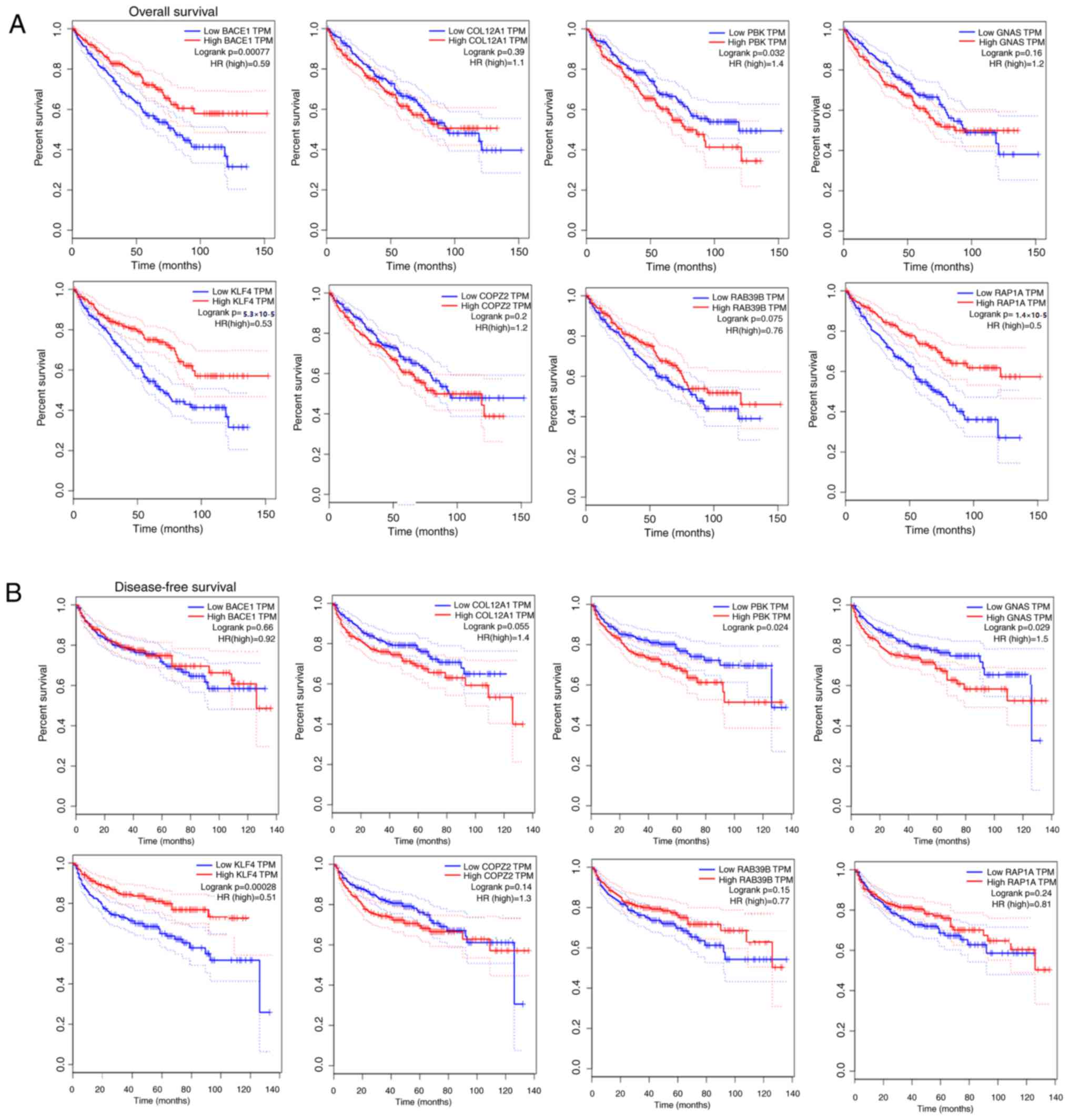
survival analysis of the hub genes was performed using a
Kaplan-Meier curve analysis. Patients with PCa and PBK, RAP1A,
GNAS, coatomer protein complex subunit ζ 2 (COPZ2), β-secretase 1
(BACE1) and collagen type XII α-1 Chain (COL12A1) upregulation
demonstrated decreased overall survival (Fig. 3A). Patients with PCa and PBK,
RAP1A, GNAS, COPZ2, BACE1 and COL12A1 upregulation exhibited
decreased disease-free survival (Fig.
