Introduction
Bone marrow-derived mesenchymal stem cells (BMSCs)
can differentiate into Schwann cells (key factors for nerve
regeneration) under different conditions, such as during sciatic
nerve transection and nerve regeneration chamber to promote the
regeneration of damaged nerves (1–4).
BMSCs transplanted in vivo to damaged areas can
differentiate into the corresponding Schwann cells (1,2,4–6),
which suggested that following injury, multiple factors may be
produced in the local in vivo microenvironment that can
induce BMSCs to differentiate into Schwann cells to repair the
injured tissue; however, the factors involved in this process are
unclear.
Exosomes are small cell-derived vesicles that
commonly are found in human body fluids, including blood, urine and
tissue fluids (7,8). Exosomes serve different functions in
the body, including participating in intercellular signaling
transduction, cell waste secretion and cell fusion (8). Following nerve injury, Wallerian
degeneration and Schwann cell proliferation occurs at the distal
end of the nerve, forming a regenerative Büngner zone that promotes
and guides proximal nerve regeneration (9). During this process, exosomes secreted
by Schwann cells promote the regeneration of peripheral nerves and
guide the direction of growth of regenerating axons (10,11);
thus, there are large numbers of exosomes in the microenvironment
following injury, which serve an important role in the recovery of
nerve damage. In addition, previous studies have demonstrated that
BMSCs participate in the myelination of spinal axons after being
cultured with rat dorsal root ganglion cells, and Schwann cell
co-culture can induce BMSCs to differentiate into Schwann cells
(12–14). These studies suggest that during
nerve regeneration within the microenvironment, in which Schwann
cells are present, certain factors can induce BMSCs to
differentiate into Schwann cells and participate in axonal
regeneration. Due to exosomes being involved in the repair of nerve
damage, it was hypothesized that exosomes may also have an
important role in stem cell differentiation following injury. In
the present study, the effect of Schwann cell-derived exosomes on
the differentiation of BMSCs into Schwann cells was
investigated.
Materials and methods
Animal studies
All animal procedures were conducted in accordance
with The Guidelines for Ethical Conduct in the Care and Use of
Animals of the Chinese National Health and Medical Research
Council, and were approved by the Animal Experimentation Ethics
Committee of Capital Medical College (Beijing, China; protocol no.
AEEI-2016-135).
BMSC culture and differentiation into
Schwann cells
Rat BMSCs were obtained from the bone marrow of one
eight-week-old male Wistar rat, which weighed between 230–270 g, as
previously described (3,15). The rat was housed in a
pathogen-free climate-controlled facility with food and water
available ad libitum. BMSCs were cultured in α-MEM growth
media (HyClone; GE Healthcare Life Sciences) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.). The third passage of
cells was used for subsequent experiments. Rat Schwann cells
(RSC96) and fibroblasts were purchased from Shanghai Fuxiang
Company (www.xiangbio.com/timemodel/product/2010-03-08/8421285.html).
BMSCs were subsequently divided into three experimental groups: i)
BMSCs induced to differentiate into Schwann cells using the
Dezawa's induction method, as previously described (3); ii) BMSCs induced to differentiate
into Schwann cells by Schwann cell-derived exosomes, in which BMSCs
were cultured and passaged for 7 days prior to the addition of 200
µg Schwann cell-derived exosomes to the media (the media was
changed every 3 days, with the exosomes being replenished each
time); iii) BMSCs induced to differentiate into Schwann cells by
fibroblast-derived exosomes, in which BMSCs were cultured and
passaged for 7 days prior to the addition of 200 µg
fibroblast-derived exosomes to the media (the media was changed
every 3 days, with the exosomes being replenished each time). RSC96
cells were used as the positive control group, and untreated BMSCs
were used as the negative control group. A preliminary study was
conducted to predetermine the dosage of Schwann cell-derived
exosomes required to induce BMSCs; out of 50, 100 or 200 µg, 200 µg
in 10 ml culture media induced fusiform shape change, in BMSCs.
Therefore, the present study used 200 µg exosomes for the induction
of BMSCs (Fig. S1).
Exosome extraction, identification and
quantification
Exosomes were extracted from fibroblasts (fb exo)
and RSC96 (RSC96 exo) cells using the ExoQuick-TC kit (System
Biosciences, LLC). The culture media containing the BMSCs was
centrifuged (3,000 × g; 15 min; 4°C) to remove apoptotic cells and
cell debris. A total of 3.3 ml exosome precipitation solution was
added to 10 ml culture supernatant and incubated at 4°C overnight.
The mixture was centrifuged (10,000 × g; 30 min; 4°C) and the
supernatant was discarded. The isolated exosomes were re-suspended
in PBS and the protein concentration was quantified using a
bicinchoninic assay kit. Briefly, the standards were diluted to
different concentrations (0, 0.2, 0.4, 0.6, 0.8 or 1.0 µg/µl). The
samples and standards were loaded separately and incubated with the
working solution for 30 min at 37°C in the dark. The optical
density values were measured using a microplate reader and the
sample concentration was determined according to the standard curve
generated. The amount of culture medium and exosomes required was
calculated to a final concentration of 20 µg/ml and used in the
subsequent experiments. CD63, CD81 and calnexin were detected by
western blotting to confirm the isolation of exosomes.
