Introduction
Breast cancer (BC) is the second leading cause of
female cancer-related mortality worldwide, and remains among the
five most commonly diagnosed cancers, with an incidence of 11.6%
(1,2). Despite the numerous approaches to the
treatment of BC, including surgery, chemotherapy, radiotherapy and
molecular-targeted therapy, the 5 year survival rate of patients
with advanced BC is only 22% (3).
Additionally, the heterogeneity and variability in treatment and
survival response of BC highlight the need to elucidate the
biological mechanisms driving this malignancy. Therefore,
identification of critical molecules involved in BC progression may
provide a breakthrough for targets and biomarkers of antitumor
drugs against this malignant disease.
Long non-coding RNAs (lncRNAs) are a set of RNAs
with a length of >200 nucleotides that lack protein translation
ability (4). Aberrant expression
of lncRNAs, which has been identified in a wide spectrum of
cancers, has been proven to play a key role in a number of cellular
processes, including cancer initiation, development, invasion and
metastasis (4–6). Due to their tissue specificity,
lncRNAs may be applied as diagnostic biomarkers and therapeutic
targets for certain types of cancer. A long intergenic non-coding
RNA 00312 (LINC00312), also referred to as NAG7, which is located
on chromosome 3p25.3, has been identified as a new putative
tumor-suppressor gene in different types of cancer, including
bladder (7), lung (8–12),
colorectal (13) and
hepatocellular (14) cancers. In
addition, a previous study also confirmed that LINC00312 may serve
as a potential biomarker for the progression, metastasis and
prognosis of nasopharyngeal carcinoma (15). However, the exact role of LINC00312
in BC progression and its mechanism of action remain unclear.
The aim of the present study was to investigate the
expression status of LINC00312 in human BC tissues and cell lines,
determine its effect on the proliferation, colony formation,
migration and invasion of BC cell lines and elucidate the
underlying mechanisms, in order to confirm whether LINC00312 may be
used as a possible novel target for the treatment of BC.
Materials and methods
Tissue samples from patients with
BC
Twenty-six pairs of BC tissues and corresponding
adjacent normal tissues were collected during surgery from BC
patients aged 32–69 years during October 2017 and February 2018 at
the Shanghai Tenth People's Hospital. All patients provided
informed consent according to procedures approved by the Shanghai
Tenth People's Hospital Institutional Review Board (certificate no.
SHSY-IEC-KY-4.0/17-23/01). All tissue specimens were rapidly frozen
in liquid nitrogen and then stored at −80°C until extraction of
RNA.
Cell culture
Normal human mammary epithelial cells (HMECs)
MCF-10A and human BC cell lines, including MCF-7, T47D, BT549,
MDA-MB-231 and SKBR3 were purchased from the American Type Culture
Collection (ATCC; Rockville, MD, USA). The MCF-10A, MCF-7,
MDA-MB-231 and T47D cell lines were incubated in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin (Gibco; Thermo
Fisher Scientific, Inc.). The BT549 and SKBR3 cell lines were
incubated in Dulbecco's MEM (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS and 1% penicillin-streptomycin. All
cells were maintained in an incubator at 37°C with 5%
CO2 in a humidified atmosphere.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was first isolated from tissues or cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
and reverse-transcribed to complementary DNA using a PrimeScript RT
Reagent kit (Takara). For the analysis of miRNA, qPCR was performed
using the TaqMan MicroRNA Reverse Transcription kit and TaqMan
Universal PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions, and
U6 small nuclear RNA (snRNA) was used as normalization control. The
primer sequences were as follows: miR-9 forward,
TCTTTGGTTATCTAGCTGTATGA and reverse, TGGTGTCGTGGAGTCG; U6 forward,
CTCGCTTCGGCAGCACA and reverse, AACGCTTCACGAATTTGCGT. For analysis
of mRNA, RT-qPCR was performed using SYBR® Premix ExTaq™
(Takara) according to the manufacturer's instructions, and GAPDH
was used as the normalization control. PCR was conducted in a total
volume of 20 µl under the following conditions: 95°C for 30 min and
60°C for 30 sec, for 30 cycles. The 2−ΔΔCq method
(16) was used to calculate the
relative levels of mRNA. The primer sequences were as follows:
LINC00312 forward, TCTGGCTGTTGTTGTGTTGGA and reverse,
GCTTATTGGCTTGGTTCGCT; and GAPDH forward, GCTGGCGCTGAGTACGTCGTGGAGT
and reverse, CACAGTCTTCTGGGTGGCAGTGATGG.
Transfection and lentivirus
transduction
The miR-9 mimic and its negative control (NC) were
purchased from RiboBio, and oligonucleotide transfection was
performed using Lipofectamine 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). The human LINC00312-cDNA was inserted
into the pCDH-CMV-MCS-EF1-coGFP vector (System Biosciences) to
construct a LINC00312-carrying lentiviral vector (Lv-LINC00312).
The empty lentiviral vector was used as control (Lv-control).
Lv-LINC00312 or Lv-control along with the packaging plasmid VSVG
and psPAX2 were transfected into 293T cells to produce recombinant
lentivirus, followed by infection of the MDA-MB-231 and SKBR3 cell
lines using Polybrene (Sigma-Aldrich; Merck KGaA).
Cell proliferation analysis
Cell Counting Kit-8 (CCK-8) and colony formation
assays were performed to detect the proliferation of MDA-MB-231 and
SKBR3 cells transfected with Lv-LINC00312 or Lv-control. For CCK-8
assays, the cells were inoculated into 96-well plates at a density
of 2,000 cells/well in 100 µl of complete medium. After 1, 2, 3, 4
and 5 days, 10 µl of the CCK-8 solution (Dojindo Molecular
Technologies, Inc.) was added into each well, and the absorbance
was then measured at 450 nm using a microplate reader after 2 h of
additional incubation. For colony formation assays, cells were
seeded into 12-well plates at a density of 1,000 cells/well and
incubated in fresh medium for ~2 weeks to allow colony formation.
