Introduction
Tumor metastasis is characterized with the spread of
tumor cells to different tissues and organs from the primary tumor
site, resulting in the formation of secondary new tumors (1). Hence, tumor metastasis is a major
reason for cancer-related high mortality. Several approaches have
been directed to interrupt the metastasis of primary tumors for
cancer treatment (1). Considering
the complexity and multi-steps involved in tumor metastasis, the
detailed mechanisms underlying this process are yet to be
completely understood. The process of tumor metastasis broadly
comprises three sequential events, namely, invasion, intravasation,
and extravasation (2). During
invasion, the tumor cells dissociate from the primary tumor cells,
dissolve the basal membrane and extracellular matrix, and initiate
the upregulation or downregulation of proteins controlling tumor
cell motility and migration (3).
MicroRNAs (miRNAs) are small-noncoding RNA molecules
that play regulatory roles in multiple cellular functions by
downregulating the expression of certain genes (4). Recent studies have identified miRNAs
as a new class of molecules associated with tumor metastasis and
revealed their roles in the regulation of tumor metastasis
(5). In gastric cancer, several
miRNAs have been recognized as regulatory molecules involved in
tumor cell invasion and metastasis. miR-337-3p expression has been
revealed to be downregulated in metastatic tissues when compared to
primary gastric cancer tissues (6). Furthermore, the overexpression of
miR-337-3p in gastric cancer cells has been revealed to result in
the reduction of their invasion ability (6). Recent evidence indicates that
miR-337-3p may bind to the promoter region and suppress the
transcription of matrix metalloproteinase 14 (MMP14) in both
gastric tumor and neuroblastoma cells (7,8). The
mechanism of the function of miR-337-3p in invasion is not yet
clear. Whether miR-337-3p targets cytoskeleton-associated molecules
in invasive gastric cancer is also unknown. In the present study,
the major target molecules of miR-337-3p in tumor invasion were
investigated.
ARHGAP10, also known as ARHGAP21, was originally
identified as a gene located on the human chromosome 10. ARHGAP10,
a cytoskeletal regulatory protein, is known to encode a
Rho-GTPase-activating protein owing to its Rho-GAP domain (9). It negatively regulates the small G
protein Rho- and Cdc42-mediated downstream signal transduction
(10,11). ARHGAP10 overexpression has been
revealed to disrupt stress fiber formation in some cells (12). A recent study suggested that
ARHGAP10 is involved in the regulation of tumor cell migration
(13). In the present study,
ARHGAP10 was identified as a molecular target of miR-337-3p and its
role was demonstrated in mediating the regulatory effects of
miR-337-3p on gastric cancer cell migration and invasion.
Materials and methods
Cell culture and transfection
Gastric cancer SGC-7901 cells, originally purchased
from the Cell Bank of the Chinese Academy of Sciences, were
maintained in Roswell Park Memorial Institute (RPMI)-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific), 2 mM
L-glutamine (Gibco; Thermo Fisher Scientific), 100 U/ml penicillin
and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific). 293T
cells, originally purchased from the Cell Bank of the Chinese
Academy of Sciences, were maintained in high glucose-Dulbecco's
Modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific)
with no glutamine and supplemented with 10% FBS, 2 mM L-glutamine,
penicillin (100 U/ml), and streptomycin (100 µg/ml). Both cell
lines were cultured in 6-well plates in humidified air supplemented
with 5% CO2 at 37°C.
SGC-7901 cells were seeded in 6-well plates and
transfected with miRNAs and cDNA after reaching 80% confluency.
Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for the transfection of miRNAs and cDNA.
For the expression of miRNAs, miR-337-3p or miR-337-3p inhibitor
and respective control miR-NC and inhibitor NC (Shanghai GenePharma
Co., Ltd.) at a final concentration of 50 nM were used for
transfection. The sequences were as follows: miR-337-3p mimics,
sense, 5′-CUCCUAUAUGAUGCCUUUCUUC-3′ and antisense,
5′-AGAAAGGCAUCAUAUAGGAGUU-3′; miR-NC, sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miRNA-337-3p inhibitor,
5′-GAAGAAAGGCAUCAUAUAGGAG-3′; and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′.
ARHGAP10 cDNA was subcloned in the expression vector
pcDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) and confirmed
by sequencing. For the expression of cDNA, 2.5ng/well of ARHGAP10
cDNA or pcDNA3.1 were added. The cells were incubated for 24–48 h
in humidified air supplemented with 5% CO2 at 37°C.
Then, the transfected cells were analyzed.
Small interfering RNA (siRNA) specific for ARGHAP10
(siRNA-ARHGAP10; 20 µmol/l; Guangzhou Ribobio Co., Ltd.) was
transfected into SGC7901 cells using Lipofectamine 2000. The
si-ARHGAP10 targeting sequence was 5′-CTGCTACTGTAGCGGACAA-3′.
Negative control siRNA (siRNA-NC; cat. no. siN0000001-1-5;
Guangzhou Ribobio Co., Ltd.) was used as a control. At 24–48 h
following transfection, the transfected cells were analyzed.
Cell viability and cell cycle
analysis
SGC-7901 cells were seeded in 96-well plates and
transfected with miRNAs after reaching 80% confluency. After 24 h,
the cells were incubated with Cell Counting Kit-8 (CCK-8; Biosharp)
for 4 h and the absorbance at 450 nm was assessed using a plate
reader. For cell cycle analysis, SGC-7901 cells were seeded in
6-well plates and transfected with miRNAs (miR-337-3p, miR-337-3p
inhibitor, miR-NC and inhibitor NC.). After 24 h of transfection,
the cells were harvested and washed with cold phosphate-buffered
saline (PBS; HyClone; GE Healthcare Life Sciences). The cells were
fixed with cold 95% ethanol at 4°C overnight, followed by staining
with 50 µg/ml propidium iodide (CWBio) and 10 µg/ml RNAse (Takara
Biotechnology Co., Ltd.) at 37°C in the dark for 30 min. Cell cycle
analysis was carried out with flow cytometry. Data were analyzed
using FlowJo version 7.6 (FlowJo LLC).
Wound healing assay
The transfected SGC-7901 cells were seeded into
6-well culture plates. The cells reached ~80% confluency after 24 h
of cutlure. A wound was gently created by scratching the monolayer
with a 10 µl pipette tip. After scratching, the cells were washed
twice with medium to remove the detached cells. Images were
captured. The cells were incubated in 1% FBS-supplemented RPMI-1640
culture medium for an additional 48 h. Images were acquired and
quantitative analysis was performed with software Image-Pro Plus
6.0 (Media Cybernetics, lnc.).
Transwell migration and invasion
assays
Gastric tumor cell migration and invasion were
assessed using a two-chamber system. The upper compartment was
inserted into the lower compartment of BD BioCoat control inserts
(Discovery Labware; BD Biosciences). The invasion assay was
conducted in a Transwell format with Matrigel in the upper chamber.
A Transwell-Matrigel insert was used for the invasion assay. The
same procedure was conducted for the migration assay without the
use of Matrigel. After transfection, the cells were cultured in
serum-free RPMI-1640 culture medium for 12 h. Cells
[5×104 in 0.1 ml of serum-free medium containing 1%
bovine serum albumin (BSA; Gibco; Thermo Fisher Scientific, Inc.)]
were seeded into the upper compartment, while the lower compartment
was filled with normal culture medium supplemented with 20% FBS.
After incubation for 24 h, the cells were wiped away from the upper
surface, and the migrated cells on the lower surface were fixed and
stained with 0.1% g/ml crystal violet (Beijing Solarbio Science
& Technology Co., Ltd.) at 25°C for 20 mins. The number of
cells that completely invaded across the filter was determined in
five random fields for each experiment using EVOS M7000 Imaging
System (Thermo Fisher Scientific, Inc.; magnification, ×100). Each
condition was assayed in triplicates, and each experiment was
repeated at least thrice.
Bioinformatic analysis
Potential targets of miR-337 were analyzed using
TargetScan version 7.2 (http://www.targetscan.org/vert_72/).
Tumor metastasis assay in an
immunodeficient mouse model
A total of 20 female NOD-Prkdcscid
Il2rgnull mice at 5 weeks of age (weighing 18–22
g) were purchased from Beijing VITALSTAR Biotechnology Co., Ltd.
The animals were maintained in an ACS OptiMICE EVC animal care
system (Animal Care Systems Inc.). All mice were housed in rooms
with a 12-h day/night cycle at 22°C and had free access to standard
chow (Sebiona). Cages, food, and bedding were sterilized by
autoclaving. All animal experiments were performed according to the
guidelines of the Laboratory Animal Ethics Committee of Huazhong
Agriculture University, approved by the Laboratory Animal Centre,
Huazhong Agriculture University (HZAHMD-2016-037). Mice were
divided into four groups (5 mice/group) for tail intravenous
injection. SGC-7901 cells were transfected with miRNAs (miR-337-3p,
miR-337-3p inhibitor, miR-NC and inhibitor NC) and harvested 12 h
after transfection. Cells (2×106 in 0.1 ml PBS) were
injected into each mouse via tail vein. Mice were maintained under
these conditions for 5 weeks, and were sacrificed after these 5
weeks by carbon dioxide asphyxiation.