Transmission electron microscopy (TEM)
of exosomes
A total volume of 50 µl exosomes were plated onto
red wax. The polyvinyl acetate/carbon-coated copper mesh was placed
in the droplets and allowed to stand at room temperature for 20
min. The copper mesh was fixed with 2% paraformaldehyde for 2 min
at room temperature, washed three times with double-distilled
H2O, and counterstained with 2% phosphotungstic acid
(XiYa Reagent; http://m.xiyashiji.com/product/info/id/4724.html)
for 1 min at room temperature. Filter paper was used to remove
excess liquid from the copper mesh, which was subsequently dried
overnight at room temperature. The extracted exosomes were observed
using TEM (magnification, ×25,000).
Cellular morphology observation
After 2, 4 and 7 days of cell differentiation,
cellular morphology was observed using a CX41 light microscope
(magnification, ×200; Olympus Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the BMSCs using
TRIzol® reagent (cat. no. R0016; Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Total RNA
was reverse transcribed into cDNA using the TransScript
First-Strand cDNA Synthesis kit (Beijing Transgen Biotech Co.,
Ltd.). The RT reaction conditions were as follows: 25°C for 10 min,
42°C for 30 min, and 85°C for 5 min. qPCR was subsequently
performed using the SYBR® Green Realtime PCR Master mix
(Toyobo Life Science; QPK-201) and the ABI PRISM™ 7500 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The following
primer pairs for each marker gene are presented in Table I. The following thermocycling
conditions were used for the qPCR: Initial denaturation at 50°C for
2 min, 94°C for 2 min, 40 cycles of 94°C for 5 sec, 60°C for 30 sec
of denaturation, annealing and elongation, 72°C for 5 min of final
extension Expression levels were quantified using the
2−ΔΔCq method (16).
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| S100 | F:
GGTTGCCCTCATTGATGTCTTCC |
|
| R:
ACCACTTCCTGCTCTTTGATTTCC |
| Sox10 | F:
GGGCAAGGTCAAGAAGGAACAG |
|
| R:
ACCAGCGTCCAGTCGTAGC |
| EGR2 | F:
CTACCCCCTACAATCCGCAC |
|
| R:
GATTCGGATGTGCCTGGTCA |
| GFAP | F:
CAAGAAACAGAAGAGTGGTATCGGT |
|
| R:
ACTCAAGGTCGCAGGTCAAGG |
| NGFR | F:
CCTGCTGCTGCTGCTGATTC |
|
| R:
GTTCACACACGGTCTGGTTGG |
| GAPDH | F:
TCAAGAAGGTGGTGAAGCAGG |
|
| R:
TCAAGAAGGTGGTGAAGCAGG |
Western blotting
Total protein was lysed from cells using lysis
buffer (Beijing Solarbio Science & Technology Co., Ltd.) and
the lysate was collected using a cell scraper. The lysate was
centrifuged (13,523 × g; 5 min; 4°C) and the supernatant was
collected and transferred to a 0.5 ml centrifuge tube. Protein
samples were mixed with 5X loading buffer, incubated at 95°C for 5
min, rapidly cooled on ice and stored at −80°C until required for
further experimentation. Total protein was quantified using a
bicinchoninic acid assay kit and 40 µg protein samples and markers
(cat. no. SM1811; Fermentas; Thermo Fisher Scientific, Inc.) were
separated via 12% SDS-PAGE. The separated proteins were
subsequently transferred onto a nitrocellulose membrane (Merck
KGaA) and blocked for 2 h at room temperature with 5% non-fat milk
diluted in TBS-Tween-20 (TBST). The membranes were incubated with
the following primary antibodies overnight at 4°C: Anti-Sox10
(1:1,000; cat. no. DF8009; Affinity Biosciences), anti-early growth
response 2 (EGR2; 1:500; cat. no. AF0480; Affinity Biosciences),
anti-S100 (1:1,000; cat. no. AF0251; Affinity Biosciences),
anti-glial fibrillary acidic protein (GFAP; 1:1,000; cat. no.
AF6166; Affinity Biosciences), anti-low-affinity nerve growth
factor receptor (NGFR; 1:2,000; cat. no. OM267104; Omnimabs),
anti-GADPH (1:1,000; cat. no. AB-P-R 001; Hangzhou Goodhere
Biotechnology Co., Ltd.), anti-CD63 (1:1,000; cat. no. DF2305;
Affinity Biosciences), anti-CD81 (1:1,000; cat. no. DF2306;
Affinity Biosciences) and anti-Calnexin (1:1,000; cat. no. AF5362;
Affinity Biosciences). Membranes were washed three times (10 min
each) with 0.1% TBST at room temperature. Following the primary
antibody incubation, membranes were incubated with a horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000;
cat. no. BA1054; Boster Biological Technology) for 2 h at 37°C on a
shaker. The membrane was thoroughly washed three times (10 min
each) with 0.05% TBST. Protein bands were visualized using an ECL
kit (CWBio), according to the manufacturer's protocol. Images of
the membrane were captured using a chemiluminescence imager
(ChemiDoc™ MP Imaging System; Bio-Rad Laboratories).
