Introduction
Mycobacterium tuberculosis (M.
tuberculosis) is the causative agent of tuberculosis (TB) and
is a major public health threat associated with high morbidity and
mortality rates worldwide (1).
Currently, >30% of the population worldwide is infected with
M. tuberculosis according to data from the World Health
Organization (2). The host immune
response against M. tuberculosis is complex and
multifaceted, thus understanding the pathogenesis and host immune
mechanisms against TB will facilitate the development of novel
diagnostic and therapeutic strategies for TB (3). Macrophages are critical immune cells
that play important roles in the host immune system by
phagocytizing M. tuberculosis to help eliminate infection,
as well as initiating the protective immune responses by presenting
antigens to T lymphocytes (4,5).
Macrophages can prevent the intracellular growth and persistence of
M. tuberculosis during various phases of TB, from primary
infection with bacillary dissemination (6). These previous studies have revealed
the functions of macrophages in M. tuberculosis infection,
but the underlying regulatory mechanisms are still not fully
understood.
Long non-coding RNAs (lncRNAs) are transcripts ~200
nucleotides in length that regulate the expression of protein
encoding genes at the transcriptional and post-transcriptional
levels (7). By dysregulating
target genes, lncRNAs can participate in the development and
progression of various human diseases, including cancer,
inflammation and autoimmune diseases (8–10).
The regulatory potential of lncRNAs-in epigenetic reprograming is
emerging as a novel mechanism to explain functional plasticity and
the diversity of immune cells including T cells, B cells, dendritic
cells and macrophages. Previous studies have shown that lncRNAs
also play an essential role in human infectious diseases such as TB
(11,12). Pawar et al (13) showed that downregulation of the
lncRNA maternally expressed 3 promotes the eradication of
intracellular mycobacterium in M. bovis Bacille Calmette
Guerin-infected macrophages.
The aim of the present study was to investigate the
regulatory mechanisms of the inflammatory response modulated by
long intergenic non-coding RNA cyclooxygenase 2 (lincRNACox2) in
M. tuberculosis H37Ra infected macrophages.
Materials and methods
Samples collection and ethical
statement
The peripheral blood samples (3 ml) were obtained
from 56 patients with active pulmonary TB (age range, 22–62 years;
median age, 36 years; sex, male: female=3:5) and 60 healthy
individuals (age range, 20–60 years; median age, 32 years;
male:female=4:6) in the China-Japan Friendship Hospital from June
2015 to June 2017. Written informed consent was obtained from all
donors prior to the study. This study was approved by the Ethics
Committee of the China-Japan Friendship Hospital (grant no.
ZRYYEC/2015/27-2).
Cell culture
THP-1 cells were purchased from the American Type
Culture Collection and cultured in RPMI-1640 (Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc) and antibiotics (100 U/ml penicillin and 100 mg/ml
streptomycin) at 37°C under the humidified atmosphere of 5%
CO2. The attenuated M. tuberculosis strain H37Ra
was provided by the China Center for Disease Control. M.
tuberculosis H37Ra was cultured on Middlebrooks 7H9 plates.
M. tuberculosis H37Ra culture and related experiments were
performed in the BSL-3 (P3) Laboratory of China Center for Disease
Control.
THP-1 cells were differentiated into macrophages
using Phorbol ester (0.01 mM; Merck KGaA) for 24 h and were then
incubated with H37Ra at a multiplicity of infection of 10 (10
bacteria:1 cell) for 24 h at 37°C.
Small interfering RNA (siRNA)
transfection and luciferase reporter assay
For gene silencing, siRNAs for lincRNACox2 were
synthesized and purchased from Invitrogen (Thermo Fisher
Scientific, Inc.). The following siRNAs were used: Mock siRNA
sense, 5′-UAAGGCUAUGAAGAGAUACUU-3′ and antisense
5′-GUAUCUCUUCAUAGCCUUAUU-3′; si-lincRNACox2 siRNA sense,
5′-GCCCUAAUAAGUGGGUUGUUU-3′ and antisense,
5′-ACAACCCACUUAUUAGGGCUU-3′; and lincRNACox2 sense,
5′-AGTATGGGATAACCAGCTGAGGT-3′ and antisense,
5′-GAATGCTGAGAGTGGGAGAAATAG-3′. Macrophages were transfected with
siRNAs (final concentration, 20 nM) using Lipofectamine RNAiMax
reagent (cat. no. 13778030; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol for 24 h. The lincRNACox2
expression vector was generated by reverse
transcription-quantitative PCR (RT-qPCR) amplification of
lincRNACox2 cDNA using RNA from macrophages and cloned into a
pcDNA3.3 vector (Thermo Fisher Scientific, Inc.). The promoter of
lincRNACox2 was amplified by PCR from human macrophage genomic DNA.
Macrophages were transfected with each reporter construct for 24 h,
followed by assessment of luciferase activity.
RT-qPCR and rapid amplification of
cDNA ends (RACE) PCR
Total RNA was extracted with TRIzol solution (cat.
no. R0016; Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. All primers for PCR analysis were designed
and synthesized by Invitrogen (Thermo Fisher Scientific, Inc.).
RT-qPCR was performed on a Master-cycler (Eppendorf Corp.) using
the SYBR Green PCR Master Mix [cat. no. R10T1; Takara Biotechnology
(Dalian) Co., Ltd.]. The PCR conditions were as follows: Initial
denaturation at 95°C for 25 sec, followed by 36 cycles at 95°C for
20 sec, 58°C for 20 sec and 72°C for 15 sec. U6 was used as an
internal control and the relative expression of genes was
calculated with the 2−ΔΔCq method (14). Primers used were as follows:
lincRNACox2 forward, 5′-AGTATGGGATAACCAGCTGAGGT-3′ and reverse,
5′-GAATGCTGAGAGTGGGAGAAATAG-3′; and U6 forward,
5′-CGCTTCGGCACATATACTA-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCA-3′.
RACE PCR was used to identify the 5′ and 3′ ends of
lincRNACox2 to localize the transcriptional start site. The SMART
RACE cDNA Amplification kit (cat. no. 634923; Clontech
Laboratories, Inc.) was used and the primers for 5′ and 3′ ends of
lincRNACox2 RACE PCR analysis were designed and synthesized by
Invitrogen (Thermo Fisher Scientific, Inc.).
ELISA assay
ELISA kits were used to detect the levels of the
cytokines tumor necrosis factor (TNF)-α (cat. no. EK0526; Wuhan
Boster Biological Technology, Ltd.), interferon (IFN)-γ (cat. no.