Histological assay
Lung and liver tissues were collected and fixed with
10% buffered formalin at 25°C for 48 h, perfused and dissected, and
paraffin-embedded. Sections (6-mm thickness) were stained with
hematoxylin and eosin (H&E) using a Hematoxylin and Eosin
Staining kit (Beyotime Institute of Biotechnology) in accordance
with the manufacturer's protocols. To evaluate the expression of
ARHGAP10 in lung tumor foci, immunohistochemical experiments were
conducted using a specific polyclonal anti-ARHGAP10 antibody
(1:1,000; cat. no. 55139-1-AP, ProteinTech Group, Inc.). All
sections were analyzed with a routine light microscope (Nikon
Corporation).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (qPCR)
For the detection of mRNA expression, total RNA was
isolated from cells and tissues using TRIzol (Invitrogen) following
the manufacturer's protocol. The quantification of the extracted
RNA was performed with NanoDrop 2000 spectrophotometer.
Approximately 1 mg of total RNA was reverse-transcribed using a
Two-Step MMLV RT-PCR kit from GeneMark, and qPCR was performed to
determine the expression level of ARHGAP10 on a Roche
LightCycler® 96 using SYBR Green Real-Time PCR Master
Mix from GeneMark according to the manufacturer's instructions. The
thermocycling conditions were denaturation at 95°C for 30 sec, then
35 cycles of denaturation at 95°C for 5 sec, annealing at 56°C for
30 sec and elongation at 72°C for 30 sec, followed by a 5 min
extension at 72°C. The relative expression of miRNAs and mRNA were
determined using the 2−ΔΔCq quantification method
(14). Three independent
experiments were replicated. The primers specific for ARHGAP10 were
5′-ACTGAAACCCTGATTAAACC-3′ (forward) and 5′-ATCTGCCTCTTGTAAATGTG-3′
(reverse), while those for glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) were 5′-CACCCACTCCTCCACCTTTG-3′ (forward) and
5′-CCACCACCCTGTTGCTGTAG-3′ (reverse).
For miRNA assessment, commercial miRcute miRNA
isolation kit, miRcute miRNA First-Strand cDNA Synthesis kit, and
miRcute miRNA qPCR detection kit (Tiagen Biotech Co., Ltd.) were
used. qPCR was performed using the mature miR-337-3p sequence as
the forward primer along with the universal reverse primer provided
with the miRNA qPCR detection kit, and small nuclear RNA U6 served
as an internal control. The specific primers for miR-337-3p
included 5′-CUCCUAUAUGAUGCCUUUCUUC-3′ and 5′-GTGCAGGGTCCGAGGT-3′,
while those for U6 included 5′-CACCCACTCCTCCACCTTTG-3′ and
5′-CCACCACCCTGTTGCTGTAG-3′.
Western blot analysis
SGC-7901 cells were lysed after transfection in 1 ml
of ice-cold tissue lysis buffer [Tris-buffered saline, 1.5% Triton
X-100, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate (SDS),
protease inhibitor cocktail, and 1 mM PMSF]. After centrifugation
(12,000 × g, 20 min, 4°C), the supernatants were collected and
protein concentrations were determined. Protein samples (15 µg per
lane) were separated on 10% SDS polyacrylamide gels and transferred
onto nitrocellulose membranes. The blots were blocked with 5%
skimmed milk (Becton, Dickinson and Company) at 25°C for 1 h and
incubated with primary antibodies specific for ARHGAP10 (1:1,000;
cat. no. 55139-1-AP) and GAPDH (1:4,000; cat. no. 60004-1-Ig; both
from ProteinTech Group, Inc.) at 4°C overnight. After developing,
each blot was washed three times in 1X TBST (Beijing Solarbio
Science & Technology Co., Ltd.). The blots were incubated with
secondary antibodies labeled with horseradish peroxidase
(ProteinTech Group, Inc.) at 25°C for 1 h. HRP-conjugated
anti-rabbit (cat. no. SA00001-2) and HRP-conjugated anti-mouse
(cat. no. SA00001-1; both from ProteinTech Group, Inc.) antibodies
were used at 1:4,000. Signals were detected by enhanced
chemiluminescence western blot reagents (Thermo Scientific
Scientific, Inc.). ImageJ version 2.1.4.7 (National Institutes of
Health) was used for densitometry.
Plasmid construction and luciferase
assays
The 3′-untranslated region (UTR) segment of ARHGAP10
predicted to specifically interact with miR-337-3p was subcloned
into pMIR-REPORT luciferase vector psiCHECKTM-2 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). 293T cells were
transfected with wild-type plasmids using Lipofectamine 2000. At 48
h after transfection, the cells were lysed and the luciferase
activity was detected with the Dual-Luciferase reporter assay
system (Promega Corporation). Firefly luciferase activity was
normalized to Renilla luciferase activity for each sample.
psiCHECKTM-2 control plasmid was used for normalization of
luciferase values. Each reporter plasmid was transfected at least
thrice, and each sample was assayed in triplicates.
Statistical analysis
The results are presented as the mean ± standard
deviation (SD) of three independent experiments. Differences
between two groups were compared using a two-tailed paired
Student's t-test; one-way analysis of variance (ANOVA) was used for
comparisons between multiple groups. The Student-Newman-Keuls test
was used as a post hoc test following ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-337-3p affects the viability of
gastric cancer cells
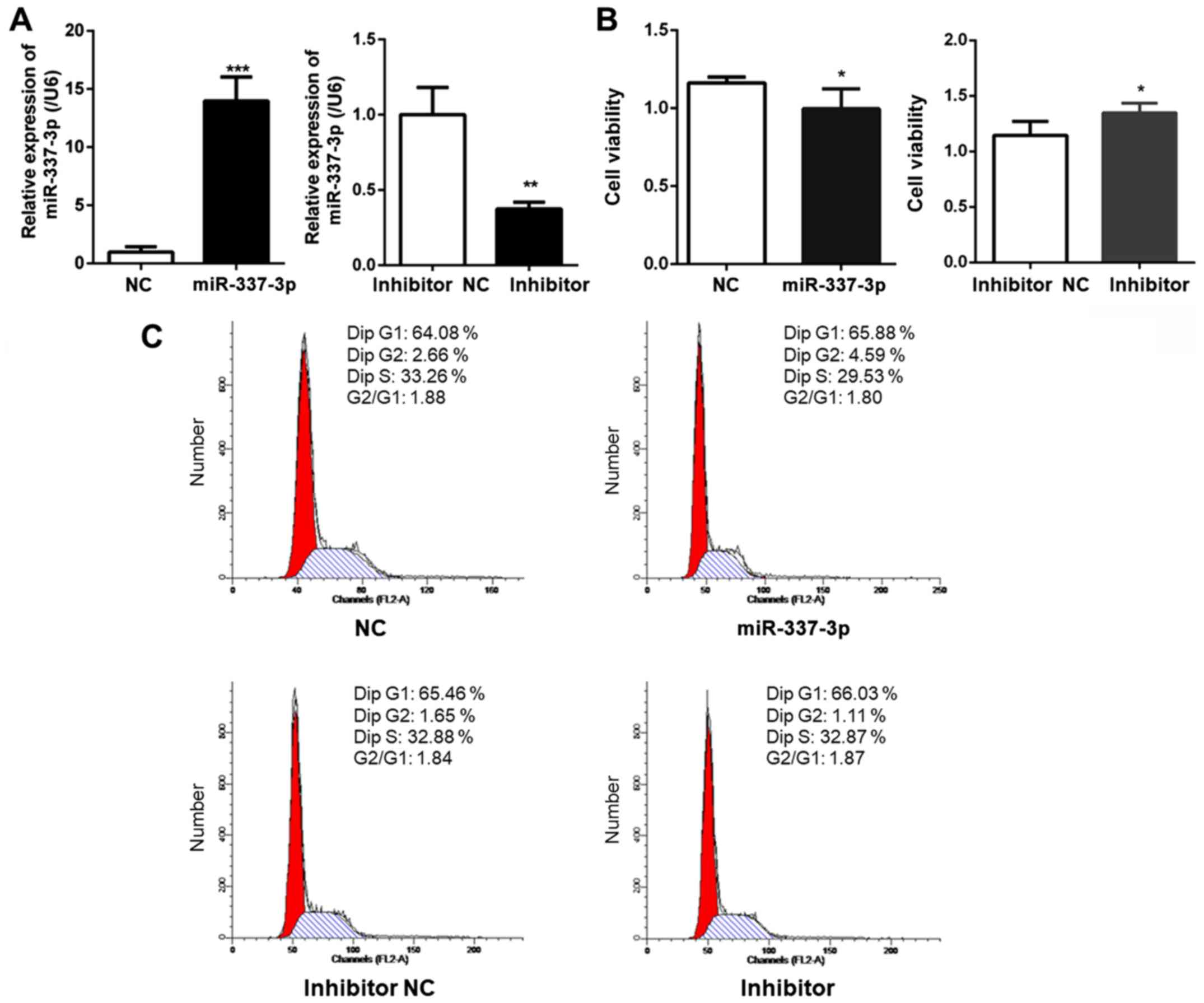
To examine the expression level of miR-337-3p after
transfection, RT-qPCR analysis was performed. The results revealed
the overexpression of miR-337-3p in the transfected cells. The
transfection of miR-337-3p inhibitor resulted in the downregulation
of miR-337-3p expression (Fig.