3B). Additionally, RAB39B and KLF4 upregulation was associated
with increased overall survival and disease-free survival. Based on
the above survival analysis, PBK and KLF4 were identified to serve
important roles in the carcinogenesis or progression of PCa.
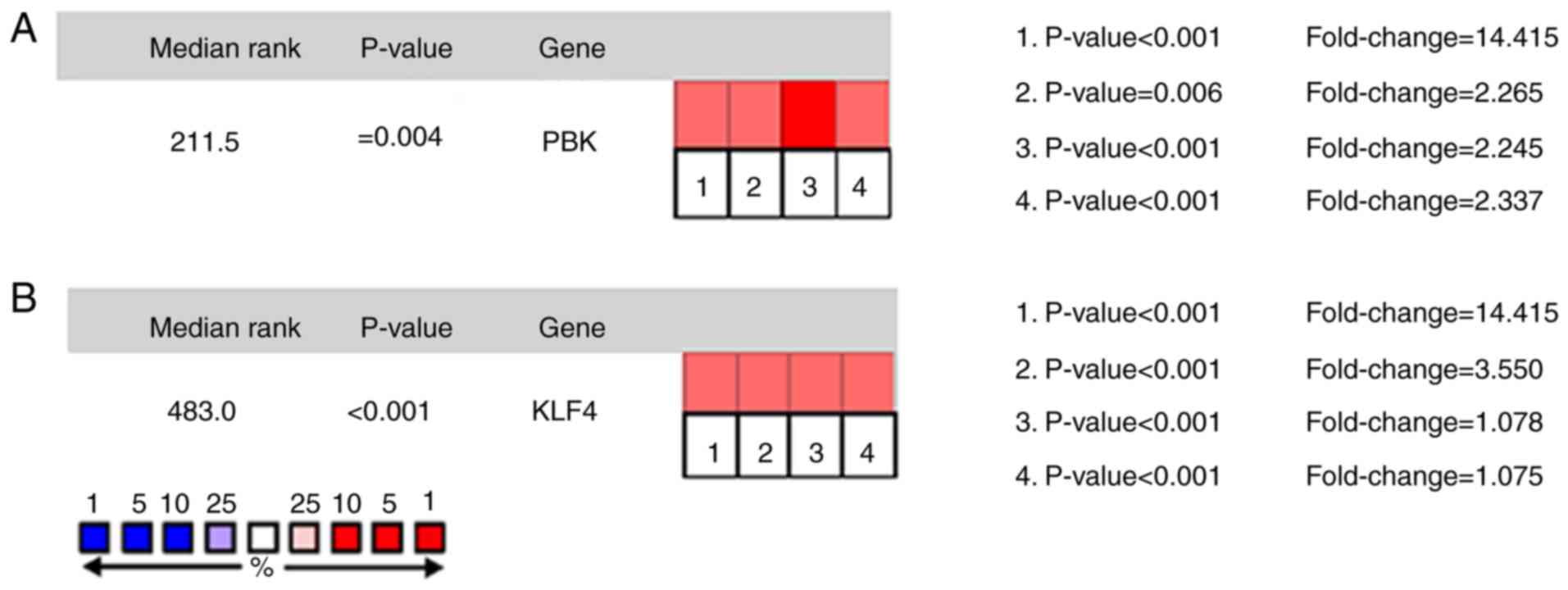
Oncomine analysis of cancer and normal tissue revealed that PBK and
KLF4 were significantly overexpressed in PCa in the different
datasets (Fig. 4A and B). In the
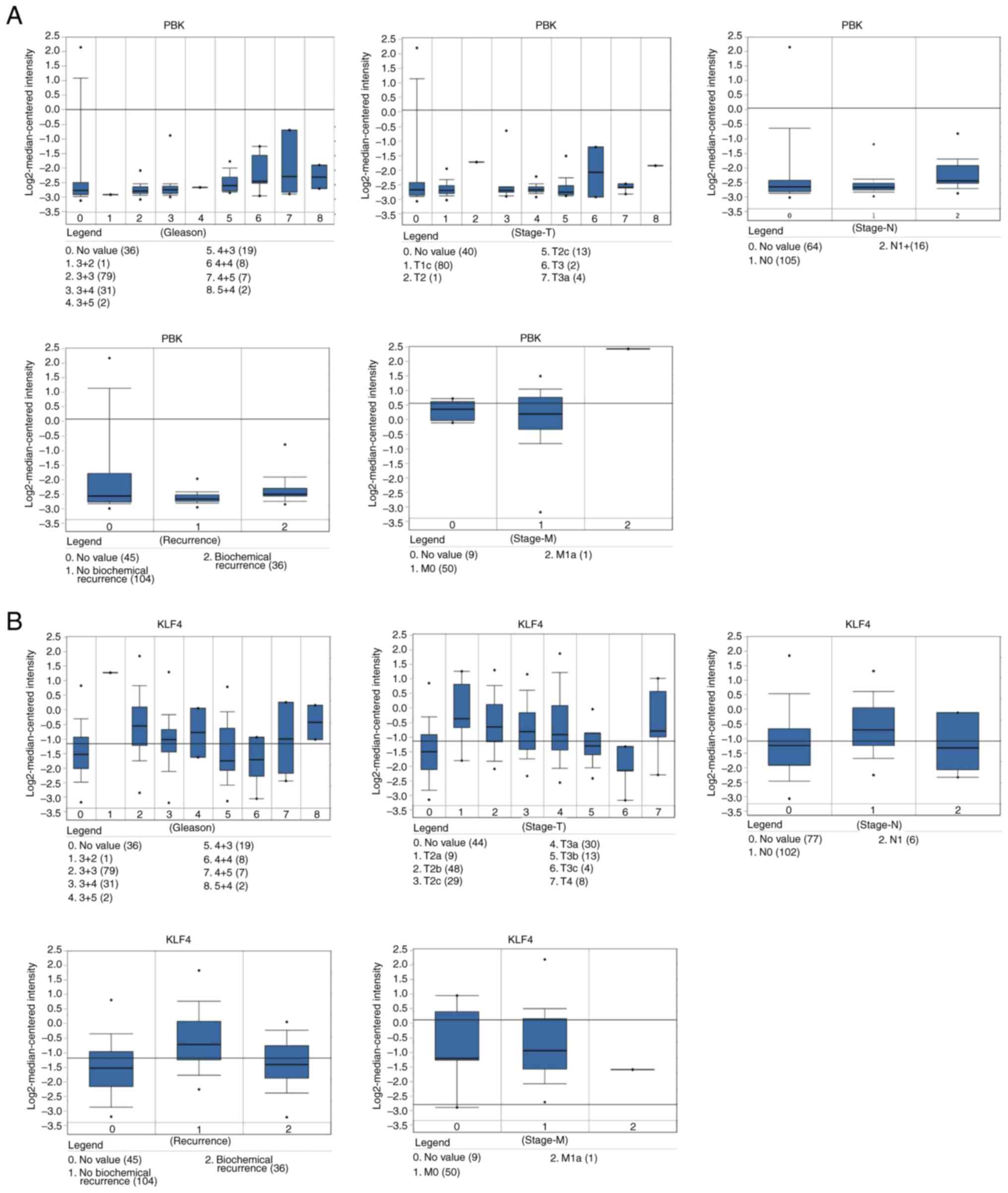
Taylor prostate of Oncomine dataset, the increased mRNA levels of
PBK were associated with TNM stage, Gleason grade and recurrence
status (Fig. 5A). In the Tatulippe
prostate of Oncomine dataset, decreased KLF4 mRNA levels were
associated with TNM stage, Gleason grade and recurrence status
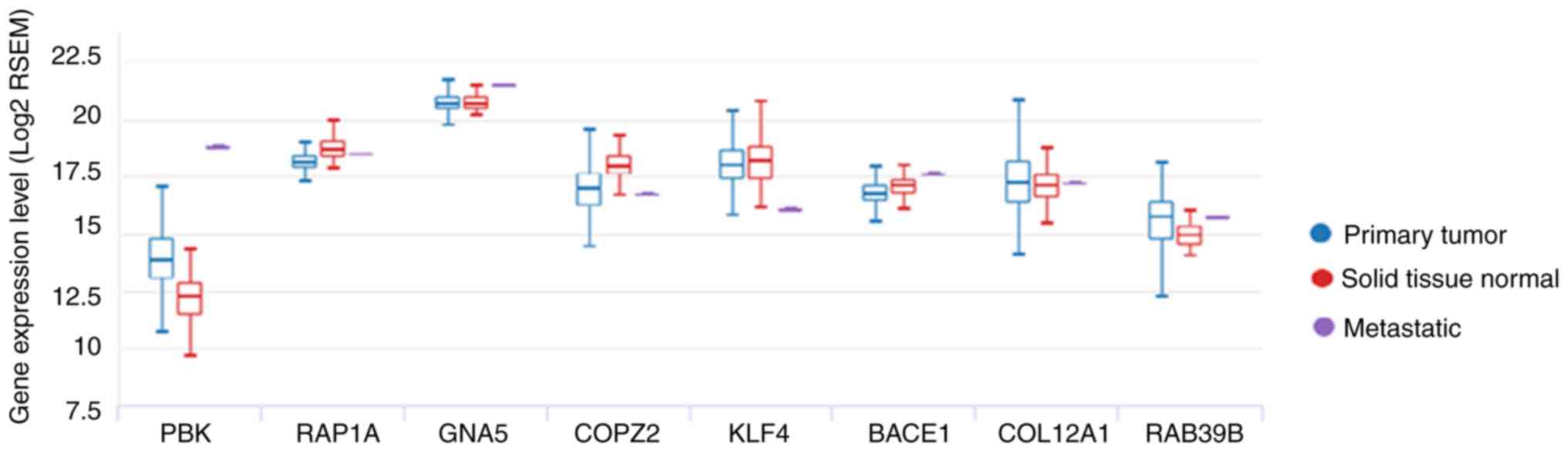
