Introduction
Melanoma is a common type of skin cancer, causing
>30% of skin cancer-related mortalities, despite accounting for
<4% of total skin cancer types in the US (1). The incidence of melanoma has
increased by ~4% in the past decade (2). Although treatment of melanoma has
improved, finding effective therapeutics with low toxicity is
urgent (3). In this regard, herbal
and dietary medicines have been considered a promising alternative
for melanoma therapy (4). Previous
studies have tested the anti-melanoma activities of natural
products isolated from fruits, vegetables and plants (5). Herb-derived polyphenols have been
intensively investigated due to their abilities to reduce the
aggressiveness of melanoma (6,7).
Protocatechualdehyde (PCA) is a polyphenol compound
found in many Chinese herbal plants, such as Salvia
miltiorrhiza, Stenoloma chusanum (L.) Ching and Ilex
chinensis Sims (8,9). PCA can also be isolated from the
butanol extract of Streptomyces lincolnensis M-20 (10), and is thought to be the main
phenolic component of Phellinus gilvus responsible for its
antioxidant effect (11). PCA has
anticancer effects against multiple cancer types, including breast
cancer (10), leukemia (12) and colorectal cancer (11). Our previous study reported that
P. gilvus-derived PCA inhibited the proliferation of human
colorectal cancer cells by inducing S phase arrest and promoting
the mitochondrial apoptotic pathway (11). Whilst the cytotoxic properties of
PCA against cancer cells have previously been shown (11), little is known about the effect and
mechanism of PCA in melanoma cells.
The present study investigated the cytotoxic
activity of P. gilvus-derived PCA in B16-F10 cells.
Whole-transcriptome sequencing was used to evaluate PCA-induced
differentially expressed mRNAs to identify the molecular mechanisms
by which PCA may inhibit melanoma development.
Materials and methods
Chemicals and reagents
Murine melanoma B16-F10 cells were obtained from the
Institute of Biochemistry and Cell Biology, Chinese Academy of
Sciences. The cells were maintained in RPMI-1640 medium
supplemented with 10% FBS (both purchased from Gibco; Thermo Fisher
Scientific, Inc.), plus penicillin (100 µg/ml) and streptomycin
(100 µg/ml). Cells were cultured at 37°C under a humidified
atmosphere with 5% CO2. MTT was obtained from
Sigma-Aldrich (Merck KGaA). Apoptosis assay kit was purchased from
Invitrogen; Thermo Fisher Scientific, Inc.
PCA preparation
PCA was isolated from the fresh fruiting bodies of
Phellinus gilvus, which was cultivated by Sericultural
Research Institute (Zhejiang Academy of Agricultural Sciences). The
purification and identification of PCA was performed as previously
described (11). High-performance
liquid chromatography (HPLC) was performed on a 1260 Infinity II LC
System (Aligent Technologies, Inc.). The separation was performed
on an Agilent ZORBAX Eclipse Plus-C18 (100×4.6 mm; 1.8 µm; Aligent
Technologies, Inc.). The binary mobile phase consisted of 0.1%
formic acid in water (A) and 0.1% formic acid in acetonitrile (B).
The flow rate was kept constant at 0.6 ml/min for a total run time
of 50 min, and the column temperature was maintained at 30°C. The
system was run with a gradient program: 0–30 min, 3–30% A; 30–50
min 30–95% A. The sample quantity was 20 µl. HPLC analysis showed
that the purity of PCA was >95%.
Cell proliferation assay
Cell proliferation was determined by an MTT-based
colorimetric assay. Cells at the exponential growth phase were
harvested and dispensed into a 96-well microplate at 100 µl/well.
After 24 h, 100 µl PCA was added to different final concentrations
(0, 0.5, 1, 2.5, 5, 12.5, 25, 50, 100 and 200 µg/ml) and incubated
at 37°C for 24, 48 or 72 h. Cells were then incubated at 37°C with
50 µl of MTT solution (1 mg/ml) for 2 h and the resulting crystals
were dissolved in DMSO. The absorbance at 570 nm was recorded to
assess the formation of formazan.
Cell cycle and apoptosis analysis by
flow cytometry
The cell cycle phase was measured by assessing the
DNA content using flow cytometry. B16-F10 cells (2×104
cells/well) were incubated at 37°C with PCA at different final
concentrations of 0, 12.5 or 50 µg/ml for 48 h. Cells were
collected and washed with ice-cold PBS, and fixed at 4°C in 70%
ethanol for 12 h. After staining with propidium iodide (PI; 0.5 ml)
at 4°C for 30 min, the proportion of cells at different phases was
determined using a flow cytometer (Cytomics FC 500 MCL; Beckman
Coulter, Inc.) and MultiCycle AV software (CXP V2.3 WIN7, C30309;
Phoenix Flow Systems, Inc.) was used for analysis. Apoptosis rate
was measured by staining at 4°C for 15 min with Annexin V-FITC (5
µl) and PI (5 µl). Annexin V−/PI+ (lower
right) cells were deemed as early apoptotic cells, Annexin
V+/PI+ (upper right) cells were defined as
late apoptotic cells, while Annexin V+/PI−
(upper left) cells were necrotic cells. All experiments were
performed in triplicate.
RNA sequencing
After treatment with PCA (50 µg/ml) and 0.1% DMSO
(Control) for 48 h, B16-F10 cells (1×107) were collected
for RNA sequencing. Total RNA was obtained from cells using a
RNAsimple Total RNA kit (Biotech Co., Ltd.). The construction of
cDNA libraries and RNA-Seq was performed by Guangzhou RiboBio Co.,
Ltd. The HISTAT2 was used to map clean reads to the genome with the
default parameters (13). The
threshold value of differentially expressed mRNA was set by
|log2FoldChange|>1 and q-value <0.02.
Bioinformatics analysis
Principal component analysis was used to assess the
differences between groups and the duplication within groups,
according to the expression level of all genes in each sample.