1A). The effects of miR-337-3p overexpression on the viability
of gastric cancer SGC-7901 cells were also examined. A CCK-8 assay
was used to assess SGC-7901 cell viability and it was revealed that
the overexpression of miR-337-3p resulted in a decrease in the
viability of gastric cancer cells to <10% (Fig. 1B). Next, the effects of miR-337-3p
expression on the cell cycle of SGC-7901 cells were examined. Flow
cytometric analysis revealed that miR-337-3p had no effect on the
cell cycle (Fig. 1C). This
observation was consistent with one previously reported, wherein
miR-337-3p did not affect the proliferation of gastric cancer cells
(4). The reduced viability
indicated that miR-337-3p may induce apoptosis in gastric cancer
cells.
miR-337-3p decreases the motility of
gastric cancer cells
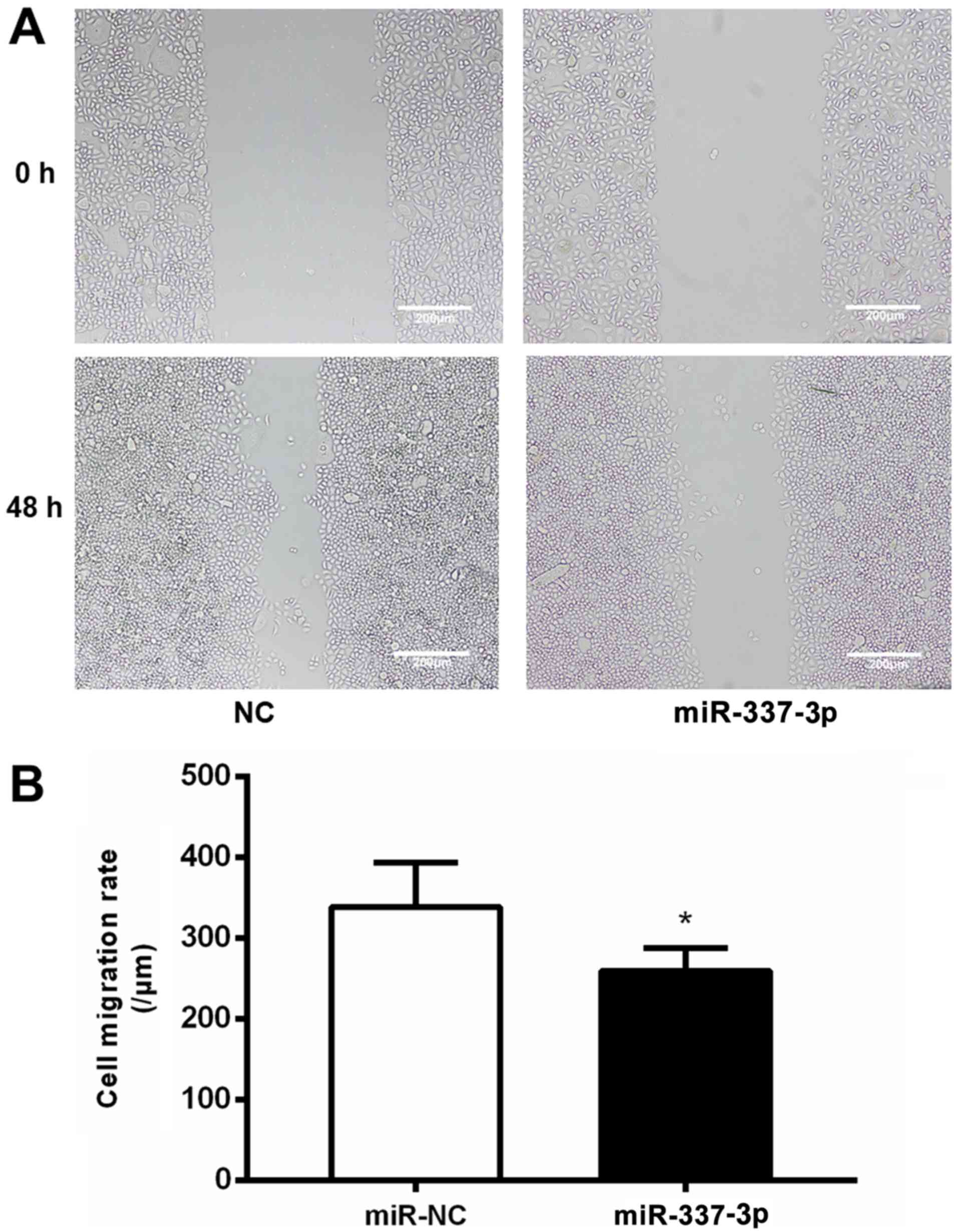
The effects of miR-337-3p overexpression on the
motility of SGC-7901 cells were examined with a wound healing
assay. SGC-7901 cells transfected with miR-337-3p exhibited lower
wound healing capacity than the control cells (Fig. 2A), indicating that miR-337-3p
inhibits the migration of gastric cancer cells (Fig. 2B). To further confirm the
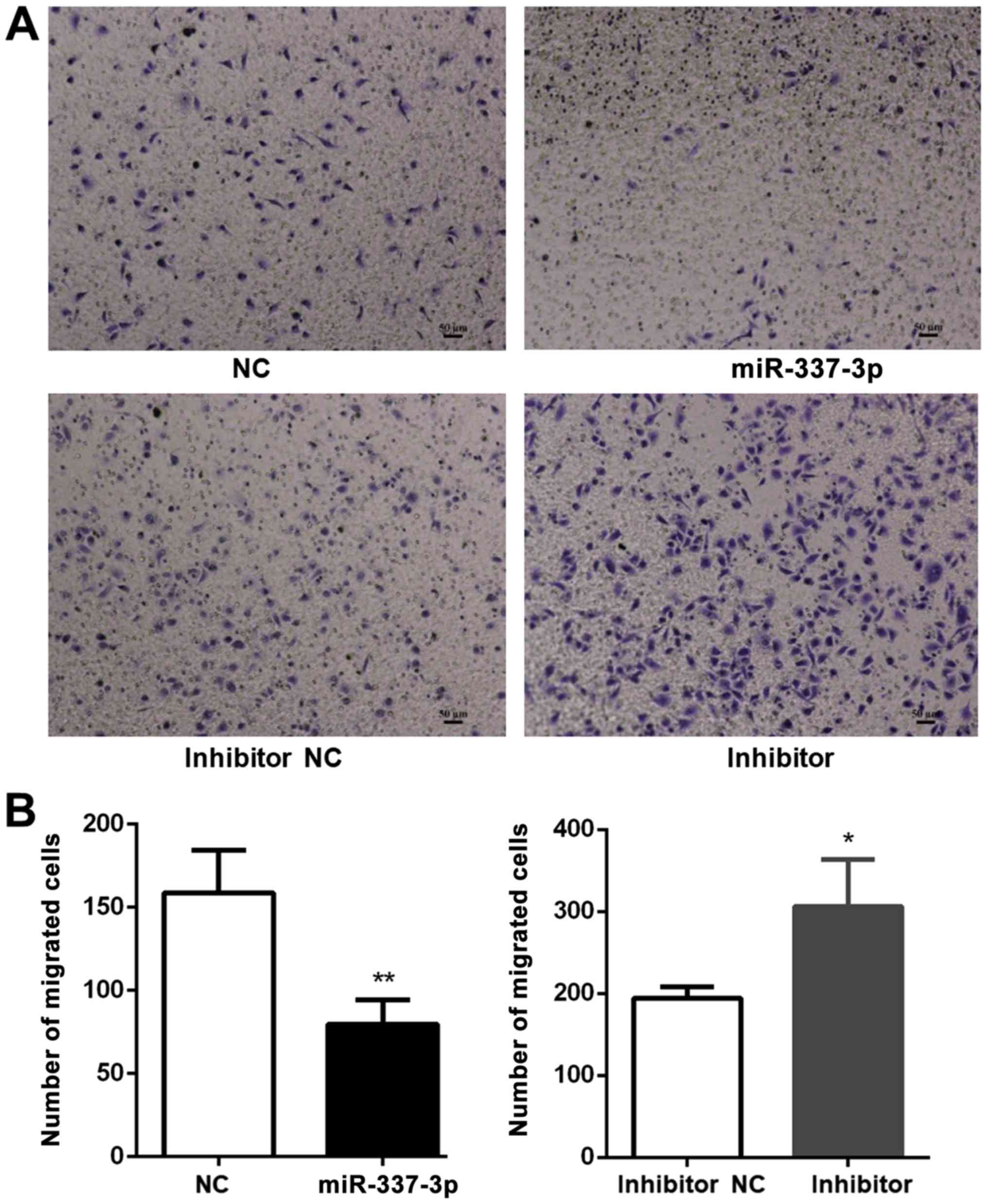
inhibitory effects of miR-337-3p on gastric cancer cell motility,
the effects of miR-337-3p overexpression on SGC-7901 motility were
investigated in a Transwell migration assay (Fig. 3A). The overexpression of miR-337-3p
in SGC-7901 cells resulted in a decrease in their migration through
the Transwell, while the inhibition of miR-337-3p expression
resulted in an increase in the Transwell migration ability
(Fig. 3B).