Immunofluorescence assay
BMSCs (2×104 cells/well) were seeded into
24-well plates; cells were divided into three groups, cultured with
different media as described above, and collected after
differentiation for 7 days. Following the removal of the culture
media, cells were washed twice with PBS and fixed with 4%
paraformaldehyde for 15 min at room temperature. Cells were washed
three times with PBS and blocked with 1% BSA (CWBio) at 37°C for 30
min and subsequently incubated with the following primary
antibodies overnight at 4°C: Anti-Sox10 (1:250), anti-EGR2 (1:100),
anti-NGFR (1:50), anti-S100 (1:100) and anti-GFAP (1:100). Cells
were washed three times with PBS for 5 min each and incubated with
goat anti-rabbit IgG H&L (Alexa Fluor® 488; 1:500;
cat. no. ab150077; Abcam) at 37°C for 30 min. The cells were washed
three times with PBS for 5 min each and incubated at room
temperature with DAPI for 5 min in the dark. The plate was mounted
with the Fluoromount-G® blocking solution containing
anti-fluorescence quencher (cat. no. 0100-01; SouthernBiotech).
Cells were visualized using an Olympus BX53 fluorescence microscope
(magnification, ×100; Olympus Corporation). To calculate the
positive rate: 5 fields were randomly selected in each plate, all
cells were counted and the expression rate of positive cells was
calculated.
Statistical analysis
Statistical analysis was performed using SPSS
version 17.2 (SPSS, Inc.) software. The data was presented as the
mean ± SD; n=3. RT-qPCR, western blotting and immunofluorescence
results were analyzed using one-way ANOVA followed by Newman-Keuls
Multiple Comparison test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Exosome extraction, isolation and
identification
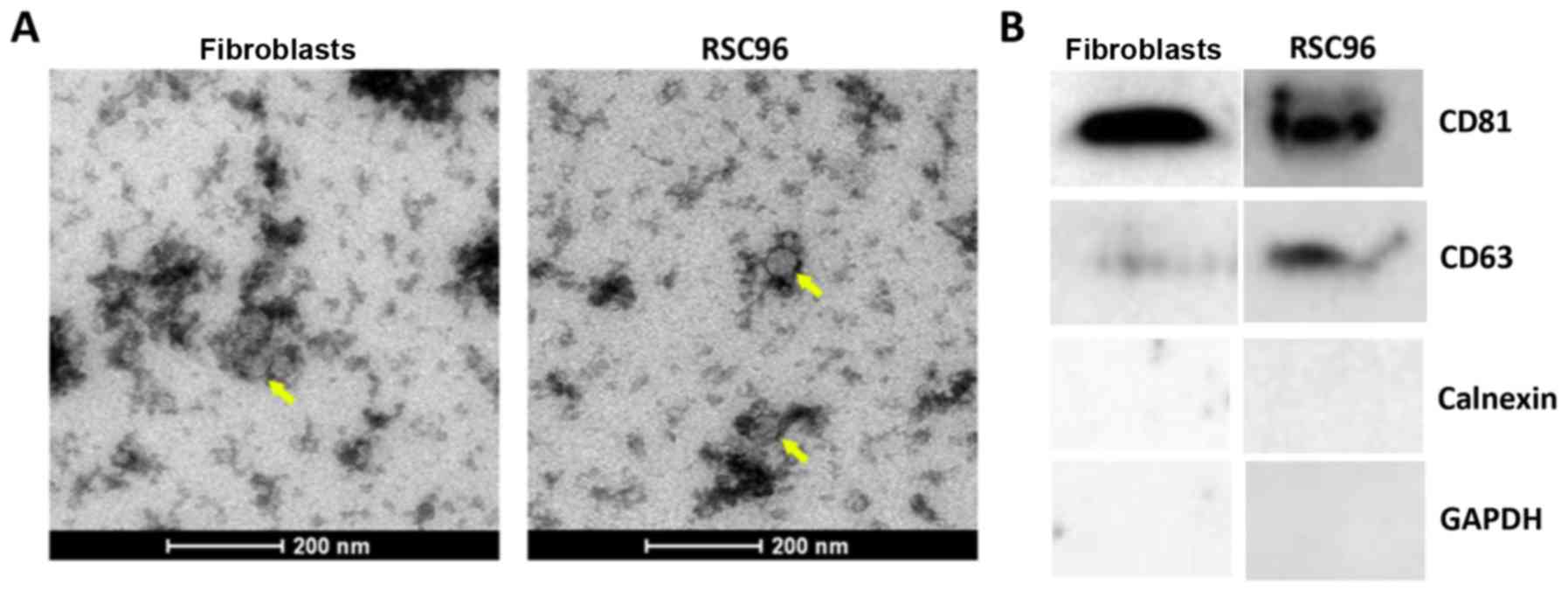
Schwann cell- and fibroblast-derived exosomes were
visualized using TEM. Observed exosomes presented as irregular
discoid vesicles with a diameter of ~50–80 nm (Fig. 1A). The concentration of protein
extracted from the exosomes of RSC96 cells and fibroblasts was 1.22
and 1.10 µg/µl, respectively. The exosomal marker proteins CD81 and
CD63 were detected by western blotting in both RSC96 cells and
fibroblasts, whereas the endoplasmic reticulum marker protein
Calnexin was not detected (Fig.
1B). Overall, this indicated that exosomes were successfully
extracted from RSC96 cells and fibroblasts.