Subsequently, the cells were fixed with 4% paraformaldehyde (PFA)
and stained with 1% crystal violet solution for 20 min. The number
of the cell colonies was automatically counted using ImageJ v1.8.0
software (National Institutes of Health, Bethesda, MD, USA) at a
size of 80-infinity.
Transwell migration and invasion
assays
Transwell migration and invasion assays were used to
evaluate the effect of LINC00312 on the migration and invasion of
MDA-MB-231 and SKBR3 cells transfected with Lv-LINC00312 or
Lv-control. For the Transwell migration assays, 2×105
cells in 100 µl of complete medium were seeded into the upper
Transwell chamber (8-µm pore size, BD Biosciences) with medium
containing 10% bovine serum albumin added to the lower
compartments. After incubation for 36 h at 37°C, the cells adhering
to the lower surface of the Transwell membrane were fixed in 20%
methanol and stained with 0.1% crystal violet solution. The number
of cells was calculated under an inverted light microscope
(magnification, ×100). For the Transwell invasion assays, the
procedure was similar to that for the migration assay, except that
the upper compartment was precoated with Matrigel (BD
Biosciences).
Luciferase reporter assay
The potential binding sequences of miR-9 and
LINC00312 wild-type (WT) and the mutant (MUT) miR-9-binding
sequences in LINC00312 were inserted into a pmirGL3-basic vector
(Promega Corp.) to construct a dual luciferase reporter plasmid.
Subsequently, the luciferase reporter vectors containing a WT or a
MUT miR-9-binding sequences in LINC00312 were co-transfected into
MDA-MB-231 and SKBR3 cells with the miR-9 mimic or NC mimic using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). After 48 h of transfection, the luciferase activity was
measured using a Dual Luciferase Reporter Assay kit (Promega
Corp.).
Western blot analysis
Cells were lysed with lysis buffer [100 mM Tris-HCl
(pH 6.8), 4% sodium dodecyl sulfate, 20% glycerol]. Total protein
(30 µg per lane) was separated by 10% SDS-PAGE and
transferred to polyvinylidene fluoride membranes. The membranes
were then blocked with 5% skimmed milk for 60 min and incubated
with primary antibodies (dilution 1:1,000) against CDH1 (cat. no.
14472, Cell Signaling Technology, Inc.), vimentin (VIM; cat. no.
5741, Cell Signaling Technology, Inc.) and GAPDH (cat. no. 5174,
Cell Signaling Technology, Inc.), followed by incubation with
fluorescence-conjugated secondary antibodies (dilution 1:1,000).
Bands were detected by a two-color infrared laser imaging system
(Odyssey; LI-COR Biosciences).
Immunofluorescence assay
MDA-MB-231 and SKBR3 cells transfected with
Lv-LINC00312 or Lv-control on coverslips were fixed with 4%
paraformaldehyde for 15 min, then permeabilized and blocked with 5%
bovine serum albumin in phosphate-buffered saline containing 0.2%
Triton X-100 for 1 h, followed by incubation with primary
antibodies against CDH1 (M168, Abcam) or VIM (RV202, Abcam) at 4°C
overnight. On the following day, the cells were incubated with the
secondary antibodies Dylight TM 488-conjugated Affinipure donkey
anti-mouse IgG (H + L) (M488, Jackson) at room temperature for 1 h,
followed by staining and sealing with ProLong Gold Antifade Reagent
plus DAPI (4,6-diamidino-2-phenylindole) (Invitrogen; Thermo Fisher
Scientific, Inc.). Finally, images of the stained cells were
captured using a laser scanning confocal microscope (magnification,
×400; CarlZeiss AG).
Statistical analysis
Data are presented as the mean ± standard deviation.
Two-way independent samples t-test for differences between two
paired groups or one-way ANOVA followed by the Bonferroni post hoc
test for multiple comparisons was performed using SPSS v.22.0 (IBM
Corp.). **P<0.01 or *P<0.05 (as indicated by asterisk(s) in
the graphical figures) were considered to indicate statistically
significant differences. All experiments were repeated at least
thrice independently.