EK0373; Wuhan Boster Biological Technology, Ltd.) and interleukin
(IL)-6 (cat. no. EK0411; Wuhan Boster Biological Technology, Ltd.)
in the plasma of patients with TB infection and in cell supernatant
of macrophages infected with H37Ra, according to the manufacturer's
protocol. Cytokine concentrations were calculated using a standard
curve.
Western blotting
Cell lysates were obtained with RIPA lysis buffer
(cat. no. P0013B; Beyotime Institute of Biotechnology) and were
quantified using a bicinchoninic acid kit (cat. no. P0012S;
Beyotime Institute of Biotechnology). Proteins (30 µg) were run on
10% SDS-PAGE and transferred to PVDF membranes (EMD Millipore).
After blocking with 5% non-fat milk for 1 h at room temperature,
the membranes were incubated overnight at 4°C with the following
antibodies according to the manufacturer's protocol: Cox2 (cat. no.
sc1745; Santa Cruz Biotechnology, Inc.; 1:800), inducible nitric
oxide synthase (iNOS; cat. no. sc-7271; Santa Cruz Biotechnology,
Inc.; 1:1,200), Stat3 (cat. no. sc-8019; Santa Cruz Biotechnology,
Inc.; 1:600), NF-κB (cat. no. sc-8414; Santa Cruz Biotechnology,
Inc.; 1:1,000) and GAPDH (cat. no. sc-47724; Santa Cruz
Biotechnology, Inc.; 1:2,000). Then, membranes were incubated with
the goat anti-rabbit immunoglobin G-horseradish peroxidase (cat.
no. sc-2004; 1:4,000; Santa Cruz Biotechnology, Inc.). Specific
protein bands were detected using the Chemiluminescent Substrate
system (Thermo Fisher Scientific, Inc.). The band intensity was
measured using ImageJ 1.47 (National Institutes of Health).
Analysis of the apoptotic rate of
H37Ra infected macrophages
Macrophages transfected with si-lincRNACox2 or
lincRNACox2 were infected with 1×105 colony forming
units (CFU) of H37Ra for 24 h at 37°C and then cultured for 24 h in
medium at 37°C. Then, cells were harvested and incubated with
Annexin-V/PI (cat. no. C1062L; Beyotime Institute of Biotechnology)
for 30 min in the dark on ice at 4°C. Apoptotic cells were detected
on a DxFLEX flow cytometer (Beckman Coulter, Inc.) and analyzed
using CytExpert version 2 software (Beckman Coulter, Inc.),
according to the manufacturer's protocol.
Proliferation analysis of H37Ra
Macrophages were transfected with si-lincRNACox2 or
lincRNACox2 for 24 h, then were infected with H37Ra
(1×105 CFU) for 24 h. For titration of intracellular
H37Ra, macrophages were lysed with 0.025% SDS, neutralized with
0.5% albumin and grow on Middlebrooks 7H9 plates (cat. no.
GOY-PYJ0567; Shanghai Guyan Biotechnology Company). Then, H37Ra
titration was determined after 3, 6, 9, 12 and 15 days to analyze
their proliferative capacity by bacteria plate count.
Statistical analysis
Data are presented as the mean ± SEM from ≥3
independent experiments. Data were compared with an unpaired