(Fig. 5A and B). PBK gene
expression in metastatic tissue was higher compared with primary
tumor and solid tissue normal via Oncomine (Fig. 6).
 | Table III.Functional roles of 8 hub genes with
degree ≥10. |
Table III.
Functional roles of 8 hub genes with
degree ≥10.
| No. | Gene symbol | Full name | Function |
|---|
| 1 | PBK | PDZ binding
kinase | Active lymphoid
cells and support testicular functions; over expression of this
gene has been implicated in tumorigenesis |
| 2 | RAP1A | Member of RAS
oncogene family | Affect cell
proliferation and adhesion, and may play a role in tumor
malignancy |
| 3 | GNAS | Member of RAS
oncogene family | The encoded protein
regulates signaling pathways that affect cell proliferation and
adhesion |
| 4 | COPZ2 | Coatomer protein
complex subunit ζ 2 | This gene encodes a
member of the adaptor complexes small subunit family |
| 5 | KLF4 | Krüppel-like factor
4 | Control the G1-to-S
transition of the cell cycle |
| 6 | BACE1 | β-secretase 1 | This gene encodes a
member of the peptidase A1 family of aspartic proteases |
| 7 | COL12A1 | Collagen type XII α
1 chain | Modify the
interactions between collagen I fibrils and the surrounding
matrix |
| 8 | RAB39B | Member RAS oncogene
family | Encodes a member of
the Rab family of proteins |
Discussion
Previous studies have demonstrated that the
TMPRSS2:ERG fusion is significantly associated with the diagnosis
of PCa (25–27). Promoter hypermethylation and
downregulated expression of glutathione peroxidase 3 have been
observed in a variety of cancer types, including thyroid cancer,
hepatocellular carcinoma and PCa (10,26,27).