DEseq2 was used to analyze the differential expression of genes in
the control group and PCA-treated B16-F10 cells (14). Gene Ontology (GO) term enrichment
analysis of differentially expressed genes (DEGs) was implemented
by the GOseq R packages based Wallenius non-central hyper-geometric
distribution (15). KOBAS (version
2.0, http://kobas.cbi.pku.edu.cn/) was used
to test the statistical enrichment of DEGs in Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathways (16,17).
RT-quantitative (q)PCR analysis
Total RNA extraction, cDNA synthesis and RT PCR were
performed as previously described (11). Briefly, total RNA was isolated from
B16-F10 cells treated with or without PCA (50 µg/ml) for 48 h using
the TaKaRa MiniBEST Universal RNA Extraction kit (Takara Bio,
Inc.), then the cDNA was synthesized using the PrimeScript RT
reagent kit with gDNA Eraser (Takara Bio, Inc.). RT-qPCR was
performed using the SYBR® Fast qPCR Mix (Takara Bio,
Inc.) via the StepOnePlus™ Real-Time PCR system (Thermo Fisher
Scientific, Inc.). The PCR volume was 20 µl, including 0.4 µl each
primer (10 nmol/l), 10 µl SYBR Fast qPCR Mix, 0.4 µl ROX reference
dye, 2.0 µl cDNA template and 6.8 µl dilution buffer (Takara Bio,
Inc.). An initial activation at 95°C for 2 min was followed by an
amplification target sequence of 40 cycles of 95°C for 30 sec, 60°C
for 30 sec and 72°C for 30 sec. PCR primers were synthesized by
Shanghai Shenergy Biocolor BioScience & Technology Co., Ltd. as
follows: p53-induced death domain-containing protein 1 (PIDD)
forward 5′-GGGAACCAGTTGAACTTGGAC-3′, reverse
5′-CTGAGCCGCAAAAACTCCAC-3′; p21 forward 5′-CCTGGTGATGTCCGAC-CTG-3′,
reverse 5′-CCATGAGCGCATCGCAATC-3′; Cyclin E forward
5′-TCT-TCACACAGATGACACAAGC-3′, reverse 5′-CAACAGCAACCTACAACACCC-3′;
Bax forward 5′-AGACAGGGGCCTTTTTGCTAC-3′, reverse
5′-AATTCGCCG-GAGACACTCG-3′; Cyclin D forward
5′-TCAAGTGTGACCCGGACTG-3′, reverse 5′-ATGTCCACATCTCGCACGTC-3′;
growth arrest and DNA damage-inducible 45 (Gadd45) forward
5′-CCGAAAGGATGGACACGGTG-3′, reverse 5′-TTATCGGGGTCTACGTTGAGC-3′;
p53 forward 5′-CACAGCACATGACGG-AGGTC-3′, reverse
5′-TCCTTCCACCCGGATAAGATG-3′; Bcl-2 forward
5′-GCTACCGTCGTGACTTCGC-3′, reverse 5′-CCCCACCGAACTCAAAGAAGG-3′;
cell division cycle-associated protein 2 (Cdc-2) forward
5′-AGAAGGTACTTACGG-TGTGGT-3′, reverse 5′-GAGAGATTTCCCGAATTGCAGT-3′;
Cyclin B forward 5′-AAGGCCAAGGTCAGTATGGC-3′, reverse
5′-CTTGCCTGTAGCTCTTCGCT-3′; phosphatidylinositol glycan anchor
biosynthesis class S (PIGs) forward 5′-TCT-GTCCCCGATTTCCCCC-3′,
reverse 5′-AGCTGCTTCAAGACTTCCGC-3′; GAPDH forward
5′-AAGAAGGTGGTGAAGCAGG-3′, reverse 5′-GAAGGTGGA-AGAGTGGGAGT-3′. The
PCR product sizes for PIDD, p21, cyclin E, Bax, cyclin D, Gadd45,
p53, Bcl-2, Cdc-2, cyclin B, PIGs and GAPDH were 107, 103, 88, 137,
175, 121, 101, 147, 128, 131, 125 and 111 bp, respectively. Gene
expression was quantified using the 2−∆∆Cq method
(18).
Statistical analysis
Data are expressed as the mean ± SEM. Statistical
analysis was performed using the SPSS 16.0 (SPSS, Inc.). One-way
ANOVA was used to analyze statistical differences between groups
under different conditions, followed by Tukey's test. P<0.05 was
considered to indicate a statistically significant difference.