Student's t-test or one-way analysis of variance with Tukey's post
hoc test. All statistical tests were performed with GraphPad Prism
6.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate
a statistically significant difference.
Results
Expression of lincRNACox2 and
inflammatory responses were analyzed in patients with TB
lincRNACox2 is a critical gene for regulating the
expression level of NF-κB in macrophages and NF-κB is an important
protein regulating the inflammatory pathway especially in the
process of tuberculosis infection. Thus the expression of
lincRNACox2 was investigated in TB patients. The expression levels
of lincRNACox2 were determined by RT-qPCR in the plasma (Fig. 1A) and mononuclear cells of patients
with TB (Fig. 1B). It was found
that lincRNACox2 was significantly increased in plasma and
mononuclear cells of patients with TB compared with healthy
individuals. Moreover, the expression levels of TNF-α, IFN-γ and
IL-6 in the plasma of patients with TB were significantly increased
compared with healthy individuals (Fig. 1C). In addition, the expression
levels of Cox2 and iNOS in mononuclear cells of patients with TB
were significantly elevated both at mRNA (Fig. 1D) and protein levels (Fig. 1E and F).
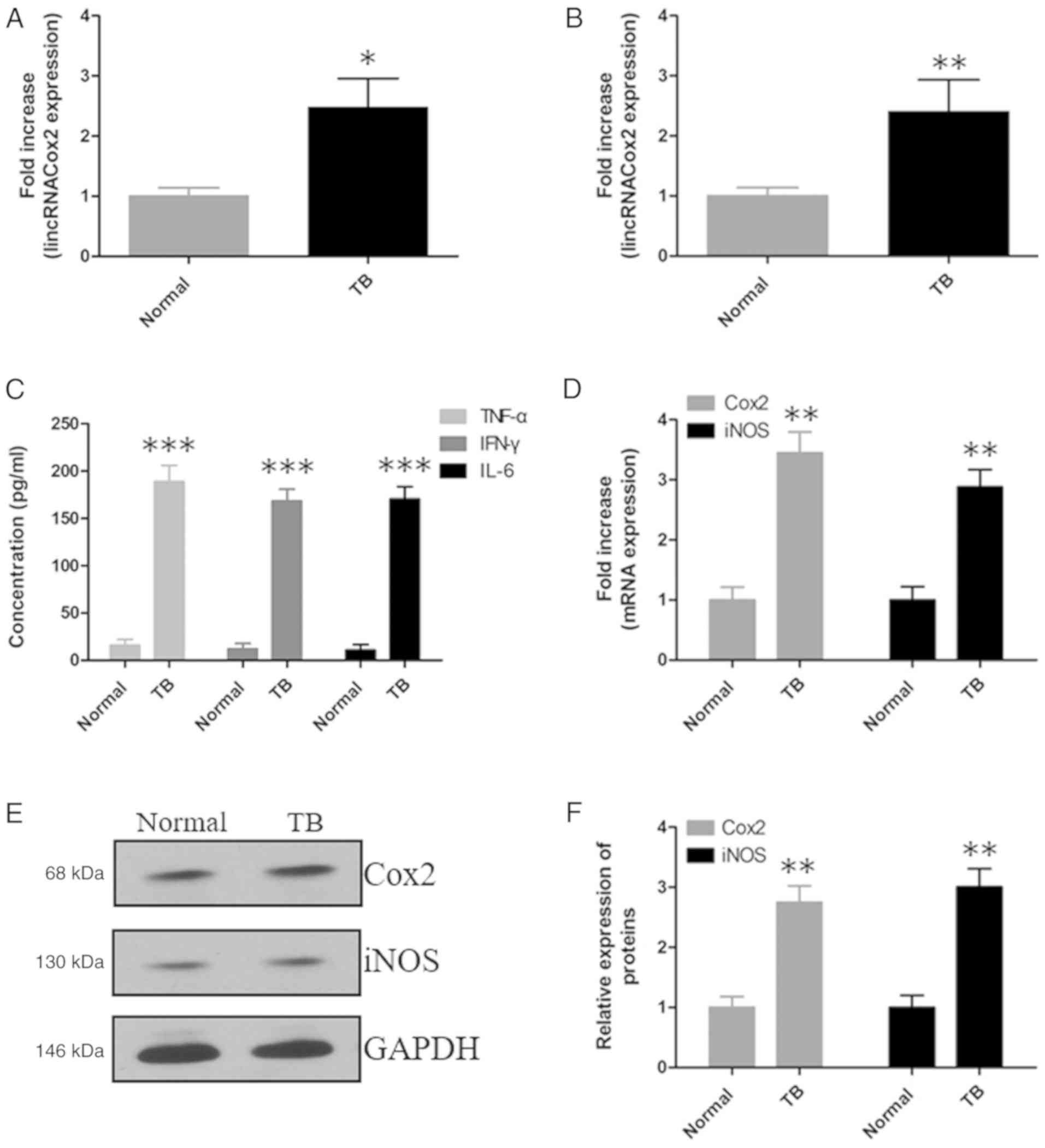 | Figure 1.Expression level of lincRNACox2 and
the inflammatory response in the peripheral blood of patients with
TB. RT-qPCR results of the expression level of lincRNACox2 in the
(A) plasma and (B) mononuclear cells of patients with TB. (C) ELISA
was used to detect TNF-α, IFN-γ and IL-6 in the plasma of patients
with TB. (D) RT-qPCR results of the mRNA expression levels of Cox2
and iNOS in mononuclear cells of patients with TB. (E) Protein
expression levels and (F) analysis of Cox2 and iNOS in mononuclear
cells of patients with TB were measured by western blotting. Data
are presented as the mean ± SEM from three independent experiments,
*P<0.05, **P<0.01 and ***P<0.001 vs. Normal. RT-qPCR,
reverse transcription-quantitative PCR; lincRNACox2, long
intergenic non-coding cyclooxygenase 2 RNA; TNF, tumor necrosis
factor; IFN, interferon; IL, interleukin; TB, tuberculosis; iNOS,
inducible NO synthase. |
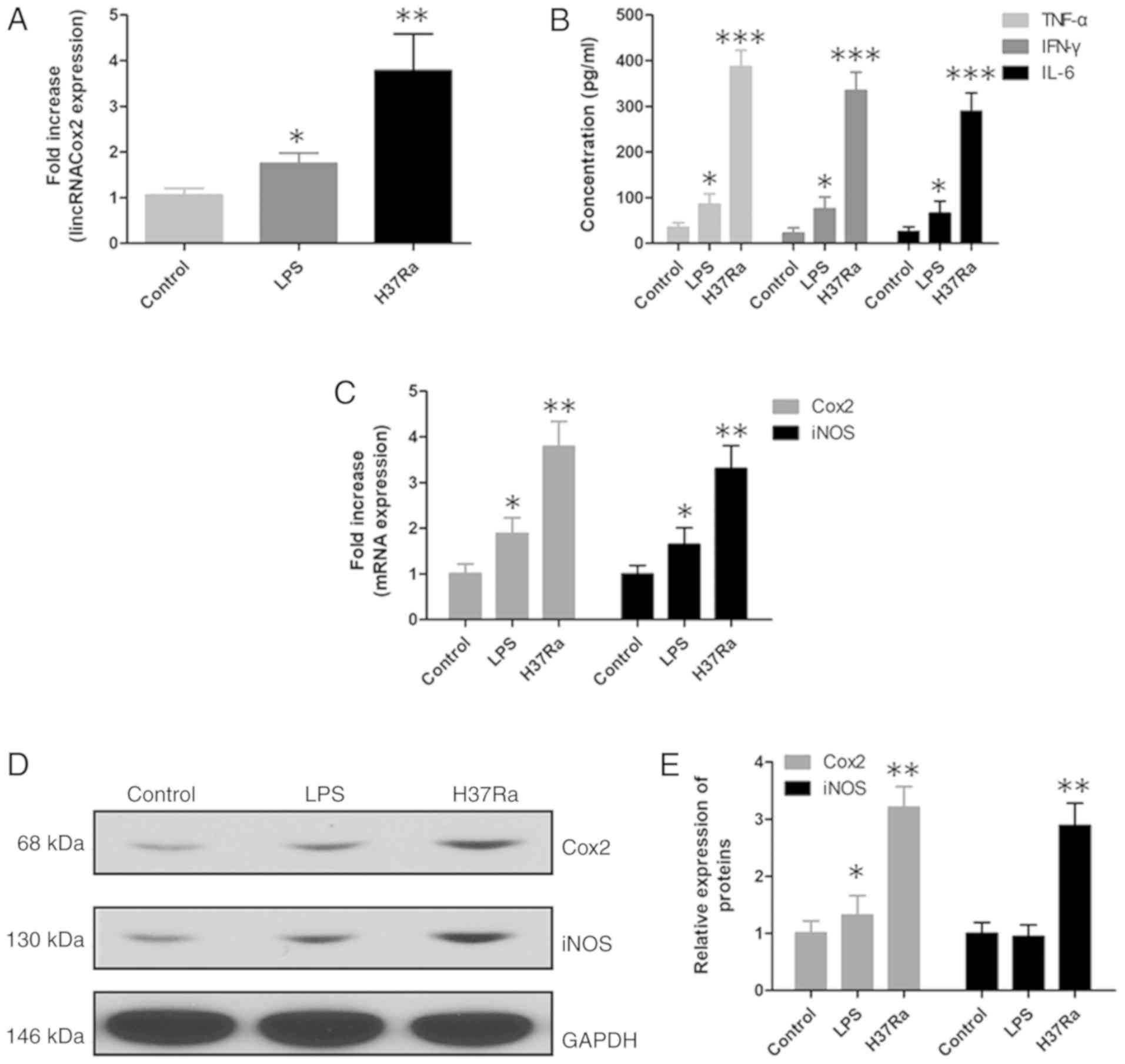
Expression of lincRNACox2 and
inflammatory responses in H37Ra infected macrophages
Previous studies have shown that lincRNACox2
increased significantly in TB patients and Cox2 and iNOS were also
promoted (15,16), so the function of lincRNACox2 in an
in vitro cell experiment was assessed. In vitro cell
experiments, RT-qPCR and western blotting were performed to assess
the expression level of lincRNACox2 in macrophages after exposure
to H37Ra. It was demonstrated that lincRNACox2 was increased in
H37Ra infected macrophages (Fig.