Yu et al (28) identified
an association between Piwi-like protein 2 (PIWIL2) gene expression
and metastatic PCa. Potential markers for use in the diagnosis and
treatment of PCa, which exhibit high efficiency, are urgently
required. To increase understanding of the molecular mechanisms of
candidate genes, GO, KEGG and PPI analyses were performed. In the
current study, the epigenetic and genetic mechanisms in PCa were
assessed using microarray technology.
A total of two mRNA microarray datasets were
selected. A total of 273 DEGs were identified, including 173
downregulated genes and 100 upregulated genes. The interactions of
DEGs were investigated using GO and KEGG analyses. DEGs were found
to be enriched in focal adhesion, regulation of the actin
cytoskeleton, tight junctions, coagulation cascades and gap
junctions. However, other studies (29,30)
have demonstrated that DEGs were enriched in a number of functional
terms, including cellular response to bone morphogenetic protein
(BMP) stimulus, response to BMP, extracellular region and pathways
that are associated with transforming growth factor-β signaling. GO
enrichment analysis revealed that changes in the most significant
modules were enriched in nucleotide binding, nucleoside phosphate
binding, small molecule binding and cytoskeletal protein binding,
while changes in KEGG were mainly enriched in focal adhesion and
regulation of the actin cytoskeleton.
A total of 8 DEGs were selected as hub genes. The
criteria for selection were as follows: MCODE scores >5, degree
cut-off=2, node score cut-off=0.2, Max depth=100 and k-score=2.
Among the 8 genes, PBK, RAP1A, GNAS and RAB39B were upregulated,
while COPZ2, KLF4, BACE1 and COL12A1 were downregulated. PBK and
KLF4 were identified to be important genes in the present study.
PBK is highly homologous to mitogen-activated protein kinase
(31,32). By virtue of target utilization, PBK
has been revealed to influence growth and differentiation (33–36).
PBK is expressed in the outer cell layer of seminiferous tubules in
primary spermatocytes (37), and
is often increased in a number of human cancer types from different
tissue sources (38,39). However, the function of PBK has not
yet been fully determined. In a previous study, the
immunohistochemical expression of PBK/T-LAK cell-originated protein
kinase (TOPK) was revealed to be significantly associated with
human bladder cancer, and was identified as a novel diagnostic
biomarker for this disease (40).
In the present study, the PPI network revealed that PBK directly
interacted with cyclin-dependent kinase 1, Rac GTPase-activating
protein 1, baculoviral IAP repeat containing 5 and protein
regulator of cytokinesis 1, indicating a key role for PBK in PCa.
The expression of PBK was subsequently assessed. Gene upregulation
in PBK were associated with reductions in overall and disease free
survival. Additionally, the KLF4 upregulation was significantly
associated with increased overall survival and disease-free
survival.
High levels of PBK were associated with advanced
stages, Gleason score ≥8 and recurrence (41). Additionally, PBK was significantly
increased in PCa (P=0.001), and the expression was higher in the
Gleason high-scoring group compared with the low-scoring group
(P=0.001) (41). PBK, Gleason
score and pathological stage are independent predictors of PCa
recurrence, and PBK has been indicated to be significantly
associated with survival of no biochemical recurrence (42). As an important mitotic kinase, PBK
has been reported to exhibit a close association with patient
clinical characteristics (43).
The current study indicated that higher mRNA levels of PBK were
associated with TNM stage, Gleason grade and recurrence status,
demonstrating the vital roles of PBK in the carcinogenesis and
progression of PCa. PBK gene expression in metastatic tissue was
higher compared with primary tumor and solid tissue normal tissue,
and PBK has been indicated to serve an important role in mitosis
(43). PBK expression and
phosphorylation are significantly increased during cell mitosis
(44). Previously, a knock-out
study of TOPK revealed that PBK can affect spindle formation
(36). When PBK is inhibited
during mitosis, the spindle (especially the central part) in
mitosis and the subsequent cells become blurred (44). The pulp division cannot be
completed smoothly and the cells will subsequently split out of the
multinucleated cells. Therefore, PBK has been identified to be
associated with the regulation of proliferation and cell cycle
changes in malignant tumor cells, and has also been revealed to
promote tumor cell transformation (33).