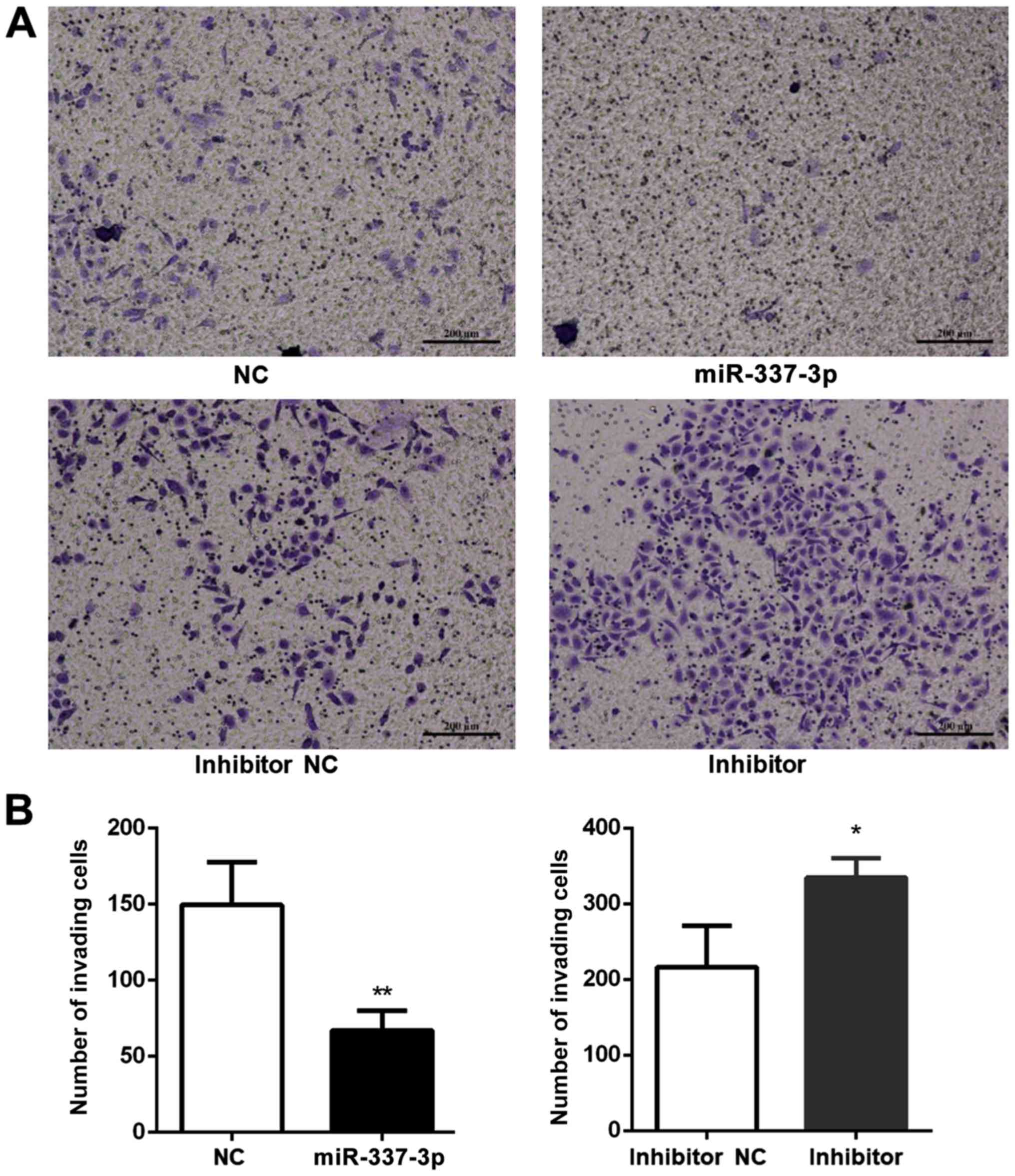
To better understand the effects of miR-337-3p on
gastric tumor metastasis, the role of miR-337-3p in SGC-7901 cell
invasion was examined (Fig. 4A).
An invasion assay was conducted in a Transwell format and Matrigel
was placed in the upper chamber. Transfection of SGC-7901 cells
with miR-337-3p decreased the number of cells in the bottom
chamber, indicating that miR-337-3p may inhibit the invasive
capacity of gastric cancer cells (Fig.
4B).
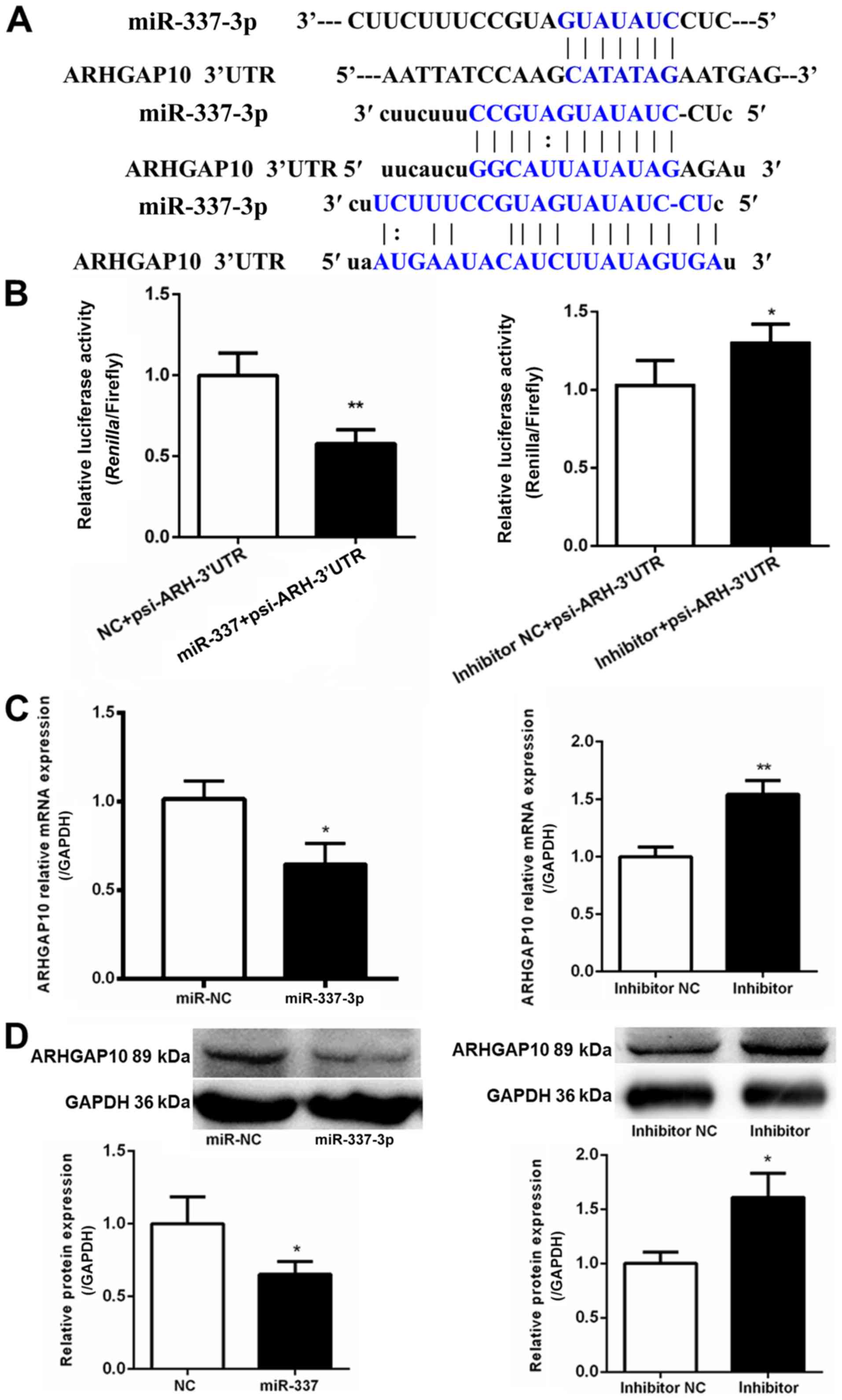
ARHGAP10 serves as a target for
miR-337-3p
To understand the mechanisms underlying the
inhibitory effect of miR-337-3p on gastric cancer cell migration,
the target molecules were explored. TargetScan (http://www.targetscan.org/vert_72/) was used to
identify ARHGAP10 as a potential target molecule. Three potential
miR-337-3p-binding sites were identified at the 3′-UTR of ARHGAP10
mRNA (Fig. 5A). The 3′-UTR of
ARHGAP10 was subcloned in a luciferase vector; the luciferase
activity assay revealed the ability of miR-337-3p to inhibit
ARHGAP10 expression (Fig. 5B). The
overexpression of miR-337-3p resulted in a decrease in the
expression of ARHGAP10 mRNA, as assessed with qPCR in SGC-7901
cells (Fig. 5C). Western blot
analysis revealed that miR-337-3p reduced the protein level of
ARHGAP10 in SGC-7901 cells (Fig.