Results
LINC00312 is downregulated in human BC
tissues and cell lines
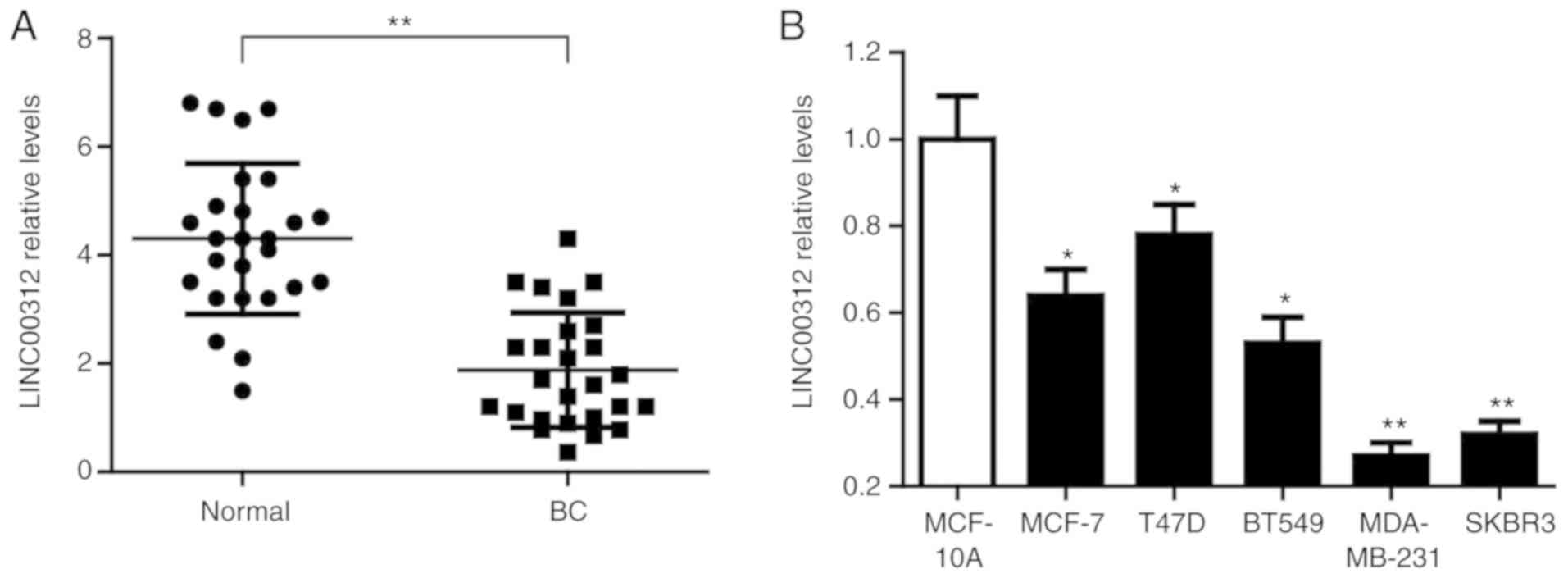
The expression levels of LINC00312 were first
determined in 26 pairs of BC tissues and adjacent normal tissues by
RT-qPCR analysis. As shown in Fig.
1A, the expression of LINC00312 was significantly downregulated
in BC compared with that in the normal tissues (P<0.01). The
expression of LINC00312 was next assessed in BC cell lines, namely
MCF-7, T47D, BT549, MDA-MB-231 and SKBR3, and compared with that in
normal human mammary epithelial cells (HMECs) MCF-10A. The results
of the RT-qPCR analysis demonstrated that the expression of
LINC00312 in all BC cell lines was significantly lower compared
with that in the normal HMECs MCF-10A (Fig. 1B). These results confirmed that
LINC00312 is downregulated in human BC.
LINC00312 inhibits proliferation,
colony formation, migration and invasion of BC cell lines
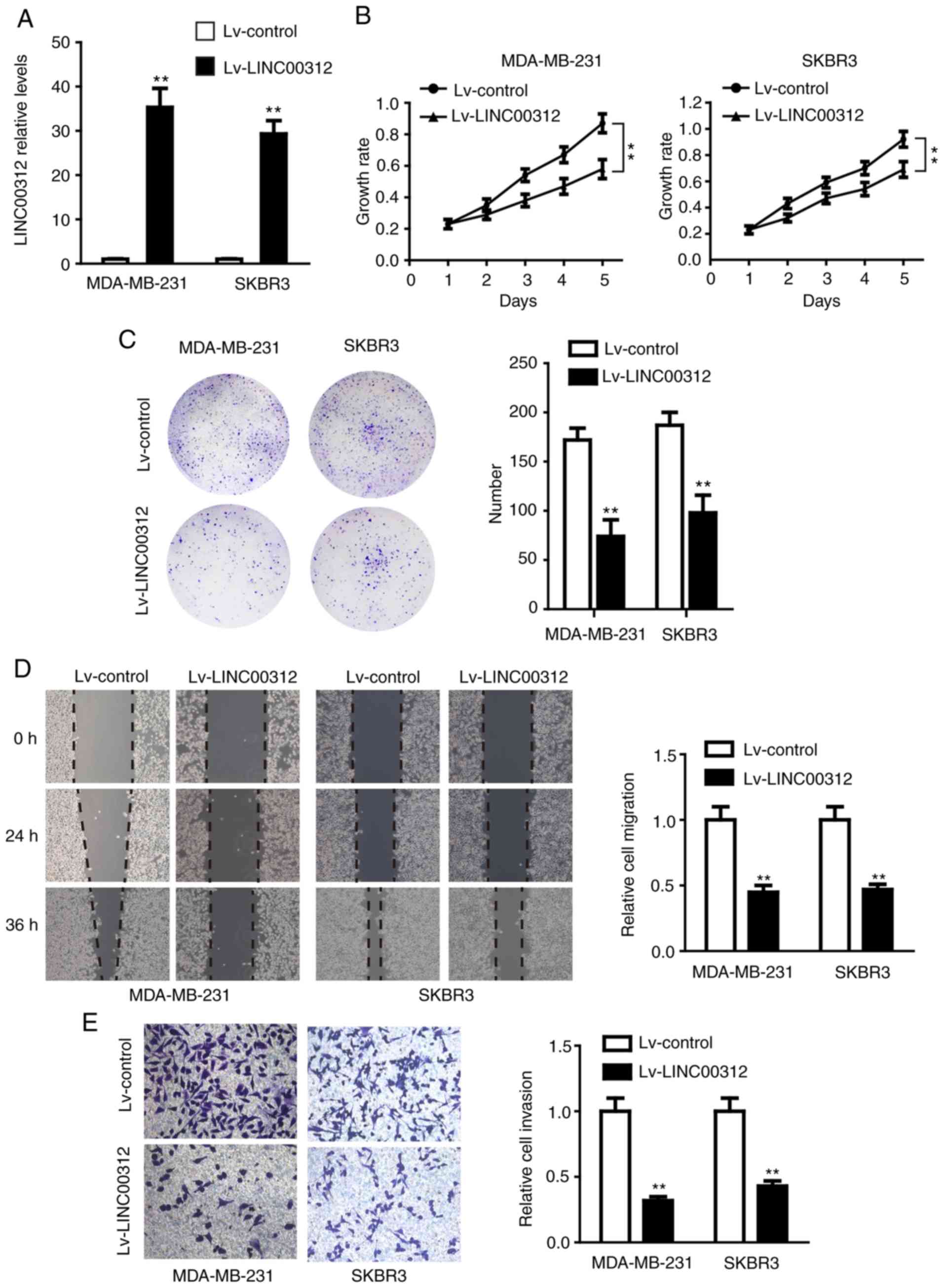
To determine the role of LINC00312 in BC, the
LINC00312-carrying lentivirus (Lv-LINC00312) or control lentivirus
(Lv-control) were transfected into the MDA-MB-231 and SKBR3 cell
lines, which express LINC00312 at low levels. The RT-qPCR results
demonstrated that the expression of LINC00312 was significantly
upregulated in both types of cells using lentivirus-mediated human
LINC00312-cDNA (Fig. 2A). The
effect of LINC00312 on cell proliferation was first examined using
CCK-8 and colony formation assays. As shown in Fig. 2B, overexpression of LINC00312 in
both MDA-MB-231 and SKBR3 cell lines significantly inhibited cell
viability compared with their corresponding controls. Consistently,
LINC00312 overexpression significantly reduced the colony-forming
ability of both MDA-MB-231 and SKBR3 cell lines (Fig. 2C). Subsequently, the effect of
LINC00312 on cell migration and invasion was assessed using
Transwell migration and invasion assays. The results demonstrated
that MDA-MB-231 and SKBR3 cells overexpressing LINC00312 exhibited
decreased migration and invasion ability compared with the negative
control (Fig. 2D and E).