KLF4 is a member of the Krüppel-like zinc finger
transcription factor family, which serves a role in regulating
important processes, including cell proliferation, differentiation
and embryo development (36). They
are also associated with numerous human cancer types, including
gastrointestinal, bladder and lung cancers (33,36,45).
A number of KLF4 targeting genes are also biomarker transcription
factors in the endothelial-mesenchymal transition (EMT) process
(45). Additionally, a previous
study indicated that the expression of E-cadherin and α-catenin in
the KLF4 overexpression treatment group was significantly higher
compared with the control group, while the mesenchymal cell marker
vimentin and the expression of vascular endothelial growth was
significantly lower compared with the control group (46). It has been shown that KLF4 protein
is negatively associated with clinical stages in patients with
meningioma, and it promotes or inhibits the EMT process by acting
on transcription factors (46).
The transcription factor KLF4 in PCa cells promotes the migration
and invasion of EMT and tumor cells in vitro (47). These results are consistent with
the results of the current study, which indicated that lower mRNA
levels of PBK were associated with TNM stage, Gleason grade and
recurrence status. KLF4 was also indicated to be downregulated in
PCa tissue with metastases. Furthermore, the stable knockdown of
KLF4 expression in PCa cells has been identified to upregulate the
expression of epithelial-related gene E-cadherin and downregulate
the expression of a variety of mesenchymal-associated genes in
vitro, and has been revealed to serve a role in the inhibition
of tumor cell migration and invasion (48). Katz et al (49) demonstrated that the expression of
KLF4 in tumor tissues was significantly decreased in patients with
PCa in the USA, and that the upregulation of KLF4 inhibited tumor
migration and invasion. Ghaleb et al (50) identified a positive feedback loop
control between KLF4 and the androgen receptor, and revealed that
the inhibition of KLF4 expression in prostatic adenocarcinoma cells
can inhibit the occurrence of EMT in vitro and serve a role
in inhibiting tumor cell migration and invasion. It has also been
indicated that KLF4 can serve the role as an oncogene or tumor
suppressor gene in a number of cellular environments (50).
In conclusion, a total of 273 DEGs and 8 hub genes
were identified as potential novel diagnostic biomarkers for PCa.
The current study identified 2 genes associated with PCa
progression, including PBK and KLF4. However, the current study is
performed based on bioinformatics methods and no experiments were
performed to confirm these conclusions. Therefore, further
experimental study is required to support the results gained from
the current analysis.
Acknowledgements
Not applicable.
Funding
The present study was supported by Special Projects
for Key R&D and Promotion (Science and Technology Breakthrough)
of Henan Department of Science and Technology (grant no.
192102310109).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SL, WBX and JH conceived and designed the study, and
the experiments were performed by SL. SL, JH and WBX analyzed the
data and wrote the manuscript. The original text was drafted and
modified by SL and WBX. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:359–386. 2015.
View Article : Google Scholar
|
|
2
|
Roobol MJ, Steyerberg EW, Kranse R,
Wolters T, van den Bergh RC, Bangma CH and Schröder FH: A
risk-based strategy improves prostatespecific antigen-driven
detection of prostate cancer. Eur Urol. 57:79–85. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bussemakers MJ, van Bokhoven A, Verhaegh
GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N and Isaacs
WB: DD3: A new prostate-specific gene, highly overexpressed in
prostate cancer. Cancer Res. 59:5975–5979. 1999.PubMed/NCBI
|
|
4
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harden SV, Sanderson H, Goodman SN, Partin
AA, Walsh PC, Epstein JI and Sidransky D: Quantitative GSTP1
methylation and the detection of prostate adenocarcinoma in sextant
biopsies. J Natl Cancer Inst. 95:1634–1637. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ilic D, Neuberger MM, Djulbegovic M and
Dahm P: Screening for prostate cancer. Cochrane Database Syst Rev.
