Introduction
Osteoporosis or bone fractures are major causes of
general bone tissue loss, and currently, no specific treatment
exists (1). With an aging
population and the need for more efficacious therapeutics with
fewer side effects, the field of bone tissue engineering is
increasing (2,3). Stem cell therapies and tissue
engineering applications are the most recent treatment options
targeted to repair damaged organizational structures and restore
physiological function of bone (4).
Human bone marrow mesenchymal stem cells (hBMSCs)
are a type of multilineage stem cell that have the potential to
differentiate into cells associated with mesenchymal tissues,
including fat, cartilage, bone, tendon, marrow stroma and muscle
(5). Over 10 years of research has
focused on efforts to evaluate and utilize hBMSCs for tissue
engineering and cell therapy applications, particularly in
cardiovascular disorders, bone and cartilage regeneration, and
neuronal damage, and through drip infusion or transplantation
within scaffolds (6). Earlier
research in the field of tissue-engineered bone regeneration
employed mesenchymal stem cells collected from the human mandible
or maxilla with substantial platelet-rich plasma. The results of
those investigations revealed that bone tissue was regenerated with
minimal invasiveness and good plasticity, and this could be used as
a clinical alternative for autogenous bone transplantation
(7,8).
MicroRNAs (miRNAs/miRs) are short non-coding RNAs
that have a key role in regulating differentiation and self-renewal
of stem cells (9). Almost all
miRNAs bind to the 3′ untranslated region of target mRNAs through
an incomplete match that inhibits their translation and stability
(10). The miR-10 family is highly
conserved and has attracted the interest of several research groups
due to its co-expression with the Hox gene development regulator
and its regulation of nearby genome localization (11,12).
miR-10a has been demonstrated to regulate numerous pathways,
including inflammation, cancer and proliferation processes
(13–15). It has been reported that miR-204
and its homolog miR-211 act as negative regulators of osteoblast
differentiation in BMSCs via the negative regulation of
runt-related transcription factor 2 (RUNX2) transcription factors
(16). A previous study has also
found that miR-206 played an inhibitory role during osteoblast
differentiation of MSCs (17).
However, to the best of our knowledge, there is no information with
regards to the specific modulatory effects of miR-10a-5p on
osteogenic differentiation of hBMSCs.
Therefore, the aim of the present study was to
investigate the relationship between hBMSCs and miR-10a-5p, and to
determine how miR-10a-5p regulated the osteogenic differentiation
process of hBMSCs in vitro and in vivo.
Materials and methods
Cell culture and osteogenic
differentiation
hBMSCs (cat. no. HUXMA-01001; Cyagen Biosciences,
Inc.) were cultured in OriCell™ human mesenchymal stem cell growth
medium (cat. no. HUXMA-90011; Cyagen Biosciences, Inc.) containing
10% human mesenchymal stem cell-qualified fetal bovine serum
(Cyagen Biosciences, Inc.), 1% penicillin-streptomycin and 1%
glutamine at 37°C in 5% CO2. The culture medium was
changed every 3 days. Once the hBMSCs reached 80–90% confluence,
they were dissociated with trypsin-EDTA and passaged. The cells
were transferred into growth medium at a concentration of
2×105 cells/cm2 in 6-well tissue culture
plates with a complete volume of 2 ml/well. Osteogenic
differentiation was induced using OriCell mesenchymal stem cell
osteogenic differentiation medium (cat. no. GUXMX-90021; Cyagen
Biosciences, Inc.) containing 10% human mesenchymal stem
cell-qualified fetal bovine serum, 1% penicillin-streptomycin, 1%
glutamine, 0.2% ascorbate, 1% β-glycerophosphate and 0.01%
dexamethasone.
RNA interference (RNAi) and miRNA
mimics
mirVana™ miRNA hsa-miR-10a-5p inhibitor (cat. no.
4464084; Ambion; Thermo Fisher Scientific, Inc.) and mimic (cat.
no. 4464066; Ambion; Thermo Fisher Scientific, Inc.) are small,
chemically modified, single-stranded (ss) and double-stranded (ds)
RNAs that inhibit and mimic endogenous miRNAs and enable miRNA
functional analysis, through down- or upregulation of miRNA
activity, respectively. The inhibitor negative control
(inhibitor-negative; cat. no. 4464076) and mimic negative control
(mimic-negative; cat. no. 4464058) were also purchased from Ambion
(Thermo Fisher Scientific, Inc.). Adherent hBMSCs (2×105
cells/well) in 6-well tissue culture plates were treated with miRNA
mimic, mimic-negative, inhibitor or inhibitor-negative diluted with
RNase-free water at 25 nM as a final concentration. Diluted
Lipofectamine® RNAi MAX Reagent (50 pmol) in 2 ml
Opti-MEM® medium (Invitrogen; Thermo Fisher Scientific,
Inc.) was added to each well and cells were incubated at 37°C in an
atmosphere containing 5% CO2 for 12 h. The medium was
replaced with OriCell mesenchymal stem cell osteogenic
differentiation medium for subsequent experiments immediately.