2A). Furthermore, the inflammatory factors TNF-α, IFN-γ and
IL-6 were increased in the supernatant of H37Ra infected
macrophages, as determined by ELISA (Fig. 2B). Moreover, the mRNA and protein
expression levels of Cox2 and iNOS were significantly increased in
H37Ra infected macrophages compared with the control group
(Fig. 2C-E).
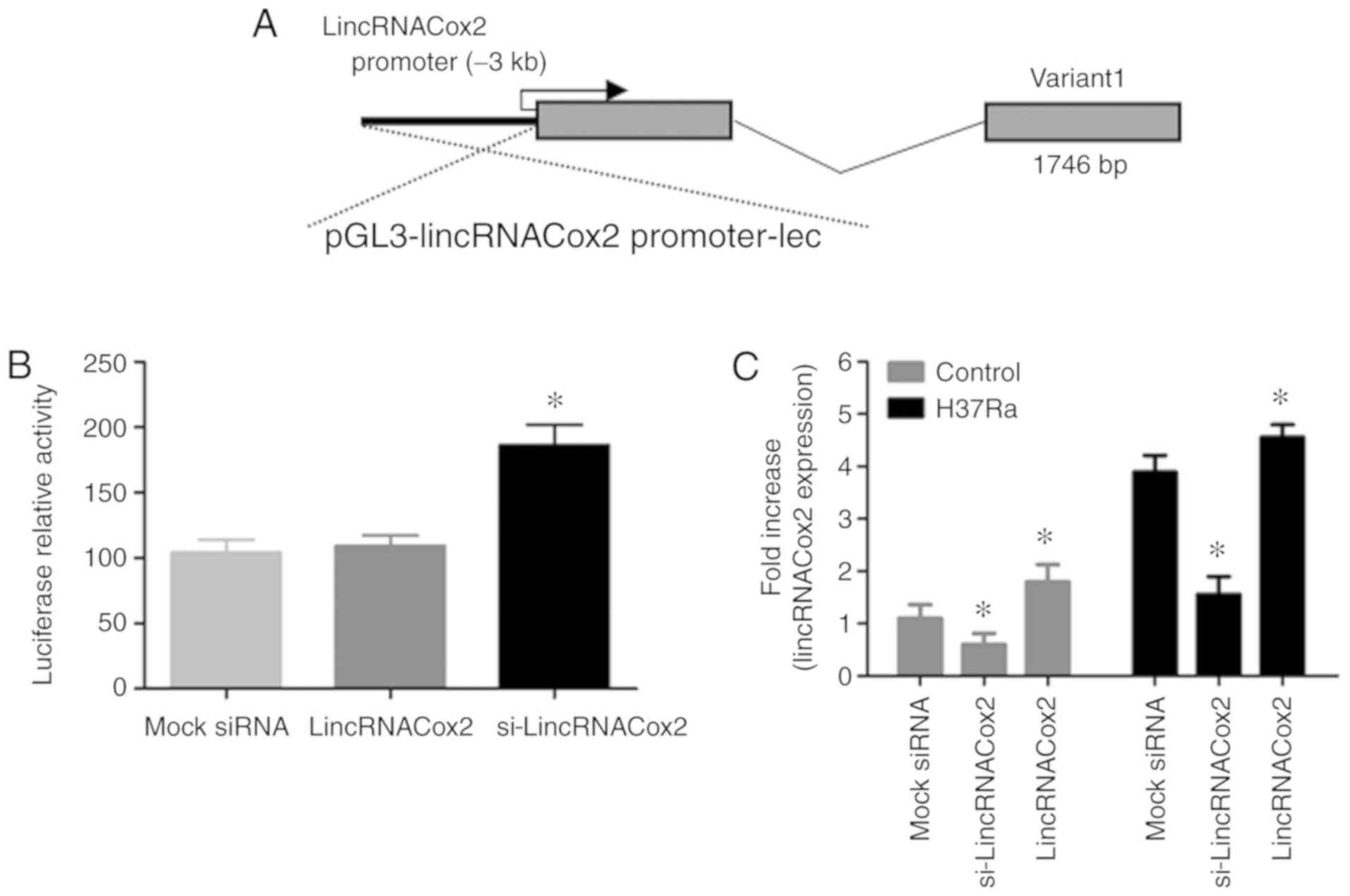
lincRNACox2 knockdown in
macrophages
The expression of lincRNACox2 was enhanced in TB
patients and TB infection macrophages in vivo and in
vitro, then the effects of the knockdown of lincRNACox2 in TB
infected host cells will be studied. The siRNA in the present study
was designed to target lincRNACox2 and lincRNACox2 was transfected
into macrophages to induce its overexpression. lincRNACox2 was
knocked down using an RNA interference approach and the expression
level of lincRNACox2 was measured by RT-qPCR. RACE PCR analysis
results identified a transcript of lincRNACox2 in macrophages using
a luciferase reporter vector (Fig.
3A). Furthermore, it was found that the expression level of
lincRNACox2 was significantly decreased when macrophages were
transfected with si-lincRNACox2 (Fig.
3B). However, H37Ra infection promoted the expression level of
lincRNACox2 in si-lincRNACox2 transfected macrophages (Fig. 3C).