Collectively, these findings confirmed that LINC00312 inhibited the
proliferation, colony formation, migration and invasion of
MDA-MB-231 and SKBR3 cells, suggesting a tumor-suppressive role of
LINC00312 in BC.
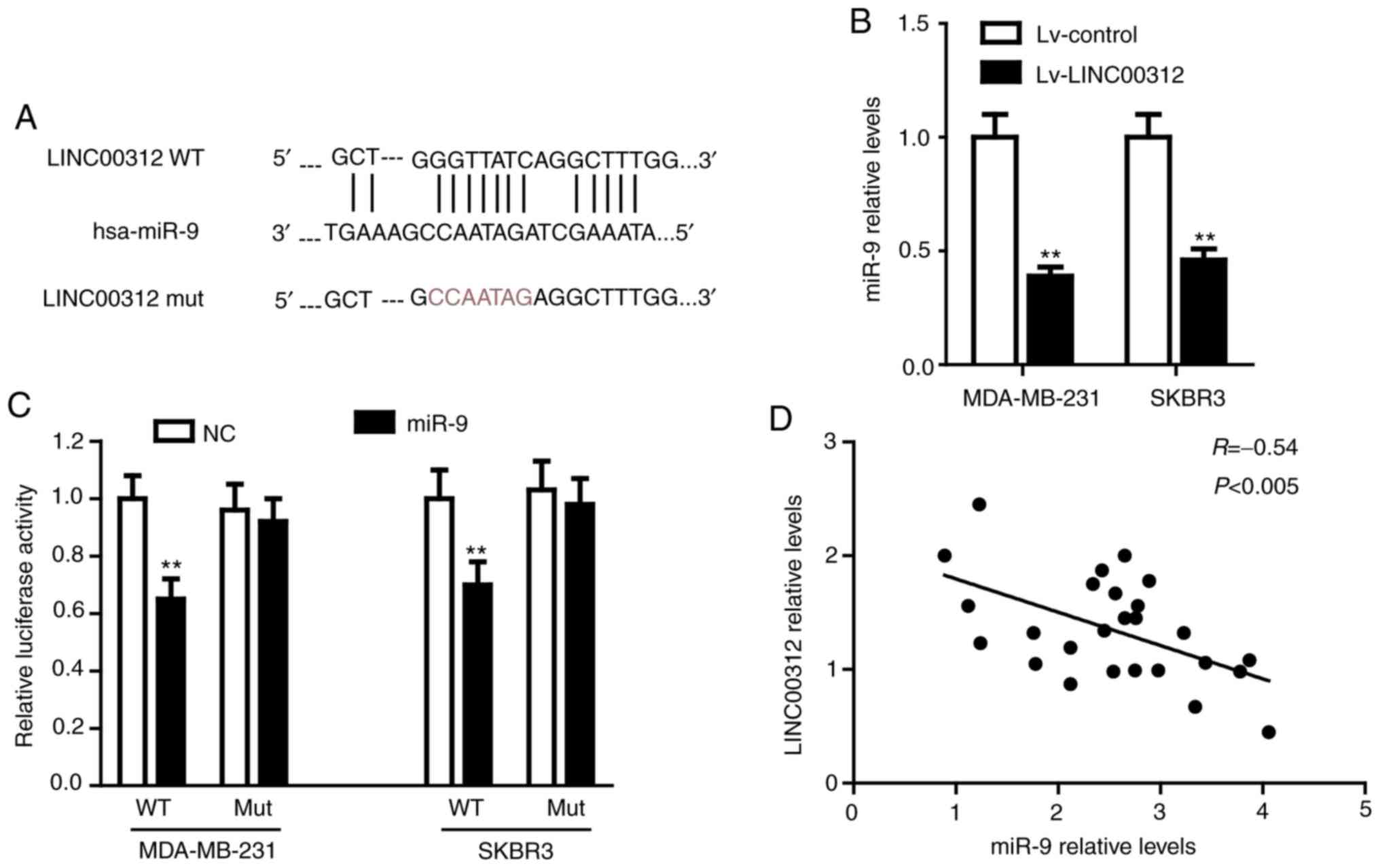
LINC00312 directly binds to miR-9 in
BC
To further explore the molecular mechanism of action
of LINC00312 in BC, two different mRNA target-predicting
algorithms, including miRcode (http://www.mircode.org/) and RNA22 (https://cm.jefferson.edu/rna22/Interactive/), were
used to predict the potential miRNAs that directly bind to
LINC00312. miR-9, the level of which was found to be upregulated in
BC cells (17), was identified as
the most promising candidate. The potential binding sequences of
miR-9 and LINC00312 are shown in Fig.