Results
PCA inhibits the viability of B16-F10
melanoma cells
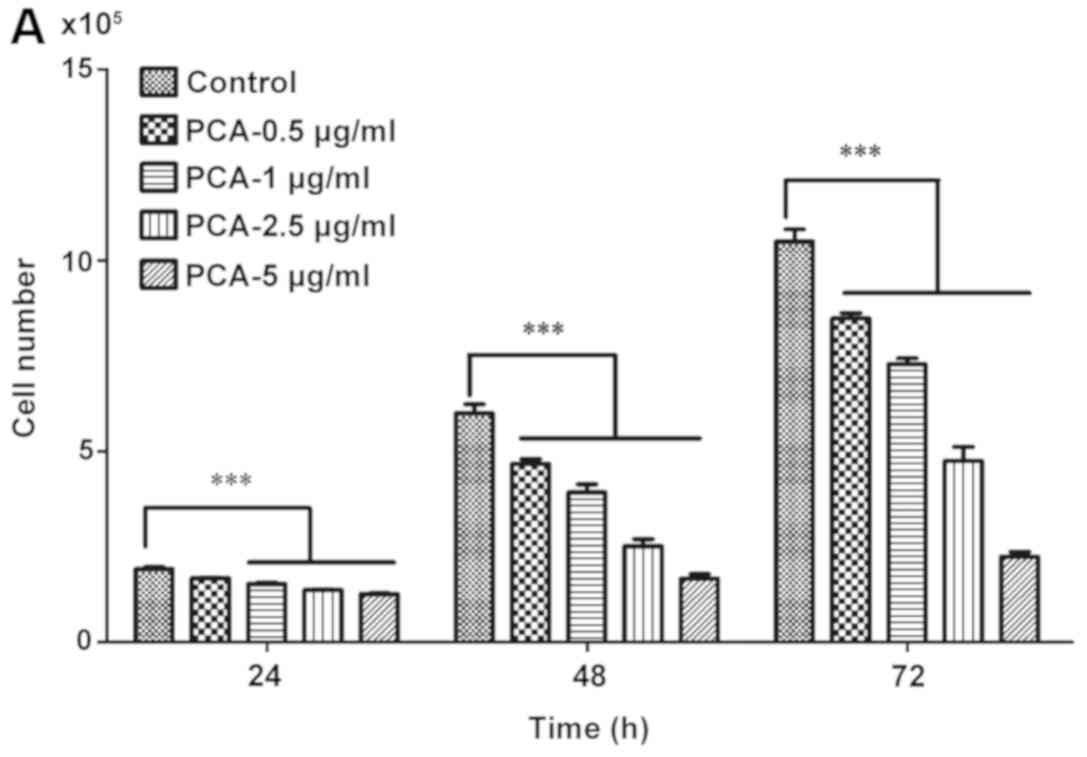
To examine the cytotoxicity of PCA on the
proliferation of melanoma cells, B16-F10 cells were treated with
different concentrations of PCA (0, 0.5, 1, 2.5, 5, 12.5, 25, 50,
100 and 200 µg/ml) for 24, 48 and 72 h, and MTT assay was
performed. Treatment with PCA for 24, 48 and 72 h significantly
decreased cell viability compared with the control group (PCA 0
µg/ml) in a dose-dependent manner (Fig. 1).
At higher concentration levels (12.5, 25, 50, 100
and 200 µg/ml), PCA significantly decreased the growth of B16-F10
cells. The present results suggested that extended treatment
periods with PCA could, to a greater extent, reduce B16-F10 cell
growth. These results suggested that PCA could be used as a cell
proliferation inhibitor for the treatment of melanoma.
PCA induces cell cycle arrest at the
G0/G1 phase
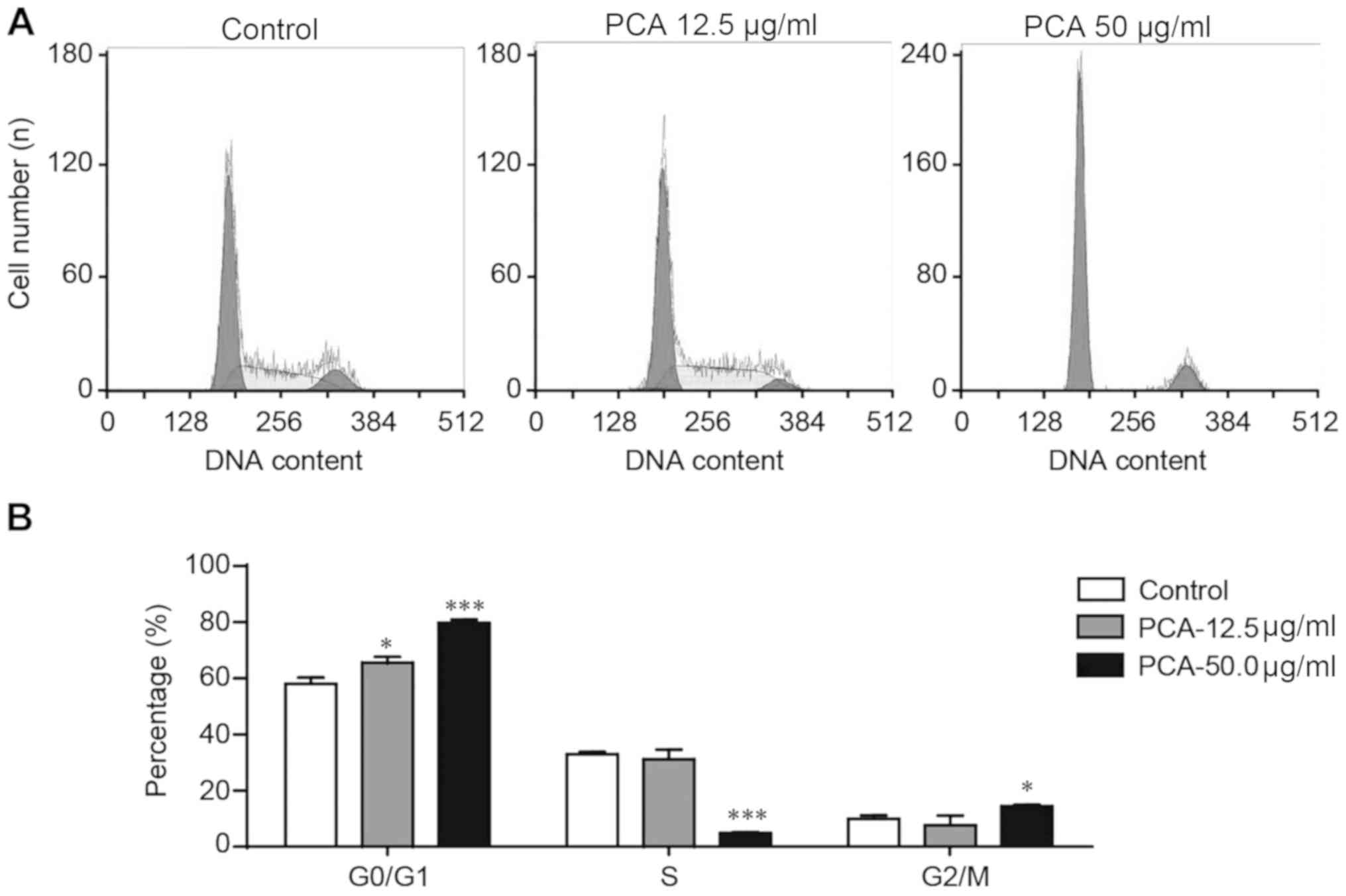
Flow cytometry was used to assess whether the
antiproliferative effects of PCA were mediated by the arrest of the
cell cycle in B16-F10 cells treated with 0, 12.5 or 50.0 µg/ml PCA
for 48 h. PCA significantly increased cell number at the G0/G1
phase and decreased cell number at the S phase in a dose-dependent
manner (Fig. 2). The influence of
PCA on the cell number at the G2/M phase was not clear in the low
dose PCA treatment group (12.5 µg/ml), but a significant increase
was observed in the high dose PCA-treated group (50.0 µg/ml). The
present results suggested that G0/G1 phase arrest may be partially
involved in the mechanism of PCA-reduced viability of B16-F10
cells.