5D). Collectively, these results indicated that miR-337-3p
downregulated ARHGAP10 expression by binding to its 3′-UTR.
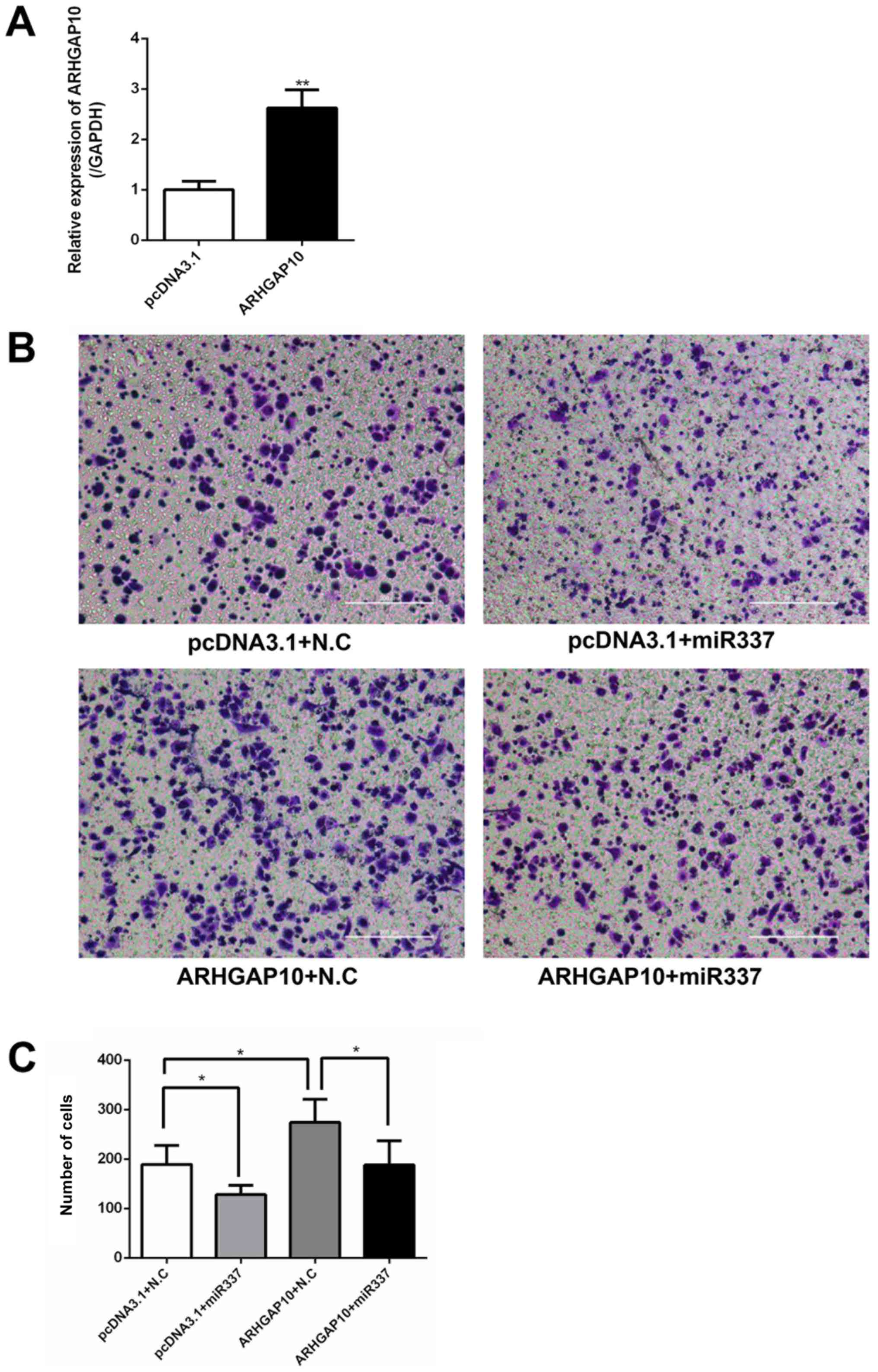
ARHGAP10 restores the migration
capacity of gastric cancer cells
Next, the effects of the overexpression of ARHGAP10
on the migration capacity of gastric cancer cells were examined by
a Transwell migration assay. SGC-7901 cells were transfected with
pcDNA3.1 or ARHGAP10 overexpression plasmid, and the ARHGAP10
plasmid induced an increase in ARHGAP10 mRNA expression (Fig. 6A). It was observed that the
overexpression of miR-337-3p reduced the Transwell migration
ability of SGC-7901 cells. Conversely, the overexpression of
ARHGAP10 increased the migration of SGC-7901 cells. Furthermore,
the co-transfection of miR-337-3p and ARHGAP10 cDNA in SGC-7901
cells abolished the inhibitory effect of miR-337-3p on cell
migration (Fig. 6B and C). These
results indicated that miR-337-3p may target and downregulate
ARHGAP10 expression to inhibit the migration of gastric cancer
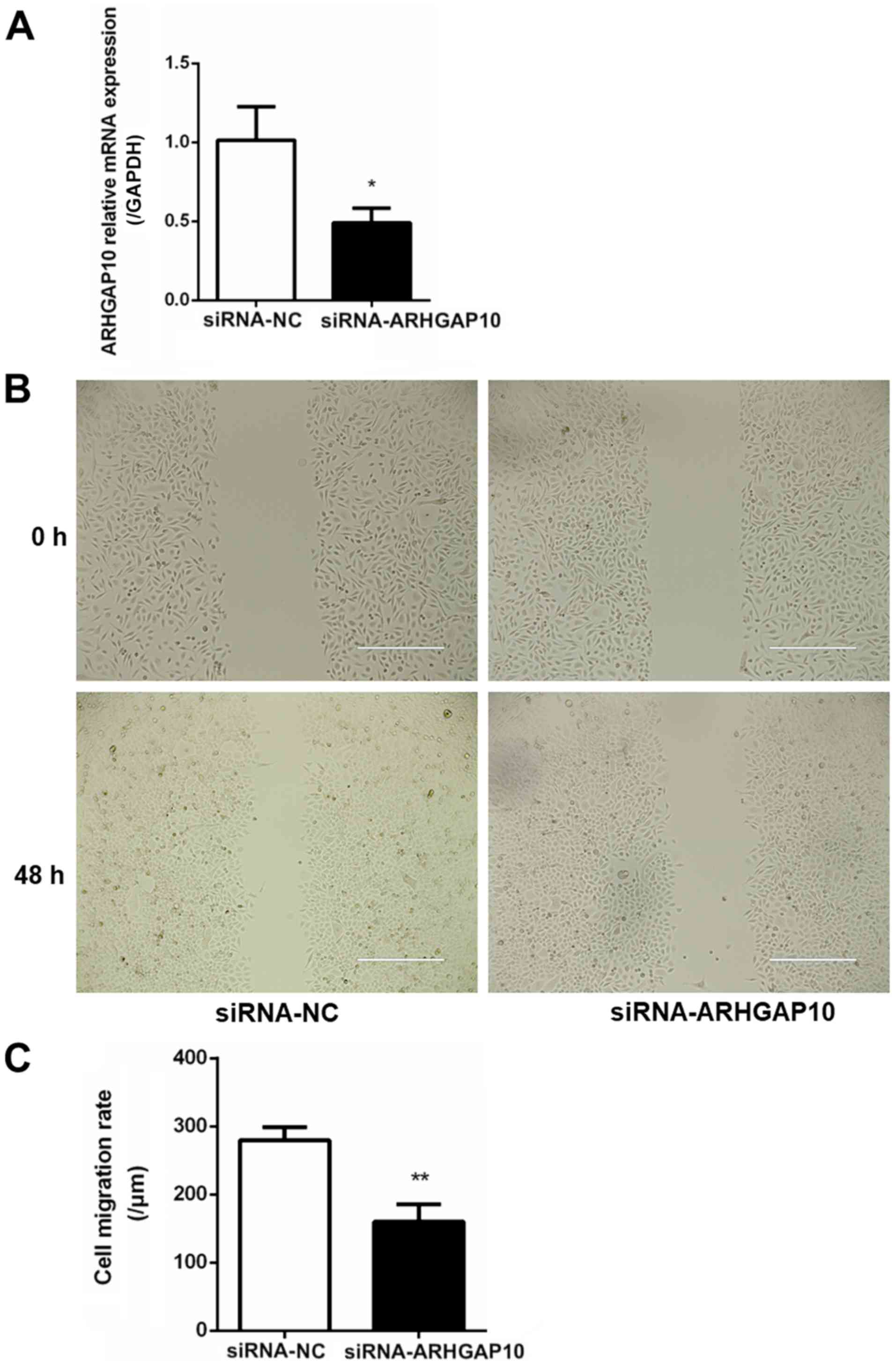
cells. To understand the influence of ARHGAP10 on gastric tumor
metastasis, SGC-7901 cells were transfected with siRNA-NC and
siRNA-ARHGAP10, and the relative expression of ARHGAP10 was
examined with qPCR (Fig. 7A).