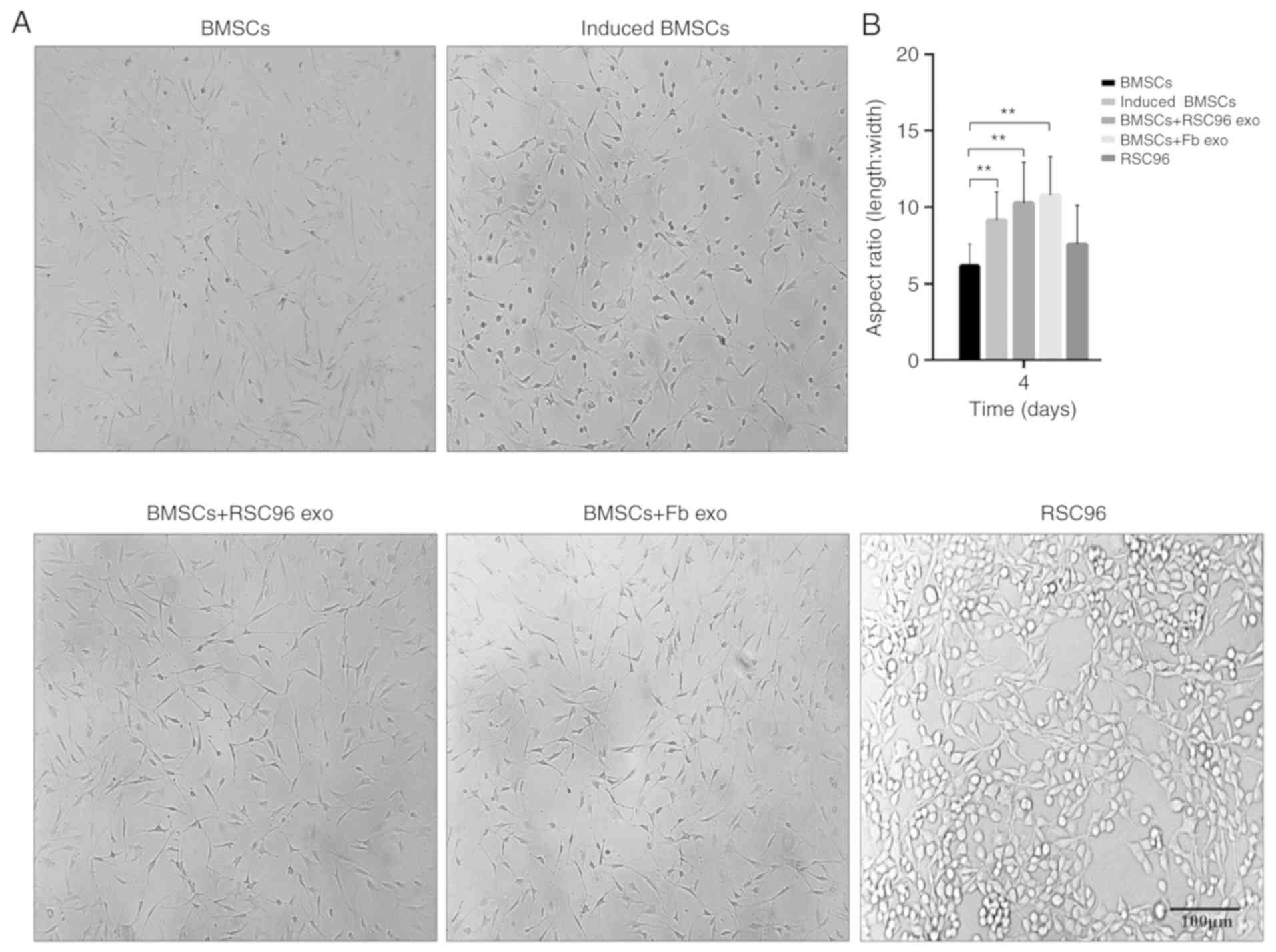
Cell morphology of BMSCs
The differentiation of BMSCs was induced according
to the experimental design aforementioned, and the morphological
features of BMSCs were evaluated 4 days post-induction. The
morphology of BMSCs gradually changed from fibroblast-like
morphology to a more fusiform shape, and the length/width ratio
significantly increased in induced BMSCs compared with control
BCSCs (Fig. 2A and B). Following
the addition of Schwann cell- or fibroblast-derived exosomes to the
cultured BMSCs, the length/width ratio of the BMSCs in these two
groups (BMSCs + RSC96 exo and BMSCs + Fb exo, respectively)
significantly increased compared with control BMSCs (Fig. 2A and B); however, there was no
significant difference in cell morphology between BMSCs + RSC96 exo
and BMSCs + Fb exo (Fig. 2B).
mRNA and protein expression levels of
Schwann cell markers in BMSCs
mRNA and protein expression levels of S100, GFAP,
Sox10, NGFR and EGR2 in induced BMSCs were significantly increased
compared with the control BMSCs (Fig.
3A-G). In addition, the expression levels of S100, GFAP, Sox10,
NGFR and EGR2 in the BMSCs + RSC96 exo group were significantly
increased compared with control BMSCs and BMSCs + Fb exo groups
(Fig. 3A-G); however, expression
levels were not significantly different compared with induced
BMSCs. The expression levels of Schwann cell markers in the BMSCs +
Fb exo group were not significantly different compared with control
BMSCs (Fig. 3A-G).