PCA promotes apoptosis in B16-F10
cells
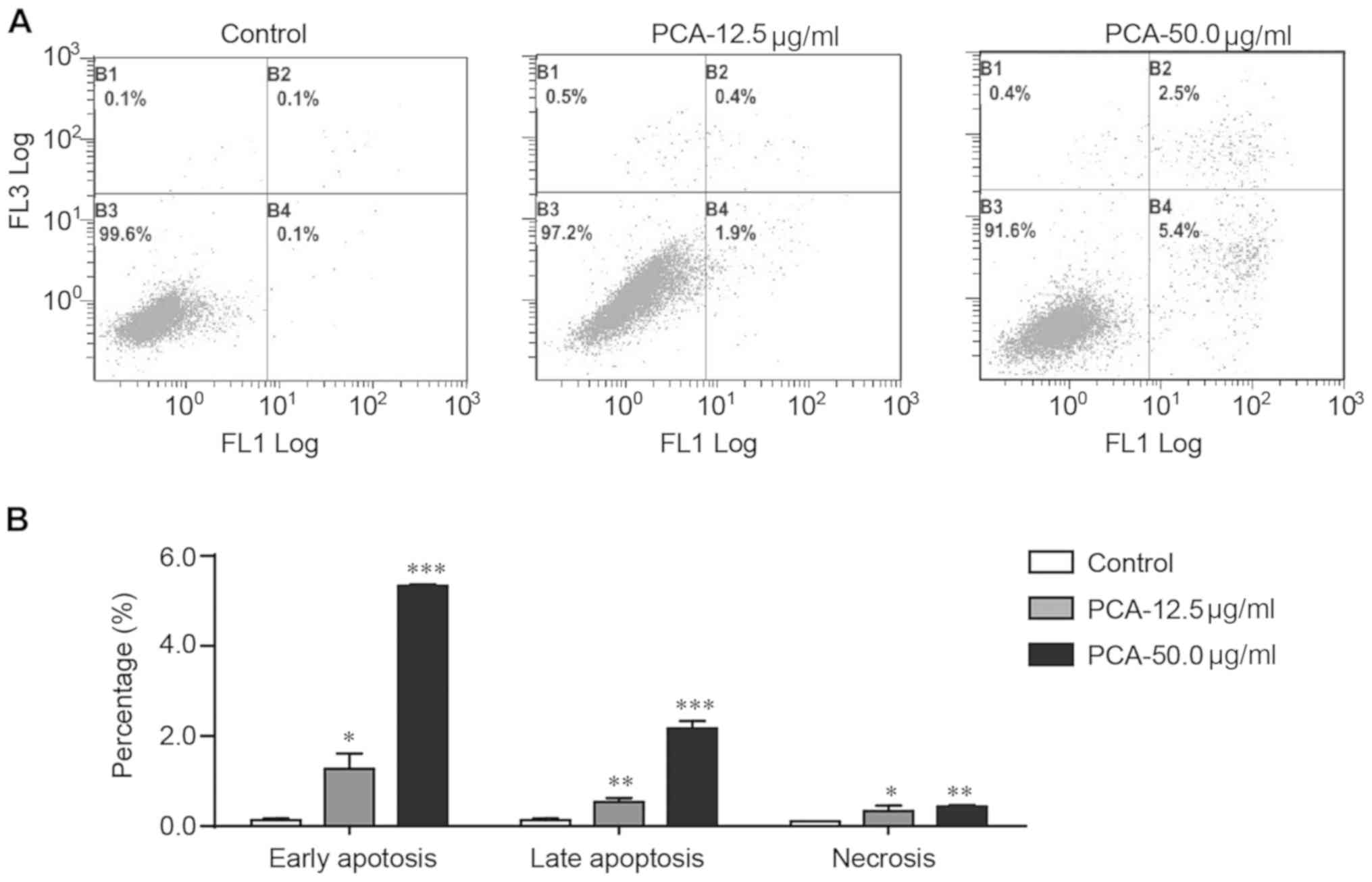
Flow cytometry with Annexin V-FITC and PI staining
was used to examine whether apoptosis was involved in the
PCA-mediated growth inhibition of B16-F10 cells. PCA significantly
increased the number of early apoptotic, late apoptotic and
necrotic cells in a dose-dependent manner (Fig. 3). Treatment with PCA significantly
increased early and late apoptosis. Treatment with 50 µg/ml PCA
increased the percentage of early and late apoptotic cells by 54-
and 25-fold, respectively.