3A. RT-qPCR was first performed to examine the regulatory
interconnection between LINC00312 and miR-9. As shown in Fig. 3B, overexpression of LINC00312 in
both MDA-MB-231 and SKBR3 cell lines significantly reduced the
miR-9 levels compared with the corresponding control (P<0.01).
Subsequently, the luciferase reporter vectors containing a WT or a
MUT miR-9-binding sequence in LINC00312 were co-transfected into
MDA-MB-231 and SKBR3 cells with the miR-9 mimic or NC mimic. A
dual-luciferase reporter assay was performed to verify the direct
interaction between LINC00312 and miR-9. As shown in Fig. 3C, miR-9 significantly decreased the
luciferase activity in both MDA-MB-231 and SKBR3 cells that were
fused to WT-LINC00312, but not MUT-LINC00312. Additionally, there
was a notable inverse correlation between the levels of LINC00312
and miR-9 in BC tissues (Fig. 3D).
Taken together, these results confirmed that LINC00312 directly
binds to miR-9 in BC.
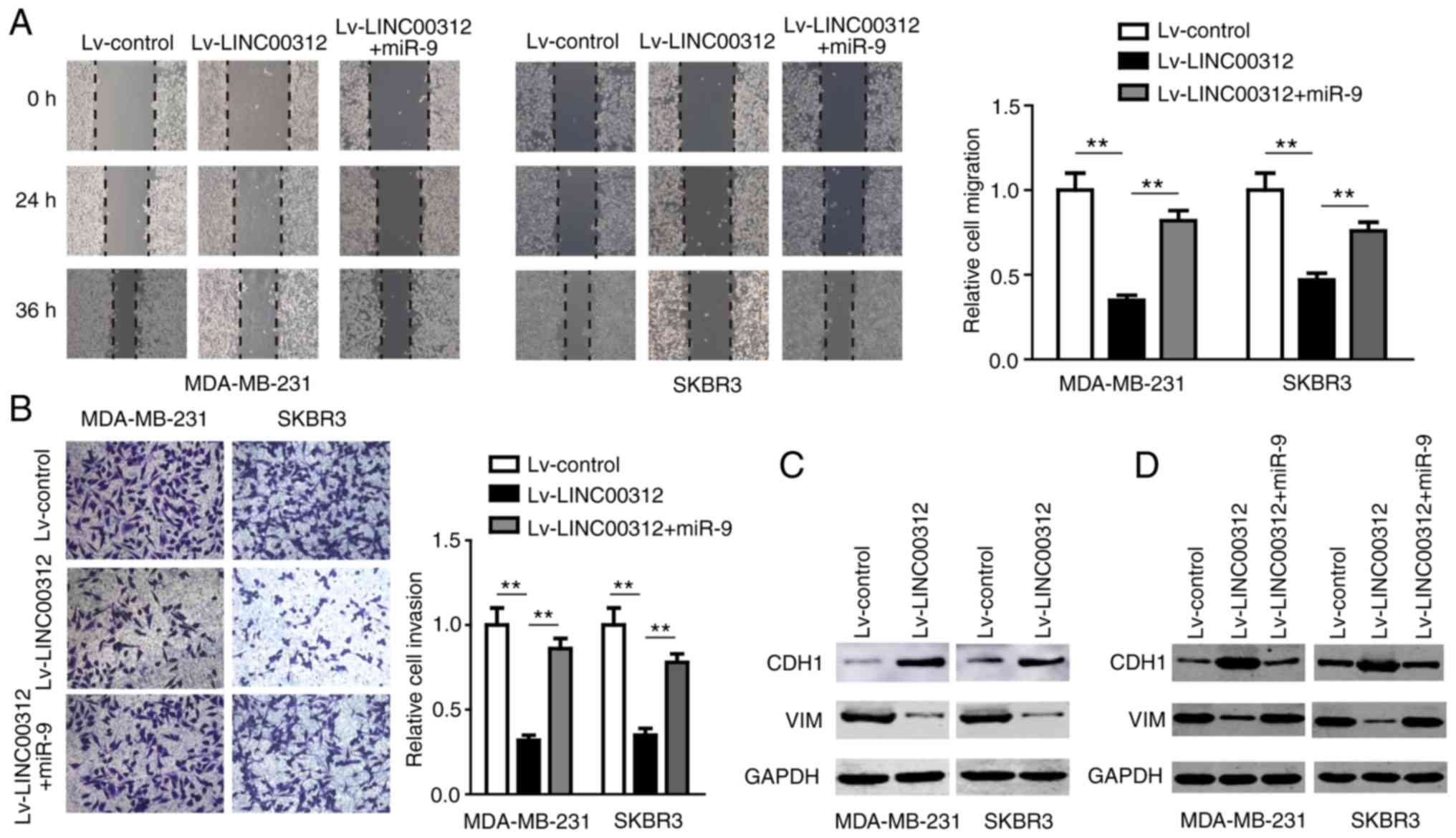
The miR-9/CDH1 axis is involved in the
anti-BC effect of LINC00312
After confirming the direct interaction between
miR-9 and LINC00312, the role of miR-9 in LINC00312-induced
suppression of BC was further investigated. The miR-9 mimic was
transfected into MDA-MB-231 and SKBR3 cells overexpressing
LINC00312, and the cell migration and invasion abilities were
assessed using Transwell migration and invasion assays,
respectively. The results demonstrated that miR-9 partly reversed
the suppressive effect of LINC00312 on cell migration and invasion
(Fig. 4A and B), suggesting that
miR-9 is involved in the anti-BC effect of LINC00312.
miR-9 has been demonstrated to directly target
CDH1, the E-cadherin-encoding mRNA, leading to
downregulation of E-cadherin and increased motility and
invasiveness of BC cells (17).