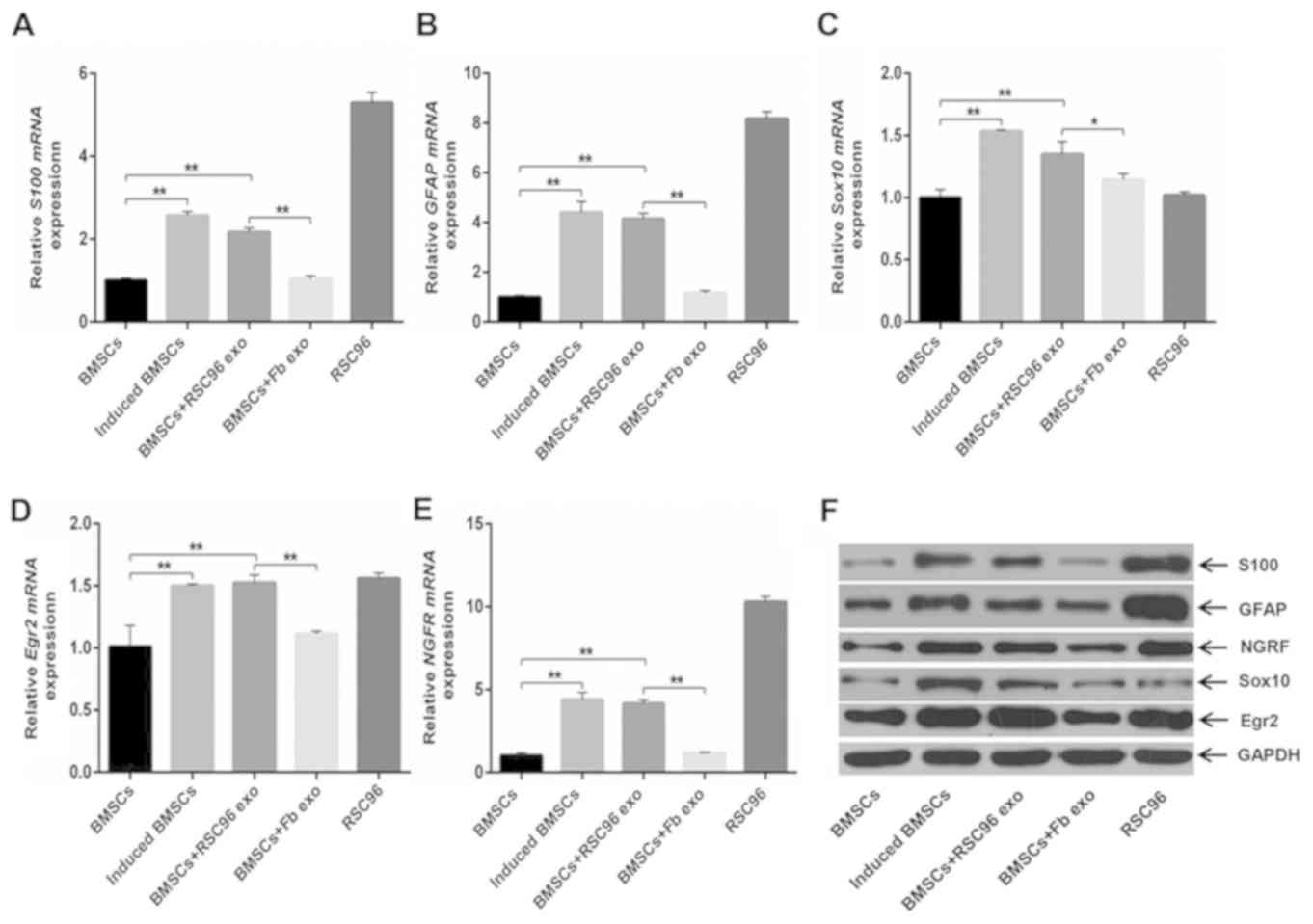 | Figure 3.mRNA and protein expression levels of
Schwann cell markers in differentially-induced BMSCs. (A-E) mRNA
expression levels of (A) S100, (B) GFAP, (C) Sox10, (D) EGR2 and
(E) NGFR in control BMSCs, induced BMSCs, BMSCs + RSC96 exo, BMSCs
+ Fb exo or RSC96 cells. (F and G) Representative immunoblot of
protein expression levels of S100, GFAP, NGRF, Sox10, EGR2 and
GAPDH in control BMSCs, induced BMSCs, BMSCs + RSC96 exo, BMSCs +
Fb exo or RSC96 cells. Data are presented as mean ± SD; *P<0.05,
**P<0.01 and ***P<0.001. BMSCs, bone marrow mesenchymal stem
cells; EGR2, early growth response 2; Fb exo, fibroblast-derived
exosomes; GFAP, glial fibrillary acidic protein; induced BMSCs,
Dezawa's induction method; NGFR, low-affinity nerve growth factor
receptor; RSC96 exo, Schwann cell-derived exosomes. |
Results from immunofluorescence investigations
demonstrated that the positive expression rates of S100, GFAP,
NGFR, Sox10 and EGR2 were significantly increased in induced BMSCs
compared with control BMSCs (Fig.