PCA affected the transcriptome of
B16-F10 cells
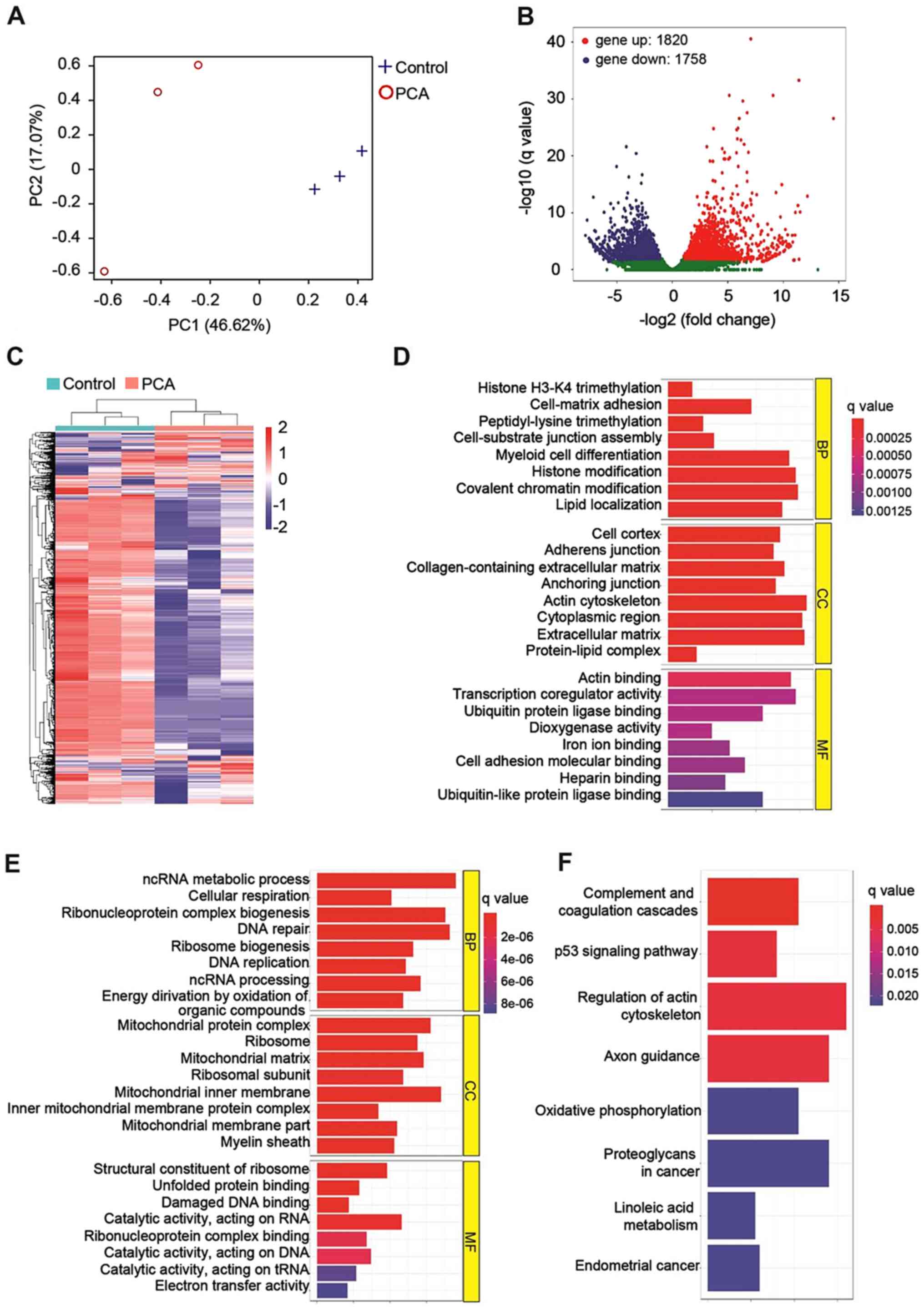
RNA sequencing was performed to monitor the gene
expression profile of PCA-treated B16-F10 cells. Principal
component analysis (Fig. 4A)
showed that PCA-treated cells were completely separated from the
vehicle-treated control cells (P<0.01), indicating that PCA
affected the whole gene expression of B16-F10 cells. In total,
3,578 differentially expressed genes were identified (Fig. 4B). Among them, 1,820 genes were
upregulated, and 1,758 genes were downregulated. The differentially
expressed genes between the PCA-treated and the control group were
clustered (Fig. 4C) and further
classified into biological process (BP), cellular components (CC)
and molecular function (MF) according to standard GO terms
(Fig. 4D and E). In the BP
category, the genes that were upregulated by PCA treatment were
found to be involved in cellular processes, such as histone
trimethylation, peptidyl-lysine trimethylation and histone
modification (Fig. 4D), while
downregulated genes were found to be associated with DNA repair and
DNA replication (Fig. 4E). In the
CC category, PCA primarily affected the expression levels of genes
located in the extracellular region or on the cytoskeleton compared
with the control cells (Fig. 4D),
while PCA decreased the expression of mitochondrial components
(Fig. 4E). Under the MF category,
PCA treatment mainly led to the upregulation of genes associated
with receptor binding (Fig. 4D)
and the downregulation of genes involved in DNA damage and protein
binding (Fig. 4E).