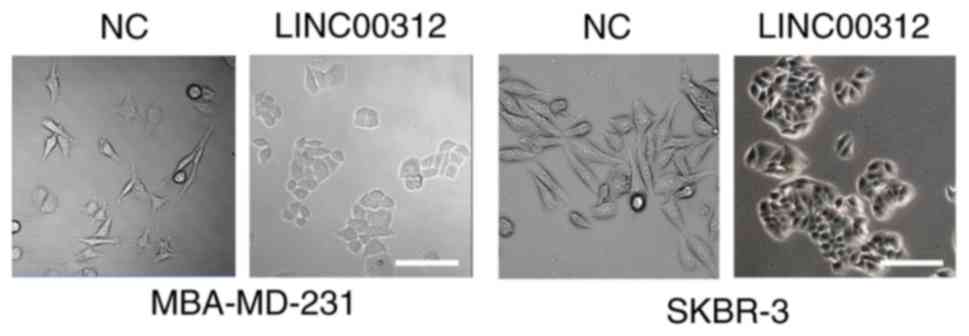
The morphological changes in BC cells transfected with Lv-LINC00312
or Lv-control were first examined using inverted microscopy. The
results demonstrated that both MDA-MB-231 and SKBR3 cells underwent
a morphological change from a spindled to a rounded or
cobblestone-like shape upon LINC00312 overexpression (Fig. 5). We then investigated whether
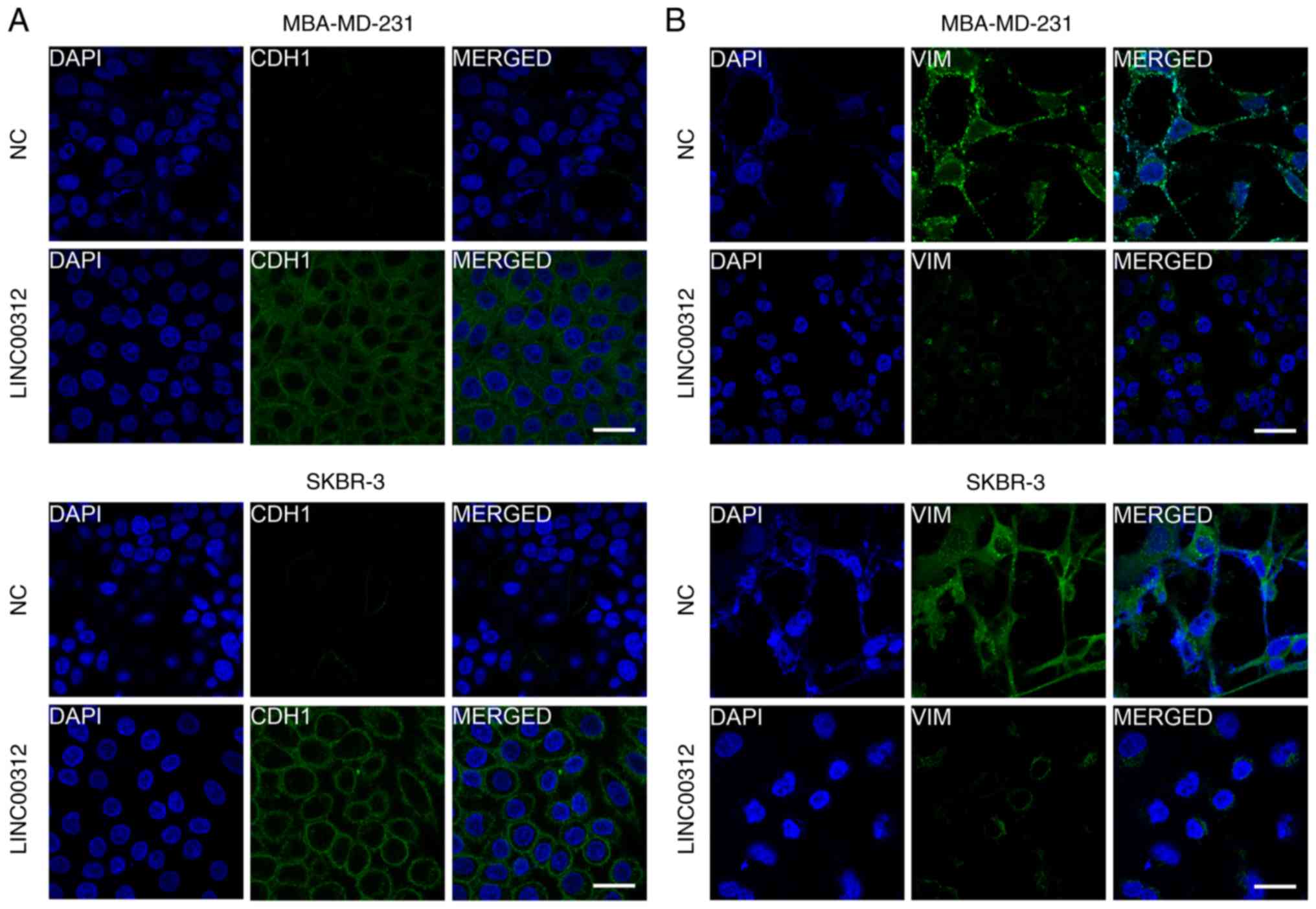
LINC00312 affects the expression of CDH1 and E-cadherin. As shown
in Figs. 4C and 6, overexpression of LINC00312 in both
MDA-MB-231 and SKBR3 cell lines significantly increased the
expression of CDH1 and decreased the expression of the mesenchymal
marker VIM. Additionally, we also observed that miR-9 partly
abrogated the upregulation of CDH1 and downregulation of VIM
induced by LINC00312 in both MDA-MB-231 and SKBR3 cells (Fig. 4D). Taken together, these results
indicate that the miR-9/CDH1 axis may be involved in the anti-BC
effect of LINC00312.