Fluorescent detection
BLOCK-iT™ Alexa Fluor® Red Fluorescent
Control (Thermo Fisher Scientific, Inc.) is an Alexa
Fluor® 555-labeled dsRNA duplex used for assessing
lipid-mediated transfection for RNAi experiments. Cells seeded at
60–80% confluence were used for transfection, and Lipofectamine
RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) was diluted in
Opti-MEM. Alexa Fluor® 555-labeled dsRNA was diluted in
Opti-MEM in a 5 ml Eppendorf tube®, and the diluted
dsRNA (25 pmol) was added to the diluted Lipofectamine RNAiMAX
reagent at a 1:1 ratio, and incubated for 5 min at room
temperature. Then, the dsRNA-lipid complex was added to the hBMSCs
for 24 h at 37°C. The nucleus was then stained using Hoechst 33342
working solution (cat. no. C1025; Beyotime Institute of
Biotechnology) for 15 min at room temperature, rinsed with 0.01
mol/l phosphate-buffered saline (PBS) 3 times for 5 min each time.
The transfection efficiency was visualized under an inverted
fluorescence microscope (Leica Microsystems, Inc.). The time
interval between transfection and visualization was 24 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
For RT-qPCR analysis, total RNA was extracted from
transfected hBMSCs with RNA Iso-Plus reagent (Takara Bio, Inc.).
RT-qPCR was performed using the primers listed in Table I (Takara Bio, Inc.). The genomic
DNA removal reaction was at 42°C for 2 min before storage at 4°C;
RT was performed using the PrimeScript RT reagent kit with gDNA
eraser (cat. no. DRR047A; Takara Bio, Inc.) in a 20-µl volume
reaction system at 37°C for 15 min, followed by inactivation at
85°C for 5 sec storage at 4°C. The expression level differences
were analyzed using Light Cycler® 480 SYBR Green I
Master (Roche Diagnostics) in a 25-µl volume in accordance with the
general manufacturer's protocol. PCR thermocycling was conducted as
follows: Pre-denaturation at 95°C for 30 sec, followed by 40 cycles
of denaturation at 95°C for 5 sec, primer annealing at 60°C for 30
sec and extension at 72°C for 1 min. The same RT-qPCR protocol was
followed for miRNA detection, primers of human miR-10a-5p and
internal control U6 were also purchased from Takara Bio, Inc.
Amplification and detection were all performed using a 7500HT Fast
Real-Time PCR system (Bio-Rad Laboratories, Inc.). Quantitative
analysis was performed with the 2−ΔΔCq method (18).
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| GAPDH | F:
GCACCGTCAAGGCTGAGAAC |
|
| R:
TGGTGAAGACGCCAGTGGA |
| ALP | F:
GCTCATGCATAACATCAGGGACA |
|
| R:
TCGTCACTCTCATACTCCACATCAG |
| RUNX2 | F:
CACTGGCGCTGCAACAAGA |
|
| R:
CATTCCGGAGCTCAGCAGAATAA |
| U6 | F:
GCTTCGGCAGCACATATACTAAAAT |
|
| R:
CGCTTCACGAATTTGCGTGTCAT |
Cell viability assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) was used to measure the effects of RNAi on
hBMSC viability. Transfected and non-transfected cells were plated
at a concentration of 2×105 cells/well in 6-well plates
with 2 ml complete medium for 12 h. The blank group was 2 ml
complete medium without cells. CCK-8 reagent was added (10
µl/well), and all groups were incubated at 37°C in 5%
CO2 for 4 h. Cell viability was measured at an optical
density (OD) of 450 nm. Cell viability (%) = [OD (transfected
cells)-OD (blank)]/[OD (non-transfected cells)-OD (blank)]
×100%.