SGC-7901 cells transfected with siRNA-ARHGAP10 revealed a decrease
in the wound healing capacity as compared with the control cells
(Fig. 7B and C).
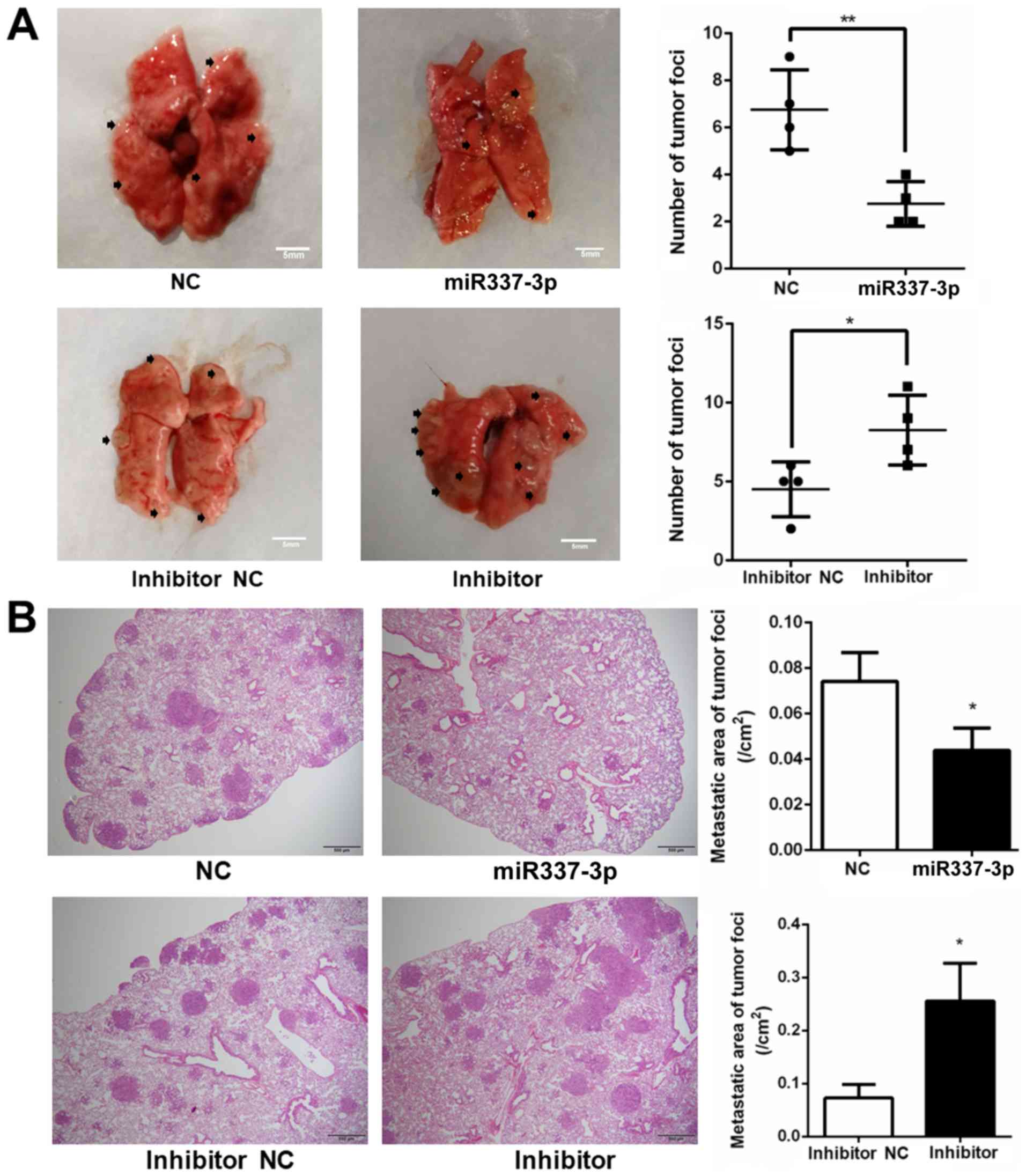
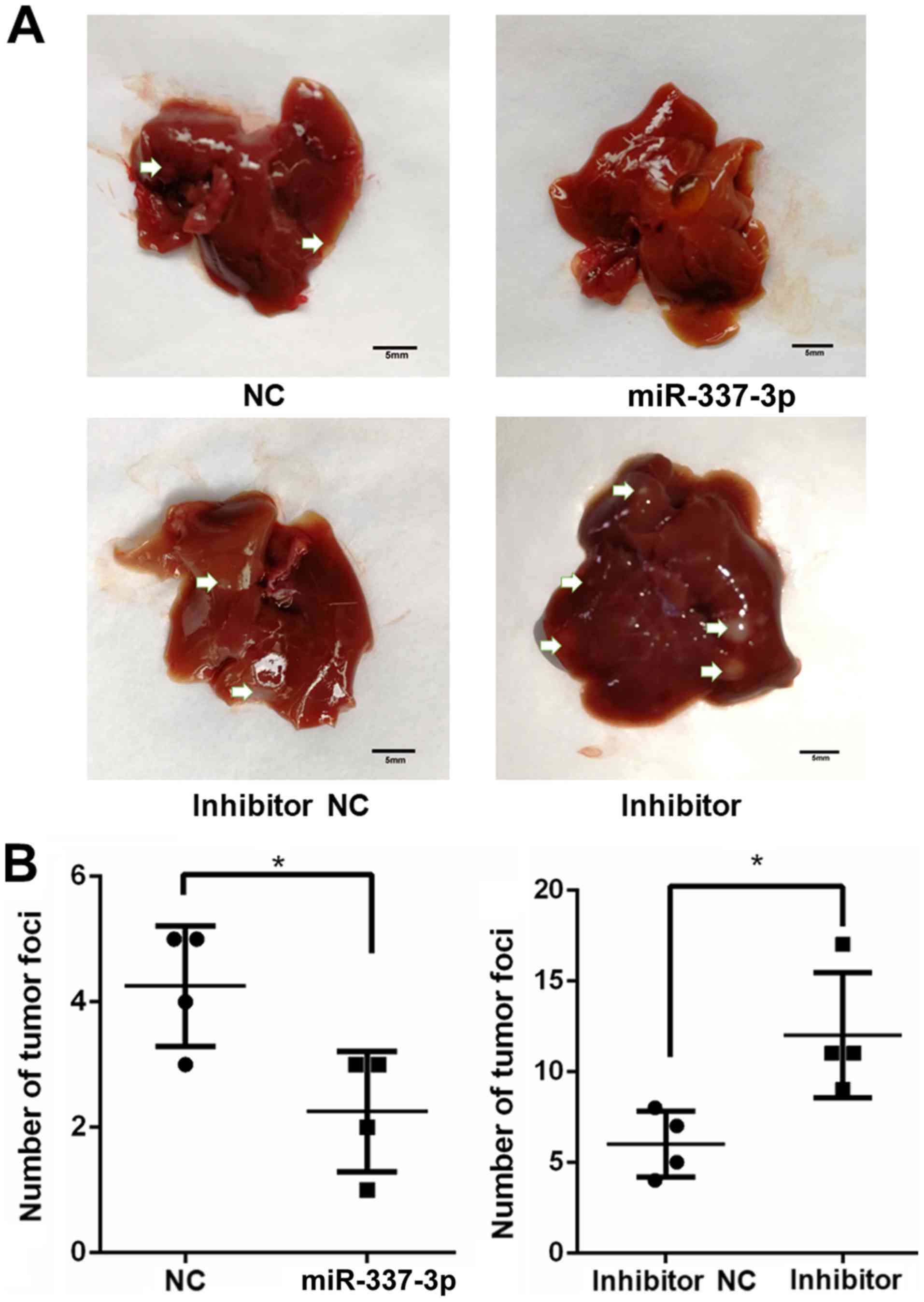
miR-337-3p decreases the in vivo
metastasis of gastric cancer cells
To understand the role of miR-337-3p in gastric
tumor metastasis under in vivo conditions, SGC7901 cells
were transfected with the miRNA and injected into
NOD-Prkdcscid Il2rgnull mice,
which lack T, B, and nature killer cells. After 5 weeks, the mice
injected with miR-337-3p-expressing SGC-7901 cells presented a
reduced number of tumor foci in the lungs and liver. Conversely,
the injection of SGC-7901 cells transfected with inhibitor resulted
in an increase in the number of tumor foci in the lungs and liver
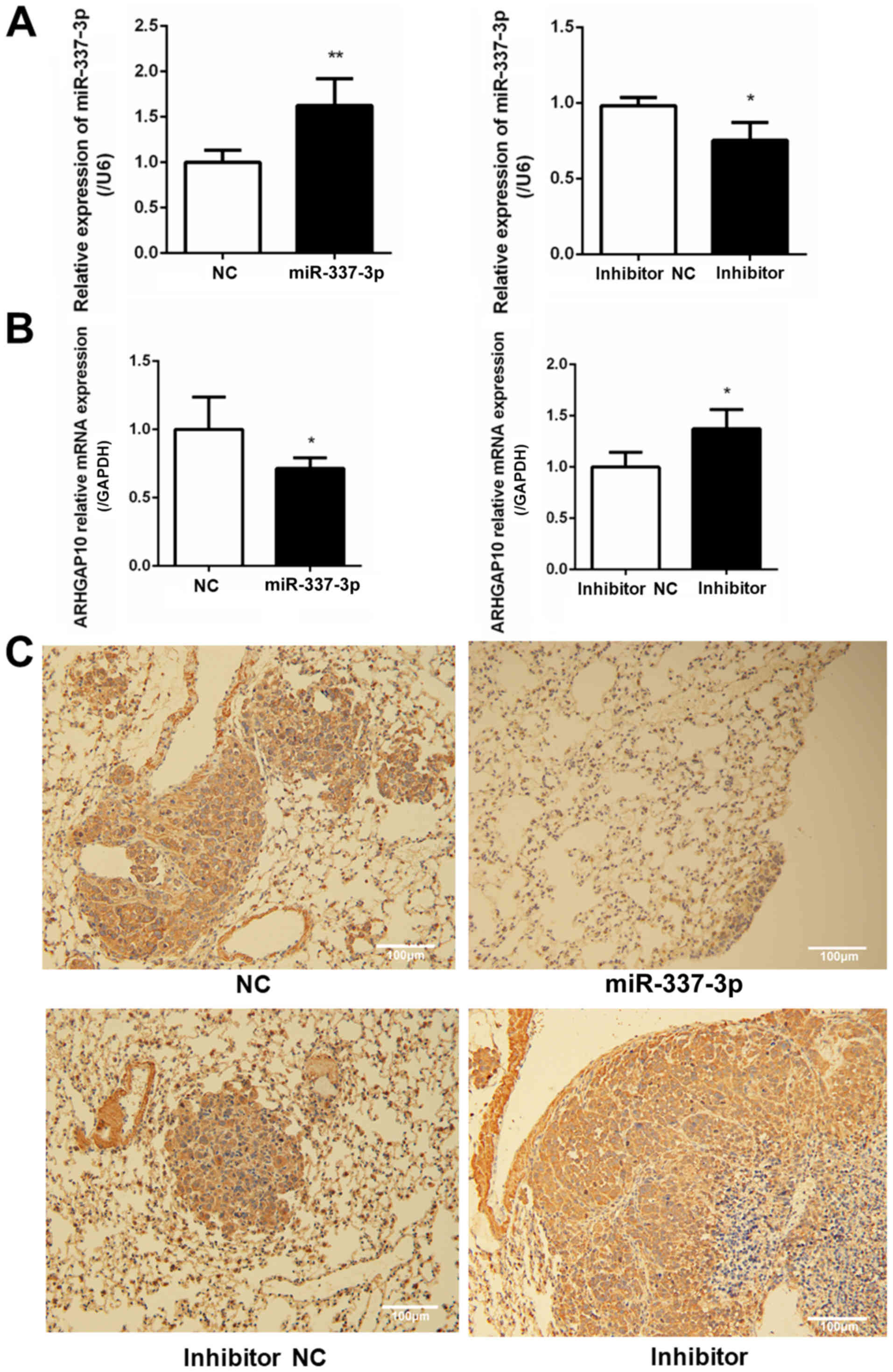
(Figs. 8 and 9). To examine the relative expression of
miR-337-3p in the lung tumor foci, RT-qPCR analysis was performed.
As a result, it was revealed that the overexpression of miR-337-3p
was evident from the high expression of miR-337-3p in the lung
tumor foci. In addition, the transfection with miR-337-3p inhibitor
downregulated the expression of miR-337-3p in the lungs (Fig. 10A). To confirm that ARHGAP10
expression is suppressed by miR-337-3p, ARHGAP10 mRNA expression
level was evaluated with RT-qPCR and histological analyses.
Overexpression of miR-337-3p decreased the expression of ARHGAP10
mRNA, as assessed by qPCR in SGC-7901 cells from the lung tumor
foci (Fig. 10B and C).
Discussion
Tumor metastasis is the leading cause of high
mortality among patients with cancer, and the control and
interruption of tumor metastasis are the key strategies in cancer
treatment (1). Recent studies have
identified several miRNAs that are related to tumor metastasis
(6,15,16).
miR-337-3p was identified as a miRNA associated with gastric tumor
metastasis (6). miR-337-3p was
revealed to suppress the binding of myeloid zinc finger 1 to MMP14
promoter and consequently suppress the progression of gastric
cancer (17). It may also play an
important role in reducing tumor metastasis. However, whether
miR-337-3p targets cytoskeleton-associated proteins in gastric
tumor cells is unclear. In the present study, ARHGAP10 was
identified as a target of miR-337-3p during the regulation of the
migration of the invasive gastric tumor cell line SGC-7901.
Several mRNAs have been identified as molecules
regulating gastric tumor metastasis. Both miR-370 and miR-301a have
been revealed to be upregulated in metastatic gastric tumors.
miR-370 is highly expressed in gastric cancer with more advanced