Discussion
Only ~1.5% of the genome is actually responsible for
protein coding (18,19). The non-coding RNAs (ncRNAs), which
represent a large group of RNAs, may be divided into two
categories, namely housekeeping and regulatory, according to their
functions (20). The regulatory
ncRNAs may be broadly classified into two major classes: Short
ncRNAs and long ncRNAs (lncRNAs) (21). The latter are one of the key
members of the ncRNA family, and have been demonstrated to be
involved in the regulation of tumorigenesis and progression as
tumor-suppressor genes or oncogenes (22–24).
LINC00312 is a long intergenic non-coding RNA, which
has been reported to be associated with the onset, progression and
prognosis of several types of cancer. The levels of LINC00312 were
previously reported to be significantly decreased in non-small cell
lung cancer (NSCLC) tissues (11,25).
LINC00312 has been demonstrated to be regulated by HOXA5, leading
to inhibition of proliferation and induction of apoptosis in NSCLC
cells (9). In colon cancer,
LINC00312 has been demonstrated to suppress the proliferation and
metastasis of cancer cells through regulation of the miR-21/PTEN
axis (13,26). In hepatocellular carcinoma,
LINC00312 has been reported to downregulate the expression of
cyclinB1 and inhibit the proliferation of cancer cells in
vitro as well as in vivo (14). In thyroid cancer, LINC00312 has
been demonstrated to inhibit the invasion and migration of cancer
cells by downregulating the PI3K/Akt signaling pathway and
microRNA-197-3p (27,28). Additionally, Zhang et al
(15) and Huang et al
(29) reported that the expression
of LINC00312 was negatively correlated with the size of
nasopharyngeal carcinoma, and that it may inhibit the invasion of
cancer cells through upregulation of the JNK2/AP-1/MMP1 pathway.
However, a recent study reported that LINC00312 promotes the
metastasis and invasion of lung adenocarcinoma cells by directly
binding to the transcription factor YBX1 (10). To the best of our knowledge, the
present study was the first to report that LINC00312 is
downregulated in human BC tissues and cell lines, and that
overexpression of LINC00312 suppresses the proliferation, colony
formation, migration and invasion of BC cell lines, suggesting that
LINC00312 may serve as a tumor-suppressor gene in BC.
miRNAs, a group of non-coding small RNA molecules,
play key roles in multiple types of cancer (30). lncRNAs have been found to suppress
the expression and biological functions of miRNAs by acting as
molecular sponges or competing endogenous RNAs (ceRNAs) (31). LINC00312 has been previously
identified as a ceRNA in several types of cancer. For example, in
thyroid and bladder cancer, LINC00312 has been reported to act as a
ceRNA for miR-197-3p (7,27). miR-9 was previously found to
promote the proliferation of tumor cells by interacting with
tumor-suppressor genes, suggesting an important oncogenic role for
miR-9 in multiple types of cancer, including BC (17). A previous study also reported that
miR-9 was upregulated in BC tissues and cells (32). We herein identified miR-9 as the
most likely target of LINC00312. Therefore, it was hypothesized
that LINC00312 may regulate BC progression by binding to miR-9. The
overexpression of LINC00312 in BC cells was found to significantly
reduce the levels of miR-9. These findings also confirm the direct
interaction between LINC00312 and miR-9 in BC. Of note, a recent
study reported a cross-talk between LINC00312 and miR-21 in colon
cancer (13). As an important
oncogenic miRNA, miR-21 is overexpressed in BC and has been
reported to be involved in transforming growth factor β1-induced
chemoresistance and invasion by targeting PTEN in BC (33). These findings suggest that other
miRNAs may also be involved in the regulation of BC by interacting
with LINC00312.
miRNAs often regulate the expression of downstream
genes by binding to the 3′-untranslated region of target mRNAs
(30). miR-9 has been demonstrated
to directly target CDH1, leading to downregulation of E-cadherin
and increased motility and invasiveness of BC cells (17). We further demonstrated that
overexpression of LINC00312 significantly increased the levels of
CDH1 and decreased the levels of VIM, a major cytoskeletal
component of mesenchymal cells. Additionally, miR-9 partly
abrogated the upregulation of CDH1 and downregulation of VIM
induced by LINC00312 in BC cells, suggesting that the miR-9/CDH1
axis may be involved in the anti-BC effect of LINC00312.
Epithelial-to-mesenchymal transition (EMT) is crucial for tumor
cells to invade and metastasize (34), and CDH1 has been proven to be an
important molecule involved in EMT. Whether LINC00312 is involved
in the EMT of BC cells through miR-9 requires further research, and
ongoing investigations on related animal models in vivo are
currently being conducted in our laboratory.
In conclusion, the data of the present study are the
first to demonstrate that LINC00312 may regulate BC progression as
a tumor-suppressor gene. Additionally, LINC00312 was shown to
increase the expression of CDH1 by directly binding to miR-9 during
the progression of BC, suggesting that LINC00312 may prove to be of
value as a novel diagnostic and therapeutic target for BC.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81503424), Guangzhou Science
and Technology Innovation Commission (grant no. 201704020171),
Shaoguan City Maternal and Child Health Care Plan (grant nos.
Y19183 and Y19184) and Guangzhou Health Science and Technology
Project (grant no. 2019A011014, 2019-2021).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XG and YC designed the experiments, analyzed data
and prepared the manuscript. YC, FQ, LH, WL, LL, CJ, XZ, LQ and ML
performed the experiments. All authors discussed the results and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All patients provided informed consent according to
procedures approved by the Shanghai Tenth People's Hospital
Institutional Review Board (certificate no.