Adipogenic differentiation
analysis
Adipogenic differentiation of transfected hBMSCs was
induced using mesenchymal stem cell adipogenic differentiation
medium (cat. no. GUXMX-90031; Cyagen Biosciences, Inc.) containing
10% human mesenchymal stem cell-qualified fetal bovine serum, 1%
penicillin-streptomycin, 0.2% insulin, 0.1% IBMX, 0.1%
rosiglitazone and 0.1% dexamethasone for 7 days. Following cell
differentiation, the adipogenic differentiation medium was removed
from the wells and cells were rinsed with 1X PBS. Cells were then
fixed with 2 ml of 4% formaldehyde solution for 30 min at room
temperature. Wells were rinsed twice with 1X PBS and cells were
stained with 1 ml oil red O (Cyagen Biosciences, Inc.) working
solution (3:2 dilution with distilled water; filtered) for 30 min
at room temperature. Finally, cells were rinsed 2–3 times with 1X
PBS, and were visualized and analyzed under a light microscope.
Western blot analysis
Transfected hBMSCs were lysed with radio
immunoprecipitation buffer (Beyotime Institute of Biotechnology)
and a Bradford kit (Beyotime Institute of Biotechnology) was used
to determine total protein concentration. Proteins (20 µg/lane)
were isolated by SDS-PAGE on 10% gels and were electrotransferred
onto nitrocellulose membranes (cat. no. IPVH00010; EMD Millipore);
membranes were then blocked with 1% BSA (Beyotime Institute of
Biotechnology) to ensure non-specific binding for 60 min at room
temperature. Membranes were incubated with anti-rabbit alkaline
phosphatase (ALP; 1:1,000; cat. no. ab75699; Abcam), anti-rabbit
RUNX2 (1:1,000; cat. no. ab192256; Abcam) and anti-rabbit GAPDH
(1:20,000; cat. no. ab181602; Abcam) primary antibodies at 4°C
overnight. Anti-HRP (1:2,000; cat. no. ab181658; Abcam) secondary
antibody was used to incubate the membrane for 1 h at room
temperature. All antibodies were diluted with dH2O
(Takara Bio, Inc.). Protein bands were observed by enhanced
chemiluminescence (Beyotime Institute of Biotechnology) and exposed
on X-ray film (Kodak). The relative grey values were analyzed by
ImageJ 1.48 software (National Institutes of Health).
Animal model construction
The present study was reviewed and approved as
appropriate and humane by the Institutional Animal Care and Use
Committee of Guangzhou Medical University (Guangzhou, China). All
research was performed in accordance with the regulations as
outlined by the National Institutes of Health (19). Animals were anesthetized before
incision and sacrificed in accordance with the council directive of
the European Community of 24 November 1986 (86/609/EEC), the Care
and Use of Animal Testing Procedures, and local laws and
regulations. A total of 4-week-old, female BALB/c mice (weight, ~20
g) supplied by Guangdong Medical Laboratory Center (Guangzhou,
China) were separated into six groups (blank, control, mimic,
mimic-negative, inhibitor and inhibitor-negative). The mice were
raised in a sterile environment with a relative humidity of 40–70%
at a temperature of 20–26°C, in a light/dark cycle of 12/12 h. The
average feed consumption per 100 g body weight was 5 g, and the
water consumption was 7 ml. Transfected and non-transfected
(control group) hBMSCs were loaded onto 10×8×2 mm3
hydroxyapatite/tricalcium-phosphate (HA/TCP) cubic scaffolds with
porosity of 30% (Department of Inorganic Material, Sichuan
University, Chengdu, China) at a ratio of 1:3 and the loaded
scaffolds were then subcutaneously implanted into the BALB/c mice.
The blank group was transplanted with HA/TCP scaffolds only
(without hBMSCs). Two longitudinal skin incisions ~1.5 cm in length
were made on the dorsal surface of each mouse and subcutaneous
pouches were made using vascular forceps. One HA/TCP implant with
cells was implanted in each pouch (two in each mouse). Surgical
sutures were used to close the incisions and the mice were placed
in separate cages until the end of the experiment. The
aforementioned step was repeated on each mouse in the study. The
graft was maintained in vivo for 4 weeks.
Preparation of specimens for scanning
electron microscopy (SEM)
The scaffolds with transfected hBMSCs and the
non-cellular scaffolds were cultured for 12 h and fixed immediately
at 1% glutaraldehyde for 12 h at 4°C. The specimens were then
rinsed twice with PBS, post-fixed in 1% osmium tetroxide for 1 h at
room temperature, subsequently rinsed twice in PBS and dehydrated
in a graded series of alcohol (40, 70, 90 and 100%) for 10 min each
and allowed to dry. The silver membrane (1,500 nm thick) was
layered on the scaffold surface and prepared for SEM at 10 kV.