clinical stage and targets transforming growth factor-β receptor II
and increases the oncogenic potential of gastric cancer cells
(18). miR-301a downregulates the
expression RUNX3 at the post-transcriptional level and promotes the
invasion of gastric cancer cells (19). In addition to miR-337-3p, miR-22,
miR-610, and miR-145 were recently reported to be downregulated in
gastric cancer, while their overexpression was revealed to inhibit
cancer invasion and metastasis. miR-22 has been revealed to target
the oncogenic gene Sp1 and inhibit the migration and
invasion of SGC-7901 and NCL-N87 gastric cancer cell lines
(20). However, its indirect
target molecules related to cell migration are unknown. In a
previous study, we revealed that miR-22 also targets vascular
endothelial cadherin in endothelial cells and interferes with the
process of angiogenesis (21).
miR-610 was revealed to suppress the actin-binding protein
vasodilator-stimulated phosphoprotein and inhibit the migration and
invasion of gastric cancer cells (22). miR-145 was reported to markedly
inhibit N-cadherin protein translation and indirectly downregulate
matrix metallopeptidase 9 expression to suppress gastric cancer
metastasis (23). It has been
reported that mir-337-3p is also able to bind to the promoter
region of MMP14 and inhibit its transcription in both gastric tumor
cells and neuroblastoma cells (7,8). To
the best of our knowledge, whether miR-337-3p targets
cytoskeleton-associated proteins in gastric tumor cells is yet
unknown.
In the present study, a cytoskeleton-regulating
protein, ARHGAP10, was revealed as a target molecule of miR-337-3p.
ARHGAP10, also known as ARHGAP21, exhibits a Rho-GAP domain and is
a cytoskeleton-associated protein. It negatively regulates the
small G protein Rho- and Cdc42-mediated downstream signal
transduction (9). There are three
potential binding sites for miR-337-3p in the 3′-UTR of ARHGAP10;
the results of the luciferase assay confirmed that miR-337-3p binds
to the 3′-UTR of ARHGAP10, and the overexpression of miR-337-3p in
SGC-7901 cells downregulates ARHGAP10 mRNA and protein levels.
These results strongly indicate that miR-337-3p targets ARHGAP10 to
regulate the motility of gastric tumor cells, and that the
downregulation of miR-337-3p expression in metastatic gastric
cancer results in the promotion of ARHGAP10 expression to
facilitate tumor cell migration and invasion.
The role of ARHGAP10 in tumor cell migration is well
known. Since ARHGAP10 negatively regulates the Rho family small
GTPases, it has been regarded as a tumor suppressor (9,13).