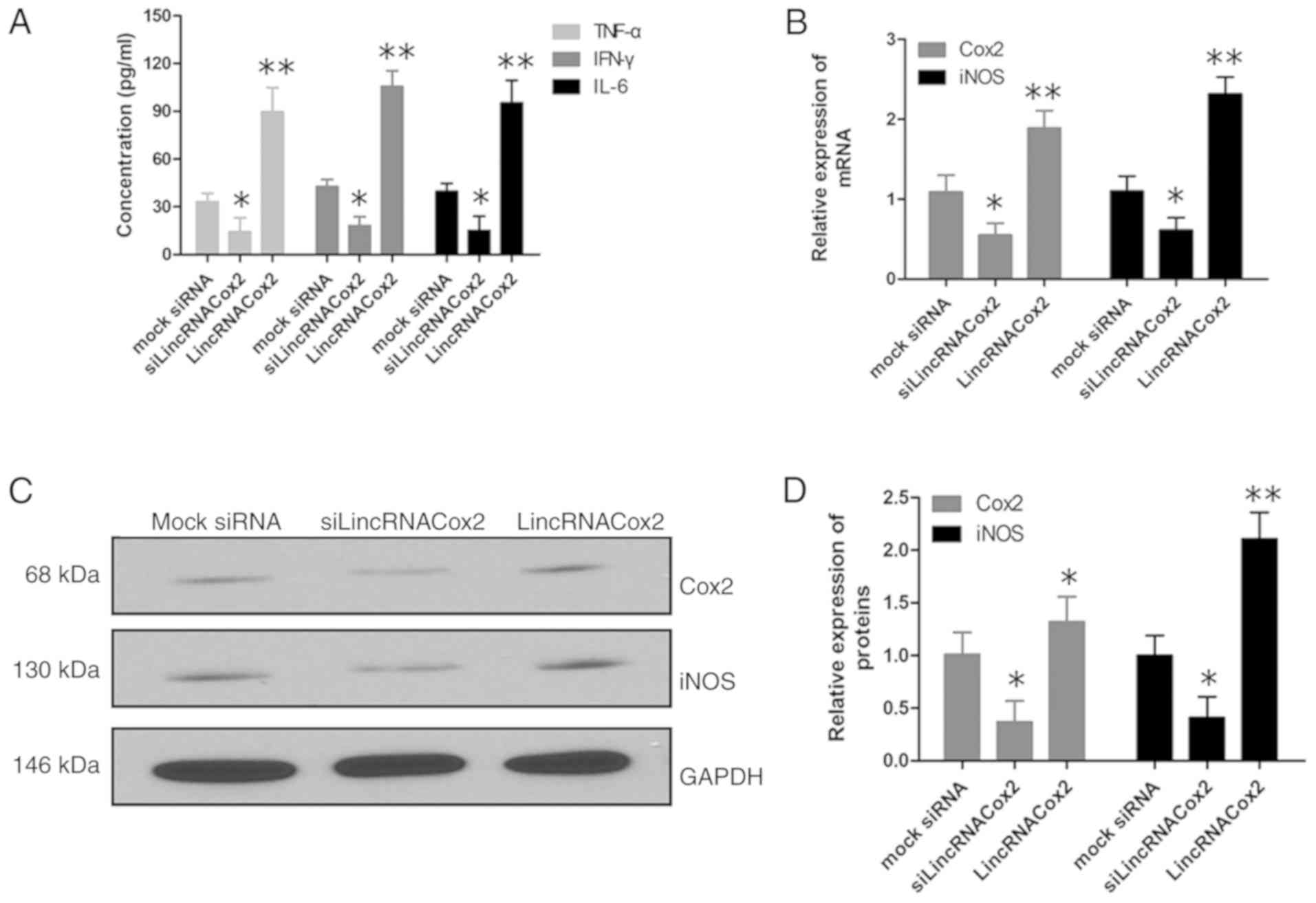
Inflammatory responses in lincRNACox2
knockdown macrophages are induced by H37Ra
As lincRNACox2 plays a very important role in the
inflammatory response pathway, so the inflammatory response of TB
infected macrophages in which lincRNACox2 was knocked down was
studied. After lincRNACox2 was silenced, macrophages were infected
with H37Ra. It was demonstrated that macrophages transfected with
si-lincRNACox2 had significantly decreased expression levels of the
inflammatory factors TNF-α, IFN-γ and IL-6 compared with other
groups (Fig. 4A). Moreover,
knockdown of lincRNACox2 reduced the mRNA and protein expression
levels of Cox2 and iNOS in H37Ra infection macrophages (Fig. 4B-D). Therefore, the present results
suggested that the knockdown of lincRNACox2 suppressed the
inflammatory response in H37Ra infected macrophages.
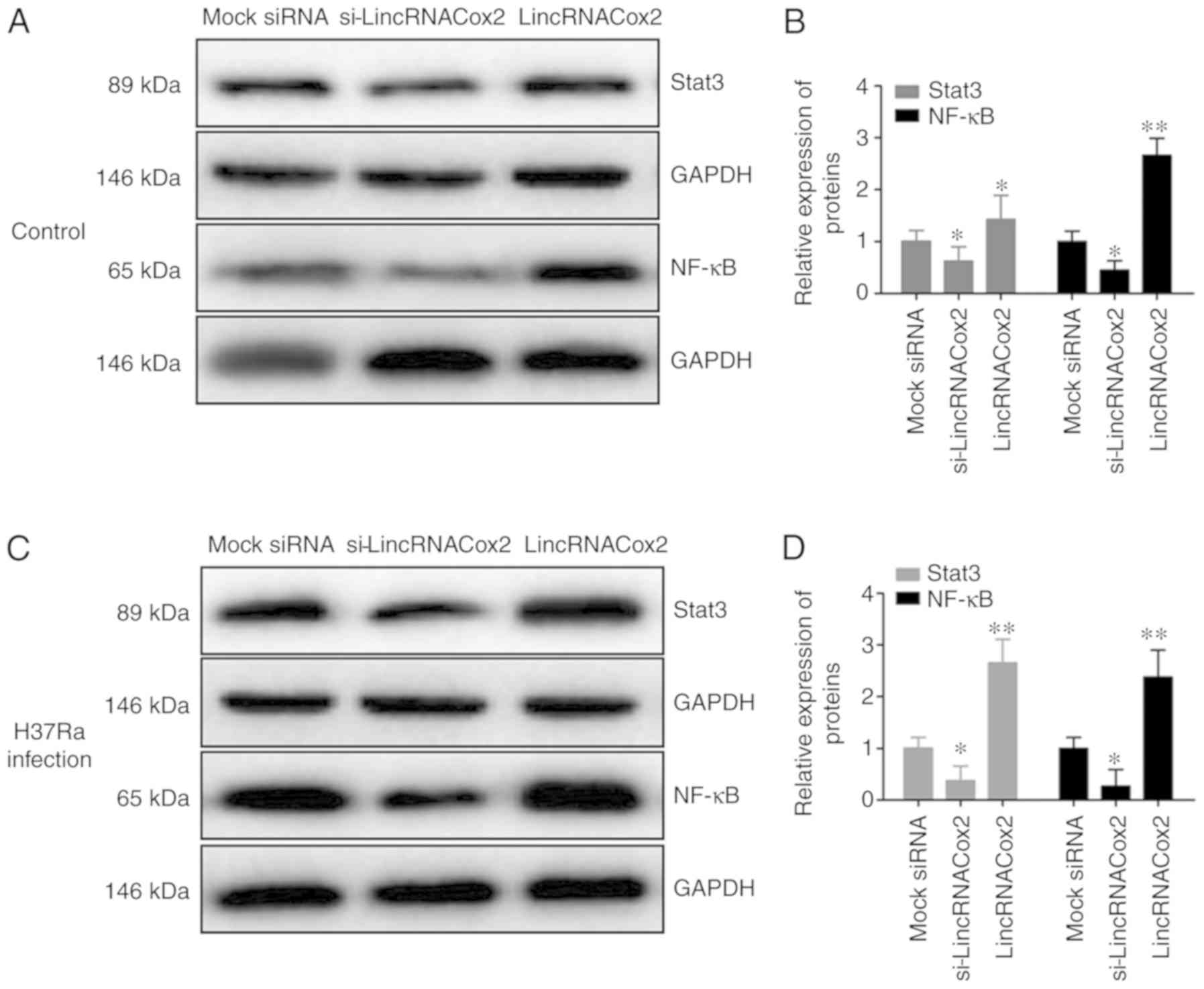
Inflammatory regulatory proteins Stat3
and NF-κB are regulated by lincRNACox2
lincRNACox2 regulates the expression of inflammatory
cytokines and inflammatory regulating proteins, so the inflammatory
signaling pathways such as Stat3 and NF-κB associated with
lincRNACox2 were investigated. It was found that the knockdown of
lincRNACox2 significantly inhibited the protein expression levels
of NF-κB and Stat3 in macrophages (Fig. 5A and B). Moreover, H37Ra
significantly suppressed the expression levels of the inflammatory
regulatory proteins Stat3 and NF-κB in lincRNACox2 knockdown
macrophages (Fig. 5C and D). Thus,
the present results indicated that the NF-κB pathway may be
triggered by lincRNACox2 in macrophages. However, H37Ra is
dependent on the expression level of lincRNACox2 to activate the
NF-κB signaling pathway to induce a cellular inflammatory
response.