Goldner's trichrome staining
After an 8-week transplantation period, the mice
were sacrificed by cervical dislocation. The implants were removed
and fixed with 4% paraformaldehyde at 4°C for 24 h. According to
the manufacturer's instructions, a plastic block with a mineralized
implant was obtained for sectioning using the Technovit 9100 methyl
methacrylate kit (Electron Microscopy Sciences). Using a heavy-duty
microtome (E300CP; EXAKT Advanced Technologies GmbH), three serial
sections (5 µm) were taken close to the surface of the block and
slices were repeated each 100 µm to obtain a total of nine levels
of implantation, by implantation depth. The Goldner's trichrome
staining (Electron Microscopy Sciences) was used for histological
analysis of heterogeneous new bone formation under light
microscopy, following the recommended protocol (20).
Micro-CT analysis of the implants
The implants were scanned with Skycan®
Micro-CT (Bruker Corporation) at 65 kV, 80 µA with an isotropic
resolution of 12 µm of three spatial dimensions, using CTAn v1.18
(Bruker Corporation) analysis for each experimental group. Bone
volume (BV) was calculated using the closed volume triangular
surface corresponding to the tetrahedron. Total bone volume was the
volume of the entire sample being examined.
Statistical analysis
Statistical analysis was performed using SPSS 16.0
(SPSS, Inc.). Experiments were repeated three times and data are
presented as mean ± standard deviation. Comparisons for experiments
with >2 subgroups were analyzed by one-way ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Transfection efficiency
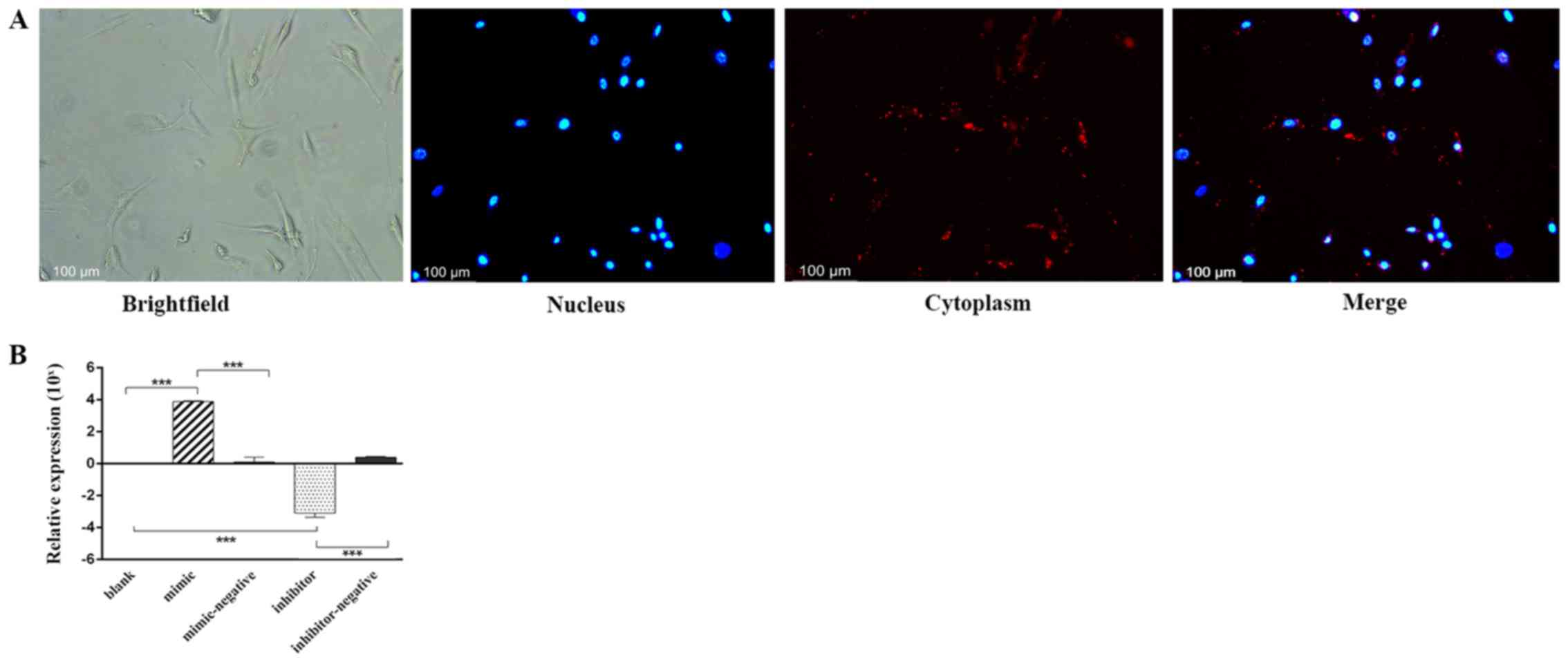
After 24 h of transfection, Alexa Fluor®
555-labeled, dsRNAs appeared in the cytoplasm as a red fluorescence
signal surrounding the nucleus (blue) under an inverted
fluorescence microscope (Fig. 1A).