The depletion of ARHGAP10 expression in glioblastoma tumor cells
was revealed to increase the migration ability of tumor cells
(11). Studies with ovarian cancer
cells have revealed that ARHGAP10 suppresses tumorigenicity
(13). However, the depletion of
ARHGAP10 expression in certain prostate cancer cells resulted in
the inhibition of tumor cell migration (24). A recent study on breast cancer
cells indicated the involvement of AHRGAP10, along with its analog
ARHGAP23, in the lateral signaling pathway, which is important in
cell migration. The depletion of both ARHGAP10 and ARHGAP23 by
siRNA transfection resulted in the inhibition of tumor cell
migration (25). In addition,
ARHGAP10 was revealed to suppress tumor progression by promoting
the p53-mediated apoptosis and autophagic cell death of gastric
tumor cells (26). The expression
level of ARHGAP10 was downregulated in lung cancer, and ARHGAP10
overexpression resulted in the suppression of the migration,
proliferation, and invasion of tumor cells (27). These studies indicated that
ARHGAP10 may play different roles in different types of tumor
cells. Different cell lines used may have also contributed to the
variations in results. Furthermore, the redundant functions of
ARHGAP10 and ARHGAP23 may further complicate this issue. ARHGAP10
may act in a GAP-independent mechanism to regulate Rho or other
signaling pathways and bind to a-catenin and regulate the
recruitment of a-catenin at cell-cell contact (12). In addition, ARHGAP10 is known to
interact with the C-terminal region of focal adhesion kinase and
regulates its activity (11). The
interaction of ARHGAP10 with a-tubulin results in the regulation of
acetylation of tubulin during epithelial-mesenchymal transition
(28). On the other hand, the
interaction between ARHGAP10 and b-arrestin was revealed to inhibit
GAP activity and attenuate stimulated stress fiber formation
(29). Although our data clearly
demonstrate the regulatory role of both miR-337-3p and ARHGAP10 in
gastric tumor cell migration, further studies are required to
elucidate the mechanism underlying the miR-337-3p-regulated
ARHGAP10 function in the metastasis of gastric tumors.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Anhui
Province National Science Foundation of China (1508085MH146, ZW)
and the Huazhong Agricultural University Scientific &
Technology Self-innovation Foundation (Program no. 2012RC011,
GL).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
ZiW, LY, GL, WG, YL, BH, MW, JuW and HZ contributed
to the conception or design of the work, and the acquisition of
data. QW completed the data analysis. GL, LY, BH, MW, and JuW
drafted the manuscript and revised it critically for important
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal experiments were performed according to
the guidelines of the Laboratory Animal Ethics Committee of
Huazhong Agriculture University, approved by the Laboratory Animal
Centre, Huazhong Agriculture University (HZAHMD-2016-037).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
3′-UTR
|
3′-untranslated region
|
|
MMP14
|
matrix metalloproteinase 14
|
References
|
1
|
Jiang WG, Sanders AJ, Katoh M, Ungefroren
H, Gieseler F, Prince M, Thompson SK, Zollo M, Spano D, Dhawan P,
et al: Tissue invasion and metastasis: Molecular, biological and
clinical perspectives. Semin Cancer Biol. 35 (Suppl):S244–S275.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alizadeh AM, Shiri S and Farsinejad S.;
Metastasis review, : From bench to bedside. Tumour Biol.
35:8483–8523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palmer TD, Ashby WJ, Lewis JD and Zijlstra
A: Targeting tumor cell motility to prevent metastasis. Adv Drug
Deliv Rev. 63:568–581. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Slezak-Prochazka I, Durmus S, Kroesen BJ
and van den Berg A: MicroRNAs, macrocontrol: Regulation of miRNA
processing. RNA. 16:1087–1095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kaboli PJ, Rahmat A, Ismail P and Ling KH:
MicroRNA-based therapy and breast cancer: A comprehensive review of
novel therapeutic strategies from diagnosis to treatment. Pharmacol
Res. 97:104–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Z, Wang J, Yang Y, Hao B, Wang R, Li
Y and Wu Q: Loss of has-miR-337-3p expression is associated with
lymph node metastasis of human gastric cancer. J Exp Clin Cancer
Res. 32:762013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H,
Jiao W, Zhao J, Huang K, Zheng L and Tong Q: miRNA-337-3p
suppresses neuroblastoma progression by repressing the
transcription of matrix metalloproteinase 14. Oncotarget.
6:22452–22466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng L, Jiao W, Mei H, Song H, Li D,
Xiang X, Chen Y, Yang F, Li H, Huang K and Tong Q: miRNA-337-3p
inhibits gastric cancer progression through repressing myeloid zinc
finger 1-facilitated expression of matrix metalloproteinase 14.
Oncotarget. 7:40314–40328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bassères DS, Tizzei EV, Duarte AA, Costa
FF and Saad ST: ARHGAP10, a novel human gene coding for a
potentially cytoskeletal Rho-GTPase activating protein. Biochem
Biophys Res Commun. 294:579–585. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dubois T and Chavrier P: ARHGAP10, a novel
RhoGAP at the cross-road between ARF1 and Cdc42 pathways, regulates
Arp2/3 complex and actin dynamics on Golgi membranes. Med Sci
(Paris). 21:692–694. 2005.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bigarella CL, Borges L, Costa FF and Saad
ST: ARHGAP21 modulates FAK activity and impairs glioblastoma cell
migration. Biochim Biophys Acta. 1793:806–816. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sousa S, Cabanes D, Archambaud C, Colland
F, Lemichez E, Popoff M, Boisson-Dupuis S, Gouin E, Lecuit M,
Legrain P and Cossart P: ARHGAP10 is necessary for alpha-catenin
recruitment at adherens junctions and for Listeria invasion. Nat
Cell Biol. 7:954–960. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo N, Guo J, Chen L, Yang W, Qu X and
Cheng Z: ARHGAP10, downregulated in ovarian cancer, suppresses
tumorigenicity of ovarian cancer cells. Cell Death Dis.
7:e21572016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Xu M, Ding L and Tang J: miR-27a: A
novel biomarker and potential therapeutic target in tumors. J
Cancer. 10:2836–2848. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li N, Xie C and Lu N: Crosstalk between
Hippo signalling and miRNAs in tumour progression. FEBS J.
284:1045–1055. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng LD, Jiao WJ, Mei H, Song HJ, Li D,
Xiang X, Chen YJ, Yang F, Li HH, Huang K and Tong Q: miRNA-337-3p
inhibits gastric cancer progression through repressing myeloid zinc
finger 1-facilitated expression of matrix metalloproteinase 14.
Oncotarget. 7:40314–40328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lo SS, Hung PS, Chen JH, Tu HF, Fang WL,
Chen CY, Chen WT, Gong NR and Wu CW: Overexpression of miR-370 and
downregulation of its novel target TGFβ-RII contribute to the
progression of gastric carcinoma. Oncogene. 31:226–237. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu
Y, Gu Q, Zhu Z and Liu B: Overexpressed miR-301a promotes cell
proliferation and invasion by targeting RUNX3 in gastric cancer. J
Gastroenterol. 48:1023–1033. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo MM, Hu LH, Wang YQ, Chen P, Huang JG,
Lu N, He JH and Liao CG: miR-22 is down-regulated in gastric
cancer, and its overexpression inhibits cell migration and invasion
via targeting transcription factor Sp1. Med Oncol. 30:5422013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu W, Zhan H, Zhou XY, Yao L, Yan M, Chen
A, Liu J, Ren X, Zhang X, Liu JX and Liu G: MicroRNA-22 regulates
inflammation and angiogenesis via targeting VE-cadherin. FEBS Lett.
591:513–526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Zhang J, Wu J, Luo D, Su K, Shi W,
Liu J, Tian Y and Wei L: MicroRNA-610 inhibits the migration and
invasion of gastric cancer cells by suppressing the expression of
vasodilator-stimulated phosphoprotein. Eur J Cancer. 48:1904–1913.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao L, Ren W, Chang S, Guo B, Huang S, Li
M, Guo Y, Li Z, Song T, Zhi K and Huang C: Downregulation of
miR-145 expression in oral squamous cell carcinomas and its
clinical significance. Onkologie. 36:194–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lazarini M, Traina F, Machado-Neto JA,
Barcellos KS, Moreira YB, Brandão MM, Verjovski-Almeida S, Ridley
AJ and Saad ST: ARHGAP21 is a RhoGAP for RhoA and RhoC with a role
in proliferation and migration of prostate adenocarcinoma cells.
Biochim Biophys Acta. 1832:365–374. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Luga V, Armitage SK, Musiol M,
Won A, Yip CM, Plotnikov SV and Wrana JL: A lateral signalling
pathway coordinates shape volatility during cell migration. Nat
Commun. 7:117142016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li B, Wang L, Li Z, Wang W, Zhi X, Huang
X, Zhang Q, Chen Z, Zhang X, He Z, et al: miR-3174 contributes to
apoptosis and autophagic cell death defects in gastric cancer cells
by targeting ARHGAP10. Mol Ther Nucleic Acids. 9:294–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Teng JP, Yang ZY, Zhu YM, Ni D, Zhu ZJ and
Li XQ: The roles of ARHGAP10 in the proliferation, migration and
invasion of lung cancer cells. Oncol Lett. 14:4613–4618. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barcellos KS, Bigarella CL, Wagner MV,
Vieira KP, Lazarini M, Langford PR, Machado-Neto JA, Call SG,
Staley DM, Chung JY, et al: ARHGAP21 protein, a new partner of
alpha-tubulin involved in cell-cell adhesion formation and
essential for epithelial-mesenchymal transition. J Biol Chem.
288:2179–2189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anthony DF, Sin YY, Vadrevu S, Advant N,
Day JP, Byrne AM, Lynch MJ, Milligan G, Houslay MD and Baillie GS:
β-Arrestin 1 inhibits the GTPase-activating protein function of
ARHGAP21, promoting activation of RhoA following angiotensin II
type 1A receptor stimulation. Mol Cell Biol. 31:1066–1075. 2011.
View Article : Google Scholar : PubMed/NCBI
|