4); the expression levels of these Schwann cell markers were
also significantly increased in the BMSCs + RSC96 exo group
compared to the control BMSCs. In addition, the protein expression
levels of all Schwann cell markers were significantly increased in
the BMSCs + RSC96 exo group compared with the BMSCs + Fb exo group,
which did not display significantly altered expression levels
compared with control BMSCs (Fig.
4). This was consistent with the RT-qPCR and western blotting
results (Fig. 3). These results
suggested that Schwann cell-derived exosomes may promote the
differentiation of BMSCs into Schwann cells because the expression
levels of Schwann cell marker proteins and transcription factors
increased following the induction, whereas fibroblast-derived
exosomes were not observed to promote the same function.
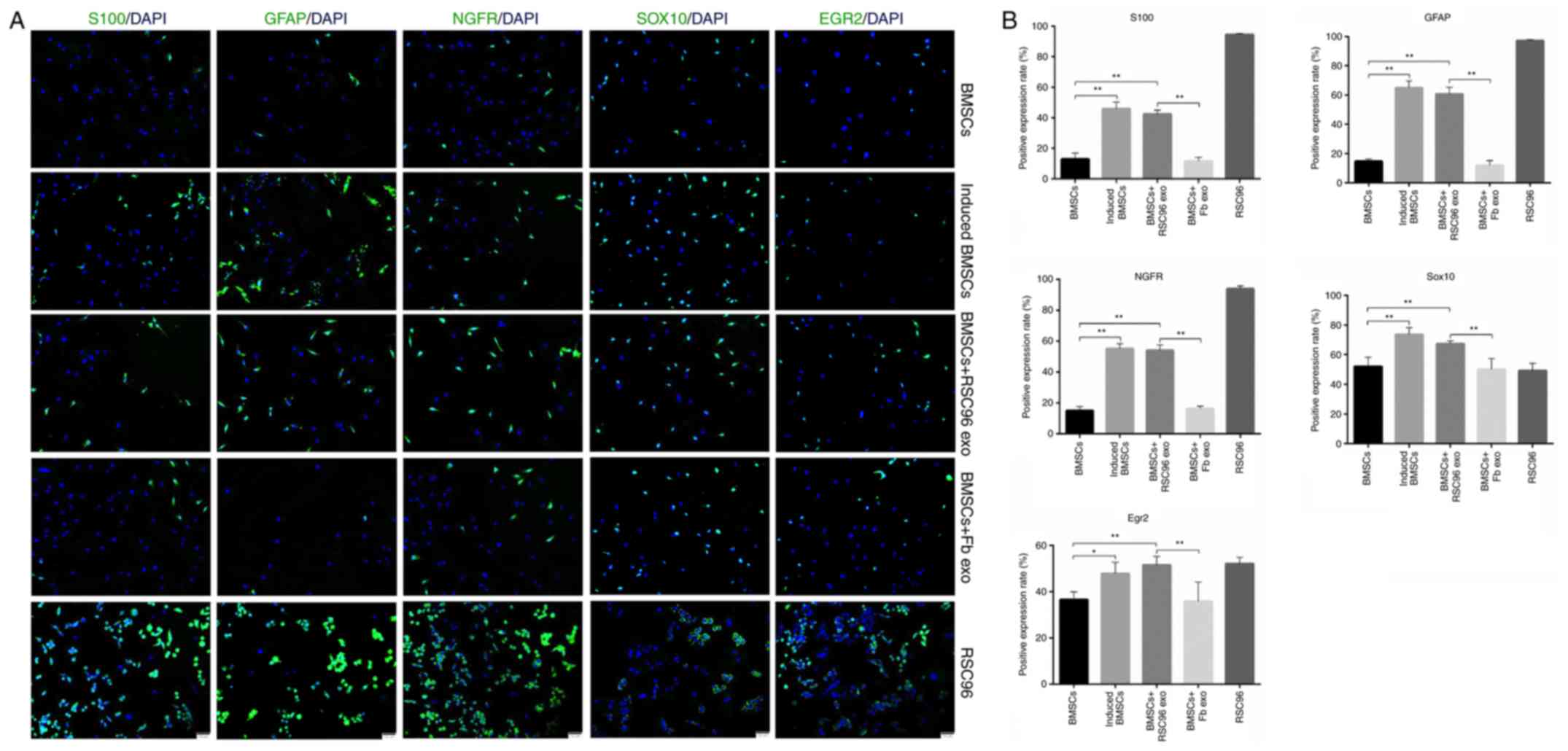 | Figure 4.Protein expression of Schwann cell
markers in differentially-induced BMSCs. (A) Representative
immunofluorescence micrographs of Schwann cell-associated marker
expressions (S100, GFAP, NGRF, Sox10 and EGR2) in control BMSCs,
induced BMSCs, BMSCs + RSC96 exo, BMSCs + Fb exo or RSC96 cells
(magnification, ×100). Scale bar, 50 µm. (B) Quantitative analysis
of the expression of Schwann cell-associated markers from (A) Data
are presented as mean ± SD; *P<0.05 and **P<0.01. BMSCs, bone
marrow mesenchymal stem cells; EGR2, early growth response 2; Fb
exo, fibroblast-derived exosomes; GFAP, glial fibrillary acidic
protein; induced BMSCs, Dezawa's induction method; NGFR,
low-affinity nerve growth factor receptor; RSC96 exo, Schwann
cell-derived exosomes. |
Discussion
Results from the present study demonstrated that
Schwann cell-derived exosomes may induce BMSCs to differentiate
into Schwann cells. During induction, BMSCs express Schwann cell
surface markers at multiple developmental stages; these include
S100, GFAP, NGRF, Sox10 and EGR2 (17–19).