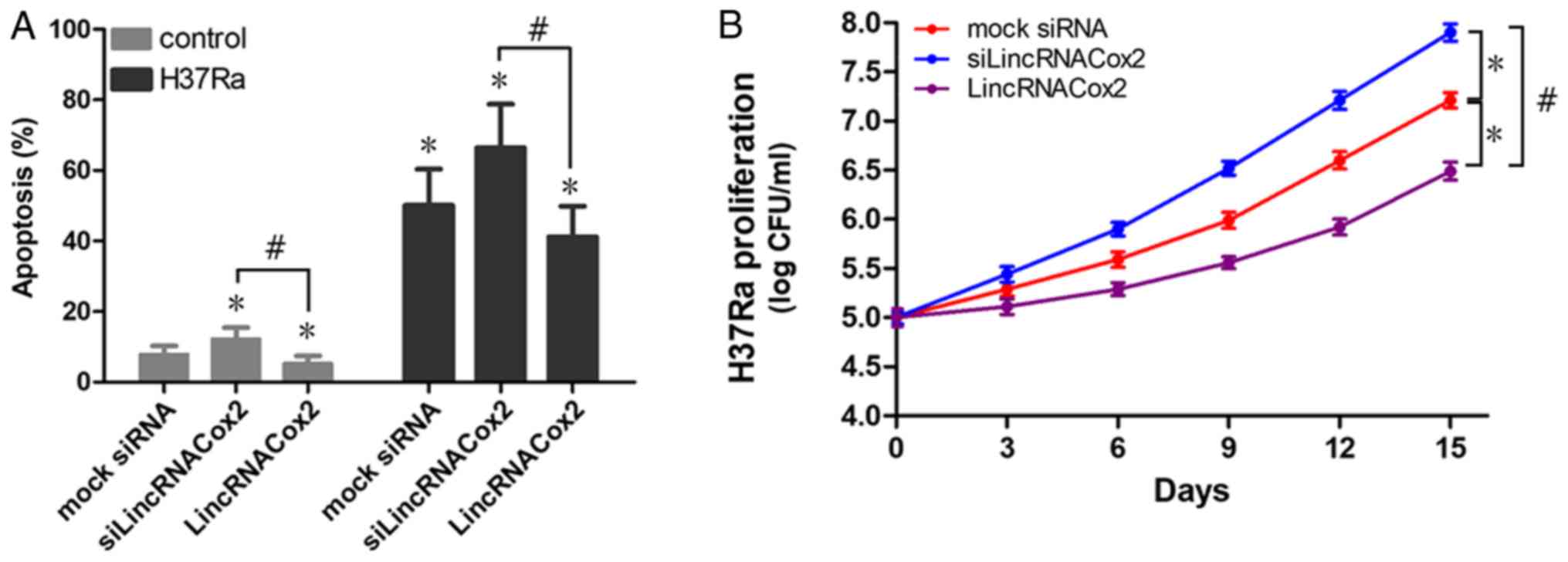
Effect of lincRNACox2 on the apoptosis
of H37Ra infected macrophages and the proliferation of H37Ra
lincRNACox2 could regulate the inflammatory response
of TB infection macrophages through Stat3 and NF-κB signaling
pathways, however the influences of lincRNACox2 on the apoptosis of
H37Ra infected macrophages and the proliferation of H37Ra remains
unknown. Macrophages transfected with si-lincRNACox2 or lincRNACox2
were infected with H37Ra and the effect of lincRNACox2 on the
apoptosis and proliferation of H37Ra infected macrophages was
investigated. It was demonstrated that knockdown of lincRNACox2
significantly increased the apoptotic rate of H37Ra infect
macrophages. However, the apoptotic rate of H37Ra infected
macrophages with lincRNACox2 overexpression was significantly
decreased compared with control groups (Fig. 6A). Moreover, knockdown of
lincRNACox2 promoted the proliferation of H37Ra extracted from
macrophages, but the overexpression of lincRNACox2 inhibited the
proliferation of H37Ra, suggesting an additive regulating effect of
lincRNACox2 on H37Ra infection (Fig.