CD0047202013.PubMed/NCBI
|
|
7
|
Wang Y, Wu N, Pang B, Tong D, Sun D, Sun
H, Zhang C, Sun W, Meng X, Bai J, et al: TRIB1 promotes colorectal
cancer cell migration and invasion through activation MMP-2 via
FAK/Src and ERK pathways. Oncotarget. 8:47931–47942.
2017.PubMed/NCBI
|
|
8
|
Yoshida A, Kato JY, Nakamae I and
Yoneda-Kato N: COP1 targets C/EBPα for degradation and induces
acute myeloid leukemia via Trib1. Blood. 122:1750–1760. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye Y, Wang G, Wang G, Zhuang J, He S, Song
Y, Ni J, Xia W and Wang J: The oncogenic role of tribbles 1 in
hepatocellular carcinoma is mediated by a feedback loop involving
microRNA-23a and p53. Front Physiol. 8:7892017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
García-Perdomo HA, Chaves MJ, Osorio JC
and Sanchez A: Association between TMPRSS2:ERG fusion gene and the
prostate cancer: Systematic review and meta-analysis. Cent European
J Urol. 71:410–419. 2018.PubMed/NCBI
|
|
11
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Varambally S, Yu J, Laxman B, Rhodes DR,
Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et
al: Integrative genomic and proteomic analysis of prostate cancer
reveals signatures of metastatic progression. Cancer Cell.
8:393–406. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu YP, Landsittel D, Jing L, Nelson J, Ren
B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al: Gene
expression alterations in prostate cancer predicting tumor
aggression and preceding development of malignancy. J Clin Oncol.
22:2790–2799. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91-103, 119–128, 244–252. 2002.
|
|
17
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast delayed enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao H, Li J, Li X, Han C, Zhang Y, Zheng
L and Guo M: Silencing GPX3 expression promotes tumor metastasis in
human thyroid cancer. Curr Protein Pept Sci. 16:316–321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao S, Yan B, Lu Y, Zhang G, Li J, Zhai W,
Guo W and Zhang S: Methylation of promoter and expression silencing
of GPX3 gene in hepatocellular carcinoma tissue. Clin Res Hepatol
Gastroenterol. 39:198–204. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu YP, Yu G, Tseng G, Cieply K, Nelson J,
Defrances M, Zarnegar R, Michalopoulos G and Luo JH: Glutathione
peroxidase 3, deleted or methylated in prostate cancer, suppresses
prostate cancer growth and metastasis. Cancer Res. 67:8043–8050.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Edelman GM and Crossin KL: Cell adhesion
molecules: Implications for a molecular histology. Annu Rev
Biochem. 60:155–190. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang E, Zhang X, Wang Q and Wang D:
Identification of prostate cancer hub genes and therapeutic agents
using bioinformatics approach. Cancer Biomark. 20:553–561. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang Y, Zhang X, Song D and Wei J: Piwil2
modulates the invasion and metastasis of prostate cancer by
regulating the expression of matrix meta lloproteinase-9 and
epithelial-mesenchymal transitions. Oncol Lett. 10:1735–1740. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gaudet S, Branton D and Lue RA:
Characterization of PDZ binding kinase, a mitotic kinase. Proc Natl
Acad Sci USA. 97:5167–5172. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abe Y, Matsumoto S, Kito K and Ueda N:
Cloning and expression of a novel MAPKK-like protein kinase,
lymphokine-activated killer T-cell-originated protein kinase,
specifically expressed in the testis and activated lymphoid cells.
J Biol Chem. 275:21525–21531. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang C, Habets G and Bollag G:
Interrogating the kinome. Nat Biotechnol. 29:981–983. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu Y, Muller M, Smith D, Dutta B, Komurov
K, Iadevaia S, Ruths D, Tseng JT, Yu S, Yu Q, et al: Kinome
siRNA-phosphoproteomic screen identifies networks regulating AKT
signaling. Oncogene. 30:4567–4577. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matsumoto S, Abe Y, Fujibuchi T, Takeuchi
T, Kito K, Ueda N, Shigemoto K and Gyo K: Characterization of a
MAPKK-like protein kinase TOPK. Biochem Biophys Res Commun.