SHSY-IEC-KY-4.0/17-23/01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cardoso F, Spence D, Mertz S,
Corneliussen-James D, Sabelko K, Gralow J, Cardoso MJ, Peccatori F,
Paonessa D, Benares A, et al: Global analysis of
advanced/metastatic breast cancer: Decade report (2005–2015).
Breast. 39:131–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nagini S: Breast cancer: Current molecular
therapeutic targets and new players. Anticancer Agents Med Chem.
17:152–163. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Daskalakis NP, Provost AC, Hunter RG and
Guffanti G: Noncoding RNAs: Stress, glucocorticoids, and
posttraumatic stress disorder. Biol Psychiatry. 83:849–865. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alvarez-Dominguez JR and Lodish HF:
Emerging mechanisms of long noncoding RNA function during normal
and malignant hematopoiesis. Blood. 130:1965–1975. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang YY, Wu ZY, Wang GC, Liu K, Niu XB, Gu
S and Meng JS: LINC00312 inhibits the migration and invasion of
bladder cancer cells by targeting miR-197-3p. Tumour Biol.
37:14553–14563. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Y, Pan Y, Ji Y, Sheng L and Du X:
Network analysis of differentially expressed smoking-associated
mRNAs, lncRNAs and miRNAs reveals key regulators in
smoking-associated lung cancer. Exp Ther Med. 16:4991–5002.
2018.PubMed/NCBI
|
|
9
|
Zhu Q, Lv T, Wu Y, Shi X, Liu H and Song
Y: Long non-coding RNA 00312 regulated by HOXA5 inhibits tumour
proliferation and promotes apoptosis in Non-small cell lung cancer.
J Cell Mol Med. 21:2184–2198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng Z, Wang J, Shan B, Li B, Peng W, Dong
Y, Shi W, Zhao W, He D, Duan M, et al: The long noncoding RNA
LINC00312 induces lung adenocarcinoma migration and vasculogenic
mimicry through directly binding YBX1. Mol Cancer. 17:1672018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tan Q, Yu Y, Li N, Jing W, Zhou H, Qiu S,
Liang C, Yu M and Tu J: Identification of long non-coding RNA 00312
and 00673 in human NSCLC tissues. Mol Med Rep. 16:4721–4729. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tian Z, Wen S, Zhang Y, Shi X, Zhu Y, Xu
Y, Lv H and Wang G: Identification of dysregulated long non-coding
RNAs/microRNAs/mRNAs in TNM I stage lung adenocarcinoma.
Oncotarget. 8:51703–51718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li G, Wang C, Wang Y, Xu B and Zhang W:
LINC00312 represses proliferation and metastasis of colorectal
cancer cells by regulation of miR-21. J Cell Mol Med. 22:5565–5572.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu J, Zhou X, Fan Y, Cheng X, Lu B and
Chen Z: Long non-coding RNA 00312 downregulates cyclin B1 and
inhibits hepatocellular carcinoma cell proliferation in vitro and
in vivo. Biochem Biophys Res Commun. 497:173–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang W, Huang C, Gong Z, Zhao Y, Tang K,
Li X, Fan S, Shi L, Li X, Zhang P, et al: Expression of LINC00312,
a long intergenic non-coding RNA, is negatively correlated with
tumor size but positively correlated with lymph node metastasis in
nasopharyngeal carcinoma. J Mol Histol. 44:545–554. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pombo A: Cellular genomics: Which genes
are transcribed, when and where? Trends Biochem Sci. 28:6–9. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Forterre P: Genomics and early cellular
evolution. The origin of the DNA world. C R Acad Sci III.
324:1067–1076. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin C and Yang L: Long noncoding RNA in
cancer: Wiring signaling circuitry. Trends Cell Biol. 28:287–301.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sullenger BA and Nair S: From the RNA
world to the clinic. Science. 352:1417–1420. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu H, Xu Q, Liu F, Ye X, Wang J and Meng
X: Identification and validation of long noncoding RNA biomarkers
in human non-small-cell lung carcinomas. J Thorac Oncol.
10:645–654. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang XM, Wang XY, Sheng SR, Wang JR and
Li J: Expression of tumor related genes NGX6, NAG-7, BRD7 in
gastric and colorectal cancer. World J Gastroenterol. 9:1729–1733.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu K, Huang W, Yan DQ, Luo Q and Min X:
Overexpression of long intergenic noncoding RNA LINC00312 inhibits
the invasion and migration of thyroid cancer cells by
down-regulating microRNA-197-3p. Biosci Rep. 37(pii):
BSR201701092017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Min X, Liu K, Zhu H and Zhang J: Long
noncoding RNA LINC003121 inhibits proliferation and invasion of
thyroid cancer cells by suppression of the
phosphatidylinositol-3-kinase (PI3K)/Akt signaling pathway. Med Sci
Monit. 24:4592–4601. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang C, Wu M, Tang Y, Li X, Ouyang J,
Xiao L, Li D and Li G: NAG7 promotes human nasopharyngeal carcinoma
invasion through inhibition of estrogen receptor alpha and
up-regulation of JNK2/AP-1/MMP1 pathways. J Cell Physiol.
221:394–401. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu M, Wang X, Gu Y, Wang F, Li L and Qiu
X: MEG3 overexpression inhibits the tumorigenesis of breast cancer
by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem
Biophys. 661:22–30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dai X, Fang M, Li S, Yan Y, Zhong Y and Du
B: miR-21 is involved in transforming growth factor β1-induced
chemoresistance and invasion by targeting PTEN in breast cancer.
Oncol Lett. 14:6929–6936. 2017.PubMed/NCBI
|
|
34
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|