Analysis using the KEGG database indicated that PCA
may inhibit the growth of B16-F10 cells via the following pathways:
i) ‘Complement and coagulation cascades’; ii) ‘p53 signaling
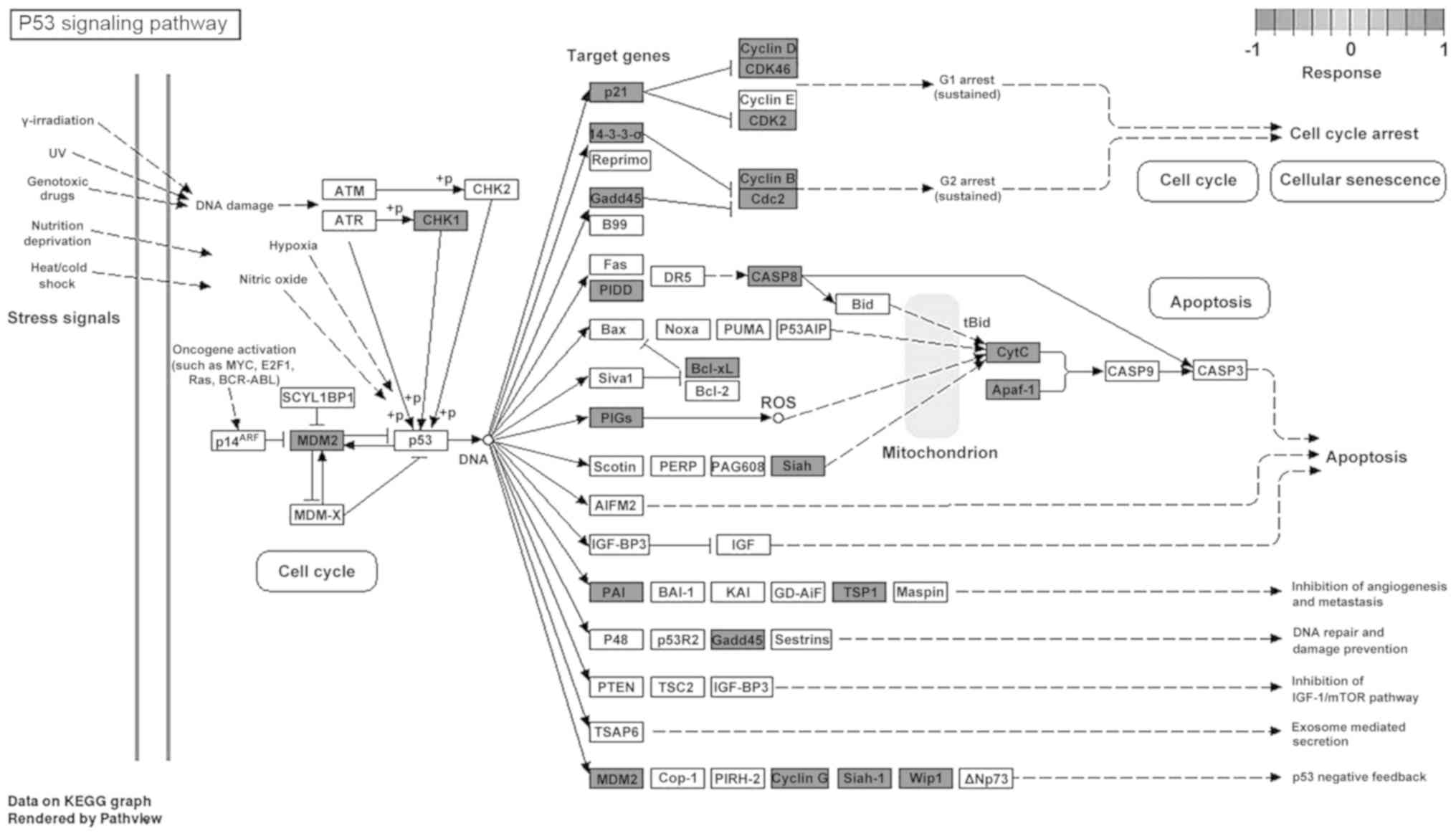
pathway’; and iii) ‘regulation of actin cytoskeleton’ (Fig. 4F). Detailed analysis of the p53
signaling pathway showed that PCA could upregulate multiple genes
involved in this pathway (Fig. 5)
(19). Furthermore, the expression
of p53-related genes in B16-F10 cells was investigated using
RT-qPCR (Fig. 6). The present
results showed that PCA significantly increased the expression
level of p21 and PIDD (Fig. 6A),
but significantly downregulated the expression of cyclin D, Gadd45,
p53, Bcl-2, Cdc-2, cyclin B and PIGs after 48 h exposure compared
with the control (Fig. 6B).
However, no detectable changes in cyclin E and Bax expression were
observed after PCA treatment (Fig.
6A). Therefore these findings suggested that PCA could regulate
the p21/cyclin D/CDK4/6 pathways, which may induce G1 phase
arrest.
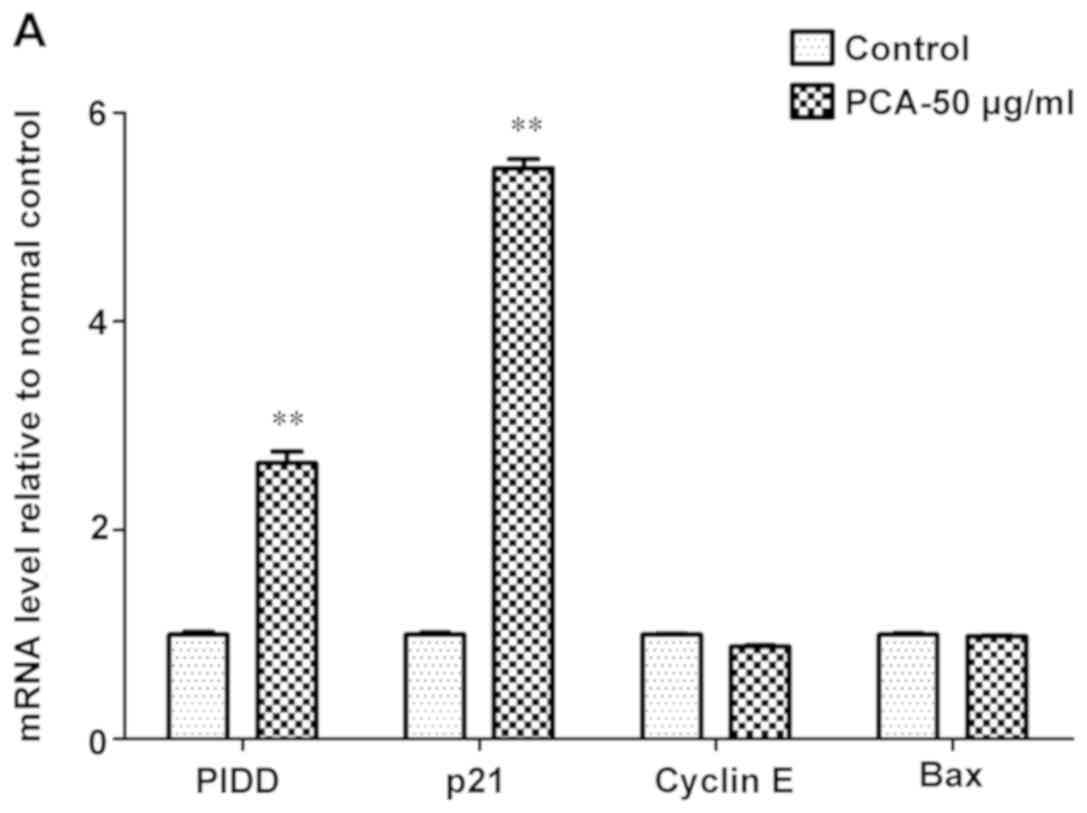 | Figure 6.mRNA expression analysis of
p53-related genes in B16-F10 cells treated with and without 50
µg/ml PCA for 48 h. mRNA expression level of the following genes:
(A) PIDD, p21, cyclin E and Bax; and (B) cyclin D, Gadd45, p53,
Bcl-2, Cdc-2, cyclin B and PIGs. Density values were normalized to
GAPDH. Data are presented as the mean ± SD from three experiments
per group. **P<0.01 vs. control. PCA, protocatechualdehyde;
Gadd45, growth arrest and DNA damage-inducible 45; PIDD,
p53-induced death domain-containing protein 1; Cdc-2, cell division
cycle-associated protein 2; PIGs, phosphatidylinositol glycan
anchor biosynthesis class S. |
Discussion
Natural products offer a source for the
identification of novel drugs for the treatment of various
diseases, including cancer. In total, >50% of pharmaceutical
drugs can be traced back to natural compounds or their analogs
(20). Previously, chemoprevention
via the dietary consumption of natural products has gained
considerable attention due to their low toxicity, patient
acceptance and effectiveness (21). Polyphenols are a large and complex
family of organic compounds that are present in various plants and
fungi (22). Polyphenols could be
used as potential anticancer agents due to their antioxidant,
anti-inflammatory, antiproliferative and anticarcinogenic
properties (23). In the present
study PCA, a natural polyphenol compound, was isolated from the
medical mushroom P. gilvus. A previous study showed that PCA
can exert antiproliferative effects in several cancer cell lines
(11). Our previous study
demonstrated that PCA inhibits the viability of human colorectal
cancer cells by inducing S phase arrest and promoting the
mitochondrial apoptotic pathway (11). However, to the best of our
knowledge, whether PCA could impact melanoma development has not
been previously reported so the present study is the first to
report that PCA could exert a growth inhibitory effect on murine
melanoma cells by inducing G0/G1 phase arrest and promoting
apoptotic cell death.