Sox10 and EGR2 are transcription factors that serve crucial roles
in Schwann cell differentiation (20,21);
Sox10 is expressed in neural crest cells and is required for the
Schwann cell phenotype and progression beyond the immature stage
(20); EGR2 is closely related to
the differentiation of myelinating Schwann cells (21). Exosomes derived from fibroblasts
were not observed to have increased expression levels of these
markers, indicating that Schwann cell-derived exosomes may be
widely involved in the differentiation of stem cells into Schwann
cells.
Previous studies have used stem cell-induced
differentiation with diverse induction methods to obtain
Schwann-like cells, including the use of cytokines (3,22)
and small molecule chemical reprogramming induction (23). BMSCS can differentiate and
participate in nerve regeneration at lesion sites (3,22,23),
suggesting that there might be a natural in vivo stem cell
induction mechanism; however, this mechanism remains relatively
unknown. Currently, the most accepted hypothesis is that BMSCs can
differentiate into Schwann cells under the control of cytokines in
the local microenvironment, which enables the newly differentiated
Schwann cells to secrete corresponding neuronal factors that
promote nerve regeneration or directly participate in the
regeneration of the myelin sheath (24,25).
It is also hypothesized that BMSCs can interact with local cells
instead of differentiating into Schwann cells to promote these
functions (26). However, to the
best of our knowledge, previous studies have not investigated
whether exosomes serve a role in the induction process. Our
previous study reported that degenerative nerve distal tissue fluid
may promote the differentiation of BMSCs into Schwann cells
following nerve injury (15),
suggesting that the source of the induced components in the
microenvironment following nerve damage may be related to Wallerian
degeneration. The present study demonstrated that exosomes derived
from Schwann cells may be involved in the differentiation of BMSCs
into Schwann cells. These results led to the hypothesis that
exosomes may induce the differentiation of BMSCs into Schwann cells
in a similar manner to cytokines, through interacting with
microRNAs, mRNAs and proteins in the exosomes. Moreover, exosomes
may also serve an important role in multiple developmental stages
of Schwann cells.
The underlying mechanisms behind exosome-induced
differentiation of BMSCs into Schwann cells remains unclear and
further investigations are required; transcriptome sequencing and
analysis may reveal the differences in the transcriptional profiles
of BMSC-derived Schwann cells and Schwann cells. In addition,
Schwann cell-derived exosomes are complex and versatile, but most
research on exosomes focuses on their role in nerve regeneration
(10,27–30).
The exosomes produced by the newly dedifferentiated Schwann cells
following nerve injury may differ from the exosomes derived from
normal Schwann cells under healthy physiological conditions.
Therefore, their function regarding the induction of Schwann cell
differentiation needs to be further investigated. In addition, it
could be that the exosome purity can affect MSCs induction, thus
the failure to evaluate the exosome purity may impact the accuracy
of the conclusions drawn from this study. In conclusion, the
present study suggested that the role of exosomes in the
differentiation of BMSCs into Schwann cells may represent a novel
target for neural regeneration; however, these results were not
validated in in vivo studies; thus, the efficiency of this
induction method requires further study.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by The National Natural
Science Foundation of China (grant. no. 81470682).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW and YJ designed and performed the research. HW,
YJ, JL and QL analyzed the data and were involved in writing the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All animal procedures were conducted in accordance
with The Guidelines for Ethical Conduct in the Care and Use of
Animals of the Chinese National Health and Medical Research
Council, and were approved by the Animal Experimentation Ethics
Committee of Capital Medical College (Beijing, China; protocol no.
AEEI-2016-135).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen X, Wang XD, Chen G, Lin WW, Yao J and
Gu XS: Study of in vivo differentiation of rat bone marrow stromal
cells into Schwann cell-like cells. Microsurgery. 26:111–115. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cuevas P, Carceller F, Dujovny M,
Garcia-Gómez I, Cuevas B, González-Corrochano R, Diaz-González D
and Reimers D: Peripheral nerve regeneration by bone marrow stromal
cells. Neurol Res. 24:634–638. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dezawa M, Takahashi I, Esaki M, Takano M
and Sawada H: Sciatic nerve regeneration in rats induced by
transplantation of in vitro differentiated bone-marrow stromal
cells. Eur J Neurosci. 14:1771–1776. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ladak A, Olson J, Tredget EE and Gordon T:
Differentiation of mesenchymal stem cells to support peripheral
nerve regeneration in a rat model. Exp Neurol. 228:242–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen J, Duan XH, Cheng LN, Zhong XM, Guo
RM, Zhang F, Zhou CP and Liang BL: In vivo MR imaging tracking of
transplanted mesenchymal stem cells in a rabbit model of acute
peripheral nerve traction injury. J Magn Reson Imaging.
32:1076–1085. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tohill M, Mantovani C, Wiberg M and
Terenghi G: Rat bone marrow mesenchymal stem cells express glial
markers and stimulate nerve regeneration. Neurosci Lett.