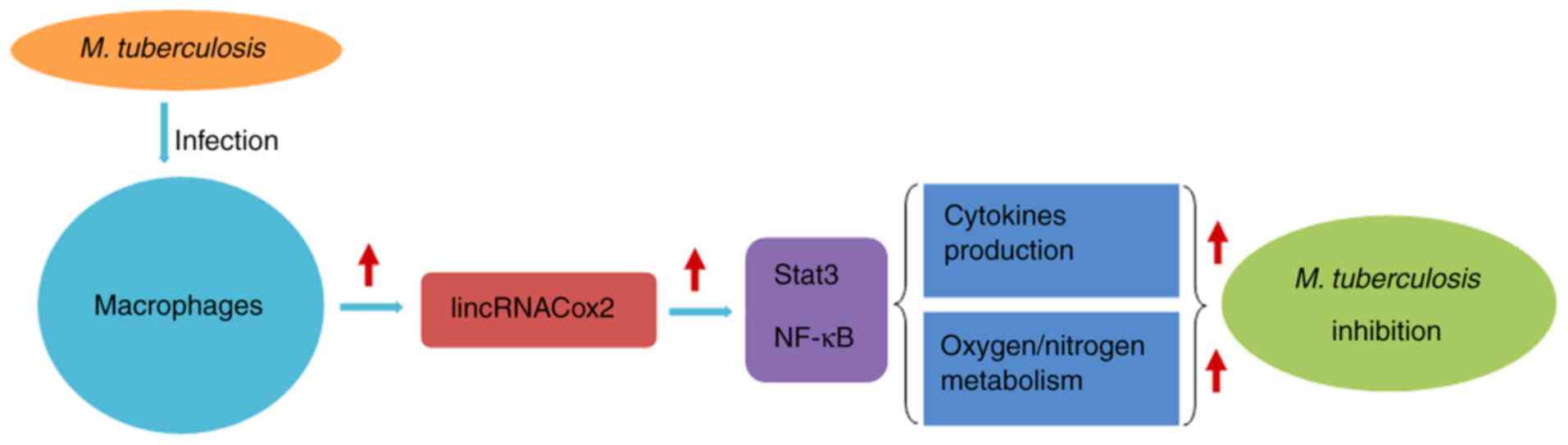
6B). Collectively, the present results suggested that the
overexpression of lincRNACox2 in H37Ra infected macrophages could
suppress H37Ra proliferation via various mechanisms, including
cytokine production and oxygen/nitrogen metabolism (Fig. 7).
Discussion
Previous studies have showed that lncRNAs play
significant roles in various biological processes, such as the
immune response, tumorigenesis and cellular infectious diseases
(17–20). Interactions between macrophages and
M. tuberculosis can alter genetic material expression
profiles, including circular RNAs expression profiles and lncRNA
expression profiles. Examining these changes can improve the
understanding of the regulation of anti-mycobacterium immunity in
macrophages (21,22). Furthermore, previous studies have
demonstrated lncRNAs can be used as biomarkers or therapeutic
targets for various diseases (23,24).
Several lncRNAs, such as miR3945HG-V1 and miR3945HG-V2, are
differentially expressed in H37Ra or H37Rv infected macrophages,
and could be used as novel diagnostic biomarkers for TB. Thus, this
would provide insight into the mechanisms of M. tuberculosis
macrophage interactions and identify potential targets for the
diagnosis and treatment of TB (25). LncRNA-TNF and HNRNPL related
immunoregulatory lncRNA in THP-1 macrophages can modulate the
expression levels of TNF-α and other inflammatory genes to regulate
inflammation (26). Moreover,
lincRNA-exopolysaccharide can regulate the inflammatory response in
macrophages exposed to microbial ligands via the regulation of
interferon regulated gene expression (27). Furthermore, lincRNACox2 can mediate
both the activation and repression of different immune genes.
The present results suggested that lincRNACox2 was
increased in patients with TB and H37Ra infected macrophages, and
that the inflammatory factors TNF-α, IFN-γ, IL-6, Cox2 and iNOS
were increased by lincRNACox2 in vivo and in vitro.
Thus, lincRNACox2 may be a critical mediator for the development of
inflammatory responses in both microglia and macrophages. The
present study knocked down the expression of lincRNACox2 in H37Ra
infected macrophages and found this could suppress the inflammatory
factors TNF-α, IFN-γ, IL-6, Cox2 and iNOS in these macrophages,
which indicated that lincRNACox2 may regulate the inflammatory
response of H37Ra infected macrophages.
lincRNACox2 expression is influenced by NF-κB
signaling and serves as a coactivator of transcriptional factors to
regulate inflammatory responses (28). NF-κB is composed of homo- or
heterodimeric complexes of NF-κB subunits including p65, RelB,
c-Rel, p50 and p52 in humans, and is an important regulator of
inflammatory and immune responses (29). However, the dysregulation of the
NF-κB signaling pathway is linked to cancer, inflammation,
autoimmune diseases and infectious diseases (30,31).
Furthermore, numerous acute and late proinflammatory genes,
critical to the pathogenesis of sepsis, are controlled by the
NF-κB-gene (32). A nuclear NF-κB
paradox has been identified during severe systemic inflammation, as
defined by a disassociation between cytosolic NF-κB activation and
nuclear NF-κB transcriptional processes (33). Previous studies have demonstrated
the inflammatory response is induced in macrophages due to
lipopolysaccharide stimulation via the NF-κB signaling pathway
(34,35).
In the present study, the inflammatory response
related signaling pathway involving Stat3 and NF-κB was analyzed in
H37Ra infected macrophages transfected with si-lincRNACox2. The
present results suggested that the expression of lincRNACox2 is
induced by the NF-κB signaling pathway, potentially via binding of
NF-κB subunits to the promoter region of the gene locus. The
induction of lincRNACox2 in patients with TB and H37Ra infected
macrophages, along with the expression of the inflammatory factors
TNF-α, IFN-γ, IL-6, Cox2 and iNOS, suggested that NF-κB-mediated
lincRNACox2 expression and its subsequent impact on the
transcription of inflammatory genes may occur in vivo during
systemic inflammation. In addition, knockdown of lincRNACox2
promoted the proliferation of H37Ra via the inflammatory pathway or
oxygen/nitrogen metabolism, thus indicating that lincRNACox2 may
have a central role in M. tuberculosis infection.
In conclusion, the present results suggested that
lincRNACox2 may be used as a novel biomarker for the diagnosis and
therapeutic target of TB. However, the function of lincRNACox2 in
regulating the immune response of macrophages infected with H37Ra
is still not fully understood.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DL and YZ designed the experiments; CG and LZ
performed the experiments and analyzed the data; DL and YZ drafted
and revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the China-Japan Friendship Hospital (grant no.
ZRYYEC/2015/27-2). Written informed consent was obtained from all
donors prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zheng L, Leung E, Lee N, Lui G, To KF,
Chan RC and Ip M: Differential microRNA expression in human
macrophages with Mycobacterium tuberculosis infection of
Beijing/W and non-Beijing/W strain types. PLoS One.