The cell cycle is strictly regulated by a complex
network of events that promote or stop the progression of cells
from one phase to the other (24).
p21 plays a crucial role in cell cycle progression and cancer
(24); it is able to mediate p53
activity and promote cell cycle arrest by inhibiting the activity
and formation of cyclin/CDK complexes (24). At high levels, p21 inhibits the
function of CDKs (25), thus
inhibiting the cyclin D/CDK4/6 complex, resulting in G1/S arrest
(26). In the present study,
treatment with PCA significantly increased the number of cells in
the G0/G1 phase and decreased the number of cells in the S phase,
which arrested the cell cycle of melanoma cells at the G0/G1 phase.
Our previous results showed that PCA inhibited the proliferation of
HT-29 cells and induced cell cycle arrest in the S phase by
downregulating the expression of cyclin A and D (11). In the present study, PCA
significantly increased p21 mRNA expression level and downregulated
Cyclin D expression in B16-F10 cells, suggesting that PCA inhibited
B16-F10 cell proliferation and blocked entry to the S phase by
regulating the p21/Cylin D/CDK4/6 pathway.
Apoptosis is a main mechanism by which anticancer
drugs can suppress tumor growth (27). p53 plays a key role in promoting
cell cycle arrest or apoptosis in response to acute DNA damage
(28). This p53-induced cell cycle
arrest allows the damaged DNA to be repaired, thereby preventing
the proliferation of cancer cells (28). Anticancer agents can trigger
apoptosis, by either directly inducing DNA damage or indirectly
inducing secondary stress-responsive signaling pathways, which
leads to the activation of the intrinsic mitochondrial apoptotic
pathway (29). The Bcl-2 family
consists of pro- and anti-apoptotic structurally related proteins
that are principal regulators of the mitochondrial apoptotic
pathway (29). Anti-apoptotic
protein Bcl-2 acts as a gatekeeper that inhibits the release of
cytochrome c from the mitochondria to the cytoplasm
(29). In the present study, flow
cytometry analysis showed that PCA significantly promoted
apoptosis, and significantly increased the number of early
apoptotic cells (54-fold) and late apoptotic cells (25-fold).
Whole-transcriptome analysis further indicated that PCA decreased
the expression levels of the genes participating in DNA repair and
replication. Furthermore, RT-qPCR results showed that PCA markedly
decreased the expression of Bcl-2, indicating that it may trigger
apoptosis via the mitochondrion-dependent pathway. The present
findings suggested that PCA stimulated cell cycle arrest and
apoptosis via upregulation of the p53 signaling pathway.
In conclusion, the present study suggested that the
P. gilvus-derived polyphenol compound PCA could effectively
inhibit the growth and proliferation of melanoma cells by inducing
G0/G1 phase arrest and promoting apoptotic cell death. These
cellular changes may be mediated by the activation of the p53
signaling pathway.
Acknowledgements
Not applicable.
Funding
This work was supported financially by The Science
and Technology Department of Zhejiang Province (grant nos.