362:200–203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Keller S, Sanderson MP, Stoeck A and
Altevogt P: Exosomes: From biogenesis and secretion to biological
function. Immunol Lett. 107:102–108. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van der Pol E, Boing AN, Harrison P, Sturk
A and Nieuwland R: Classification, functions, and clinical
relevance of extracellular vesicles. Pharmacol Rev. 64:676–705.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Niel G, Porto-Carreiro I, Simoes S and
Raposo G: Exosomes: A common pathway for a specialized function. J
Biochem. 140:13–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ching RC and Kingham PJ: The role of
exosomes in peripheral nerve regeneration. Neural Regen Res.
10:743–747. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lopez-Leal R and Court FA: Schwann cell
exosomes mediate neuron-glia communication and enhance axonal
regeneration. Cell Mol Neurobiol. 36:429–436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ravasi M, Scuteri A, Pasini S, Bossi M,
Menendez VR, Maggioni D and Tredici G: Undifferentiated MSCs are
able to myelinate DRG neuron processes through p75. Exp Cell Res.
319:2989–2999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu S, Li J, Zhu Q, Dai T, He B, Zhou X,
Xiang J and Liu X: Differentiation of human amniotic epithelial
cells into Schwann-like cells via indirect co-culture with Schwann
cells in vitro. Mol Med Rep. 11:1221–1227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei Y, Gong K, Zheng Z, Liu L, Wang A,
Zhang L, Ao Q, Gong Y and Zhang X: Schwann-like cell
differentiation of rat adipose-derived stem cells by indirect
co-culture with Schwann cells in vitro. Cell Prolif. 43:606–616.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang H, Zhang H, Liu M and Wang N: Distal
segment extracts of the degenerated rat sciatic nerve induce bone
marrow stromal cells to express Schwann cell markers in vitro.
Neurosci Lett. 544:89–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using quantitative PCR and the
2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Finzsch M, Schreiner S, Kichko T, Reeh P,
Tamm ER, Bösl MR, Meijer D and Wegner M: Sox10 is required for
Schwann cell identity and progression beyond the immature Schwann
cell stage. J Cell Biol. 189:701–712. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bremer M, Frob F, Kichko T, Reeh P, Tamm
ER, Suter U and Wegner M: Sox10 is required for Schwann-cell
homeostasis and myelin maintenance in the adult peripheral nerve.
Glia. 59:1022–1032. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jaegle M and Meijer D: Role of Oct-6 in
Schwann cell differentiation. Microsc Res Tech. 41:372–378. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Britsch S, Goerich DE, Riethmacher D,
Peirano RI, Rossner M, Nave KA, Birchmeier C and Wegner M: The
transcription factor Sox10 is a key regulator of peripheral glial
development. Genes Dev. 15:66–78. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zorick TS, Syroid DE, Brown A, Gridley T
and Lemke G: Krox-20 controls SCIP expression, cell cycle exit and
susceptibility to apoptosis in developing myelinating Schwann
cells. Development. 126:1397–1406. 1999.PubMed/NCBI
|
|
22
|
Cai S, Tsui YP, Tam KW, Shea GK, Chang RS,
Ao Q, Shum DK and Chan YS: Directed differentiation of human bone
marrow stromal cells to fate-committed schwann cells. Stem Cell
Reports. 9:1097–1108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thoma EC, Merkl C, Heckel T, Haab R,
Knoflach F, Nowaczyk C, Flint N, Jagasia R, Jensen Zoffmann S,
Truong HH, et al: Chemical conversion of human fibroblasts into
functional Schwann cells. Stem Cell Reports. 3:539–547. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Keilhoff G, Goihl A, Stang F, Wolf G and
Fansa H: Peripheral nerve tissue engineering: Autologous Schwann
cells vs. transdifferentiated mesenchymal stem cells. Tissue Eng.
12:1451–1465. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choi BH, Zhu SJ, Kim BY, Huh JY, Lee SH
and Jung JH: Transplantation of cultured bone marrow stromal cells
to improve peripheral nerve regeneration. Int J Oral Maxillofac
Surg. 34:537–542. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weimann JM, Charlton CA, Brazelton TR,
Hackman RC and Blau HM: Contribution of transplanted bone marrow
cells to Purkinje neurons in human adult brains. Proc Natl Acad Sci
USA. 100:2088–2093. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lopez-Leal R, Alvarez J and Court FA:
Origin of axonal proteins: Is the axon-schwann cell unit a
functional syncytium? Cytoskeleton (Hoboken). 73:629–639. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Gregorio C, Diaz P, Lopez-Leal R,
Manque P and Court FA: Purification of exosomes from primary
Schwann cells, RNA extraction, and next-generation sequencing of
exosomal RNAs. Methods Mol Biol. 1739:299–315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qing L, Chen H, Tang J and Jia X: Exosomes
and their MicroRNA cargo: New players in peripheral nerve
regeneration. Neurorehabil Neural Repair. 32:765–776. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Simeoli R, Montague K, Jones HR, Castaldi
L, Chambers D, Kelleher JH, Vacca V, Pitcher T, Grist J, Al-Ahdal
H, et al: Exosomal cargo including microRNA regulates sensory
neuron to macrophage communication after nerve trauma. Nat Commun.
8:17782017. View Article : Google Scholar : PubMed/NCBI
|