10:e01260182015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suárez I, Fünger SM, Kröger S, Rademacher
J, Fätkenheuer G and Rybniker J: The diagnosis and treatment of
tuberculosis. Dtsch Arztebl Int. 116:729–735. 2019.PubMed/NCBI
|
|
3
|
Baer CE, Rubin EJ and Sassetti CM: New
insights into TB physiology suggest untapped therapeutic
opportunities. Immunol Rev. 264:327–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hmama Z, Peña-Díaz S, Joseph S and Av-Gay
Y: Immunoevasion and immunosuppression of the macrophage by
Mycobacterium tuberculosis. Immunol Rev. 264:220–232. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang Z, Luo Q, Guo Y, Chen J, Xiong G,
Peng Y, Ye J and Li J: Mycobacterium tuberculosis-induced
polarization of human macrophage orchestrates the formation and
development of tuberculous granulomas in vitro. PLoS One.
10:e01297442015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Korb VC, Chuturgoon AA and Moodley D:
Mycobacterium tuberculosis: Manipulator of protective
immunity. Int J Mol Sci. 17:1312016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heward JA and Lindsay MA: Long non-coding
RNAs in the regulation of the immune response. Trends Immunol.
35:408–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Z, Zhang M, Ying J, Hu X, Zhang J,
Zhou Y, Zhou Y, Song X and Ying B: Significance of genetic
polymorphisms in long non-coding RNA AC079767.4 in tuberculosis
susceptibility and clinical phenotype in western Chinese Han
population. Sci Rep. 7:9652017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mirza AH, Kaur S, Brorsson CA and Pociot
F: Effects of GWAS-associated genetic variants on lncRNAs within
IBD and T1D candidate loci. PLoS One. 9:e1057232014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He J, Ou Q, Liu C, Shi L, Zhao C, Xu Y,
Kong SK, Loo JFC, Li B and Gu D: Differential expression of long
non-coding RNAs in patients with tuberculosis infection.
Tuberculosis (Edinb). 107:73–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu Y, Gao K, Tao E, Li R and Yi Z:
Aberrantly expressed long non-coding RNAs in CD8+ T
cells response to active tuberculosis. J Cell Biochem.
118:4275–4284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pawar K, Hanisch C, Palma Vera SE,
Einspanier R and Sharbati S: Down regulated lncRNA MEG3 eliminates
mycobacteria in macrophages via autophagy. Sci Rep. 6:194162016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carpenter S, Aiello D, Atianand MK, Ricci
EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB,
et al: A long noncoding RNA mediates both activation and repression
of immune response genes. Science. 341:789–792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin J, Jiang Y, Liu D, Dai X, Wang M and
Dai Y: Early secreted antigenic target of 6-kDa of Mycobacterium
tuberculosis induces transition of macrophages into epithelioid
macrophages by downregulating Inos/NO-mediated H3K27 trimethylation
in macrophages. Mol Immunol. 117:189–200. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moran VA, Perera RJ and Khalil AM:
Emerging functional and mechanistic paradigms of mammalian long
non-coding RNAs. Nucleic Acids Res. 40:6391–6400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Satpathy AT and Chang HY: Long noncoding
RNA in hematopoiesis and immunity. Immunity. 42:792–804. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rajaram MV, Ni B, Dodd CE and Schlesinger
LS: Macrophage immunoregulatory pathways in tuberculosis. Semin
Immunol. 26:471–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gan H, Lee J, Ren F, Chen M, Kornfeld H
and Remold HG: Mycobacterium tuberculosis blocks
crosslinking of annexin-1 and apoptotic envelope formation on
infected macrophages to maintain virulence. Nat Immunol.
9:1189–1197. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boon RA, Jae N, Holdt L and Dimmeler S:
Long noncoding RNAs: From clinical genetics to therapeutic targets?
J Am Coll Cardiol. 67:1214–1226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang X, Yang J, Wang J, Wen Q, Wang H, He
J, Hu S, He W, Du X, Liu S and Ma L: Microarray analysis of long
noncoding RNA and mRNA expression profiles in human macrophages
infected with Mycobacterium tuberculosis. Sci Rep.
6:389632016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Chao TC, Chang KY, Lin N, Patil VS,
Shimizu C, Head SR, Burns JC and Rana TM: The long noncoding RNA
THRIL regulates TNFα expression through its interaction with
hnRNPL. Proc Natl Acad Sci USA. 111:1002–1007. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Atianand MK, Hu W, Satpathy AT, Shen Y,
Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD,
Blin J, et al: A long noncoding RNA lincRNA-EPS acts as a
transcriptional brake to restrain inflammation. Cell.
165:1672–1685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu G, Liao K, Niu F, Yang L, Dallon BW,
Callen S, Tian C, Shu J, Cui J, Sun Z, et al: Astrocyte EV-induced
lincRNA-Cox2 regulates microglial phagocytosis: Implications for
morphine-mediated neurodegeneration. Mol Ther Nucleic Acids.
13:450–463. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hayden MS and Ghosh S: Shared principles
in NF-kappaB signaling. Cell. 132:344–362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Natoli G, Saccani S, Bosisio D and Marazzi
I: Interactions of NF-kappaB with chromatin: The art of being at
the right place at the right time. Nat Immunol. 6:439–445. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haddad JJ and Abdel-Karim NE: NF-κB
cellular and molecular regulatory mechanisms and pathways:
Therapeutic pattern or pseudo regulation? Cell Immunol. 271:5–14.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Böhrer H, Qiu F, Zimmermann T, Zhang Y,
Jllmer T, Männel D, Böttiger BW, Stern DM, Waldherr R, Saeger HD,
et al: Role of NFkappaB in the mortality of sepsis. J Clin Invest.
100:972–985. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schreiber J, Jenner RG, Murray HL, Gerber
GK, Gifford DK and Young RA: Coordinated binding of NF-kappaB
family members in the response of human cells to
lipopolysaccharide. Proc Natl Acad Sci USA. 103:5899–5904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Zhang W, Wang C, Fang X, Cheng H,
Liu S and Chen XL: Inhibitor of Tec kinase, LFM-A13, decreases
pro-inflammatory mediators production in LPS-stimulated RAW264.7
macrophages via NF-κB pathway. Oncotarget. 8:34099–34110.
2017.PubMed/NCBI
|
|
35
|
Chen Y, Guo S, Jiang K, Wang Y, Yang M and
Guo M: Glycitin alleviates lipopolysaccharide-induced acute lung
injury via inhibiting NF-κB and MAPKs pathway activation in mice.
Int Immunopharmacol. 75:1057492019. View Article : Google Scholar : PubMed/NCBI
|