2018C02003 and LGN18C170005), Zhejiang Medical and Health Science
and Technology Project (grant no. 2018KY250) and Key Laboratory of
Creative Agriculture, Ministry of Agriculture and Rural
Affairs.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ and YL contributed to the conception of the
study. SZ, QJ and JZ performed the experiments. YL contributed
significantly to the data analysis and manuscript preparation. TY
contributed to the data analysis. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Savoia P, Fava P, Casoni F and Cremona O:
Targeting the ERK signaling pathway in melanoma. Int J Mol Sci.
20:E14832019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mishra H, Mishra PK, Ekielski A, Jaggi M,
Iqbal Z and Talegaonkar S: Melanoma treatment: From conventional to
nanotechnology. J Cancer Res Clin Oncol. 144:2283–2302. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weinstock MA, Lott JP, Wang Q, Titus LJ,
Onega T, Nelson HD, Pearson L, Piepkorn M, Barnhill RL, Elmore JG
and Tosteson ANA: Skin biopsy utilization and melanoma incidence
among Medicare beneficiaries. Br J Dermatol. 176:949–954. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barbieri A, Quagliariello V, Del Vecchio
V, Falco M, Luciano A, Amruthraj NJ, Nasti G, Ottaiano A, Berretta
M, Iaffaioli RV and Arra C: Anticancer and anti-inflammatory
properties of ganoderma lucidum extract effects on melanoma and
triple-negative breast cancer treatment. Nutrients. 9:E2102017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Albuquerque KRS, Pacheco NM, Del Rosario
Loyo Casao T, de Melo F, Novaes RD and Goncalves RV: Applicability
of plant extracts in preclinical studies of melanoma: A systematic
review. Mediators Inflamm. 2018:67979242018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Momtaz S, Niaz K, Maqbool F, Abdollahi M,
Rastrelli L and Nabavi SM: STAT3 targeting by polyphenols: Novel
therapeutic strategy for melanoma. Biofactors. 43:347–370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen X, Chang L, Qu Y, Liang J, Jin W and
Xia X: Tea polyphenols inhibit the proliferation, migration, and
invasion of melanoma cells through the down-regulation of TLR4. Int
J Immunopathol Pharmacol. 32:3946320177395312018.PubMed/NCBI
|
|
8
|
Gao JW, Yamane T, Maita H, Ishikawa S,
Iguchi-Ariga SM, Pu XP and Ariga H: DJ-1-Mediated protective effect
of protocatechuic aldehyde against oxidative stress in SH-SY5Y
cells. J Pharmacol Sci. 115:36–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Ji Y, Kang Z, Lv C and Jiang W:
Protocatechuic aldehyde ameliorates experimental pulmonary fibrosis
by modulating HMGB1/RAGE pathway. Toxicol Appl Pharmacol.
283:50–56. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim KJ, Kim MA and Jung JH: Antitumor and
antioxidant activity of protocatechualdehyde produced from
Streptomyces lincolnensis M-20. Arch Pharm Res.
31:1572–1577. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhong S, Li YG, Ji DF, Lin TB and Lv ZQ:
Protocatechualdehyde induces S-phase arrest and apoptosis by
stimulating the p27(KIP1)-cyclin A/D1-CDK2 and mitochondrial
apoptotic pathways in HT-29 cells. Molecules. 21:E9342016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee BH, Yoon SH, Kim YS, Kim SK, Moon BJ
and Bae YS: Apoptotic cell death through inhibition of protein
kinase CKII activity by 3,4-dihydroxybenzaldehyde purified from
Xanthium strumarium. Nat Prod Res. 22:1441–1450. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11:R142010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo W and Brouwer C: Pathview: An
R/Bioconductor package for pathway-based data integration and
visualization. Bioinformatics. 29:1830–1831. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harvey AL, Edrada-Ebel R and Quinn RJ: The
re-emergence of natural products for drug discovery in the genomics
era. Nat Rev Drug Discov. 14:111–129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
DiMarco-Crook C and Xiao H: Diet-based
strategies for cancer chemoprevention: The role of combination
regimens using dietary bioactive components. Annu Rev Food Sci
Technol. 6:505–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bahramsoltani R, Ebrahimi F, Farzaei MH,
Baratpourmoghaddam A, Ahmadi P, Rostamiasrabadi P, Rasouli
Amirabadi AH and Rahimi R: Dietary polyphenols for atherosclerosis:
A comprehensive review and future perspectives. Crit Rev Food Sci
Nutr. 59:114–132. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rothwell JA, Knaze V and Zamora-Ros R:
Polyphenols: Dietary assessment and role in the prevention of
cancers. Curr Opin Clin Nutr Metab Care. 20:512–521.
2017.PubMed/NCBI
|
|
24
|
Moussa RS, Park KC, Kovacevic Z and
Richardson DR: Ironing out the role of the cyclin-dependent kinase
inhibitor, p21 in cancer: Novel iron chelating agents to target p21
expression and activity. Free Radic Biol Med. 133:276–294. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Harper JW, Adami GR, Wei N, Keyomarsi K
and Elledge SJ: The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ogryzko VV, Wong P and Howard BH: WAF1
retards S-phase progression primarily by inhibition of
cyclin-dependent kinases. Mol Cell Biol. 17:4877–4882. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Newton K, Wickliffe KE, Dugger DL,
Maltzman A, Roose-Girma M, Dohse M, Kőműves L, Webster JD and Dixit
VM: Cleavage of RIPK1 by caspase-8 is crucial for limiting
apoptosis and necroptosis. Nature. 574:428–431. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garcia PB and Attardi LD: Illuminating p53
function in cancer with genetically engineered mouse models. Semin
Cell Dev Biol. 27:74–85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hennessy EJ: Selective inhibitors of Bcl-2
and Bcl-xL: Balancing antitumor activity with on-target toxicity.
Bioorg Med Chem Lett. 26:2105–2114. 2016. View Article : Google Scholar : PubMed/NCBI
|