325:997–1004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Park JH, Nishidate T, Nakamura Y and
Katagiri T: Critical roles of T-LAK cell-originated protein kinase
in cytokinesis. Cancer Sci. 101:403–411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fujibuchi T, Abe Y, Takeuchi T, Ueda N,
Shigemoto K, Yamamoto H and Kito K: Expression and phosphorylation
of TOPK during spermatogenesis. Dev Growth Differ. 47:637–644.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Park JH, Lin ML, Nishidate T, Nakamura Y
and Katagiri T: PDZ-binding kinase/T-LAK cell-originated protein
kinase, a putative cancer/testis antigen with an oncogenic activity
in breast cancer. Cancer Res. 66:9186–9195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Singh PK, Srivastava AK, Dalela D, Rath
SK, Goel MM and Bhatt ML: Expression of PDZ-binding kinase/T-LAK
cell-originated protein kinase (PBK/TOPK) in human urinary bladder
transitional cell carcinoma. Immunobiology. 219:469–474. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim DJ, Li Y, Reddy K, Lee MH, Kim MO, Cho
YY, Lee SY, Kim JE, Bode AM and Dong Z: Novel TOPK inhibitor
HI-TOPK-032 effectively suppresses colon cancer growth. Cancer Res.
72:3060–3068. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun H, Zhang L, Shi C, Hu P, Yan W, Wang
Z, Duan Q, Lu F, Qin L, Lu T, et al: TOPK is highly expressed in
circulating tumor cells, enabling metastasis of prostate cancer.
Oncotarget. 6:12392–12404. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen JH, Fang SM and Liang YK: Expression
and clinical significance of PBK in prostate cancer tissues.
Guangzhou Pharm. 48:1000–8535. 2017.
|
|
44
|
Simons-Evelyn M, Bailey-Dell K, Toretsky
JA, Ross DD, Fenton R, Kalvakolanu D and Rapoport AP: PBK/TOPK is a
novel mitotic kinase which isupregulated in burkitt's lymphoma and
other highly proliferative malignant cells. Blood Cells Mol Dis.
27:825–829. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Simons-Evelyn M, Bailey-Dell K, Toretsky
JA, Ross DD, Fenton R, Kalvakolanu D and Rapoport AP: PBK/TOPK is a
novel mitotic kinase which is upregulated in Burkitt's lymphoma and
other highly proliferative malignant cells. Blood Cells Mol Dis.
27:825–829. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wei D, Gong W, Kanai M, Schlunk C, Wang L,
Yao JC, Wu TT, Huang S and Xie K: Drastic down-regulation of
Krüppel-like factor 4 expression is critical in human gastric
cancer development and progression. Cancer Res. 65:2746–2754. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ohnishi S, Ohnami S, Laub F, Aoki K,
Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F and Yoshida T:
Downregulation and growth inhibitory effect of epithelial-type
Krüppel-like transcription factor KLF4, but not KLF5, in bladder
cancer. Biochem Biophys Res Commun. 308:251–256. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wei D, Kanai M, Jia Z, Le X and Xie K:
Kruppel-like factor 4 induces p27Kip1 expression in and suppresses
the growth and metastasis of human pancreatic cancer cells. Cancer
Res. 68:4631–4639. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Katz JP, Perreault N, Goldstein BG, Lee
CS, Labosky PA, Yang VW and Kaestner KH: The zinc-finger
transcription factor Klf4 is required for terminal differentiation
of goblet cells in the colon. Development. 129:2619–2628.
2002.PubMed/NCBI
|
|
50
|
Ghaleb AM, McConnell BB, Nandan MO, Katz
JP, Kaestner KH and Yang VW: Haploinsufficiency of Krüppel-like
factor 4 promotes adenomatous polyposis coli dependent intestinal
tumorigenesis. Cancer Res. 67:7147–7154. 2007. View Article : Google Scholar : PubMed/NCBI
|