The relative expression levels of miR-10a-5p were analyzed by
RT-qPCR in the different experimental groups. Compared with the
blank group (classed as 0) and the mimic-negative group, miR-10a-5p
expression in the mimic group was significantly higher. Whereas,
the miR-10a-5p inhibitor group exhibited significantly lower levels
of miR-10a-5p than the blank and the inhibitor-negative group
(P<0.001; Fig. 1B).
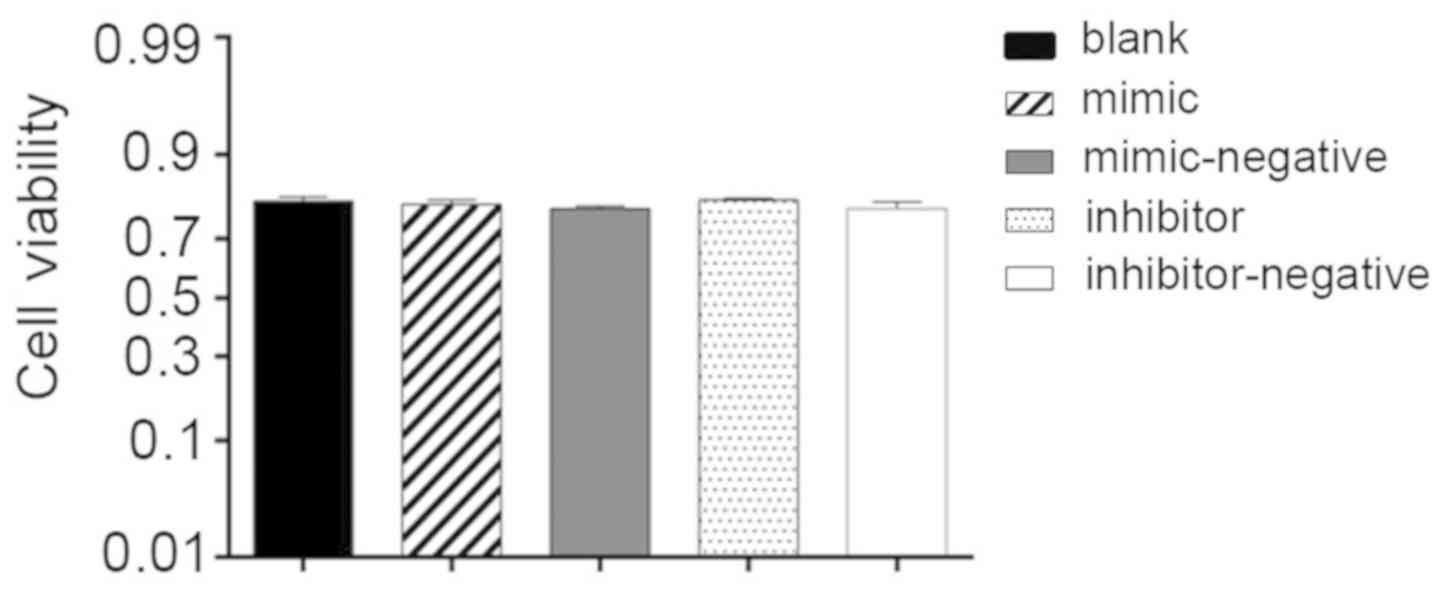
Cell viability
Additionally, 24 h post-transfection hBMSCs,
cultured in the OriCell human mesenchymal stem cell growth medium
with 10 µl CCK-8 reagent, were used to assess cell viability. The
OD values demonstrated no significant difference among the groups
studied and the average percentage of cell viability reached 79%.
Lipofectamine, miR-10a-5p mimics and inhibitors had no influence on
cell viability (Fig. 2).
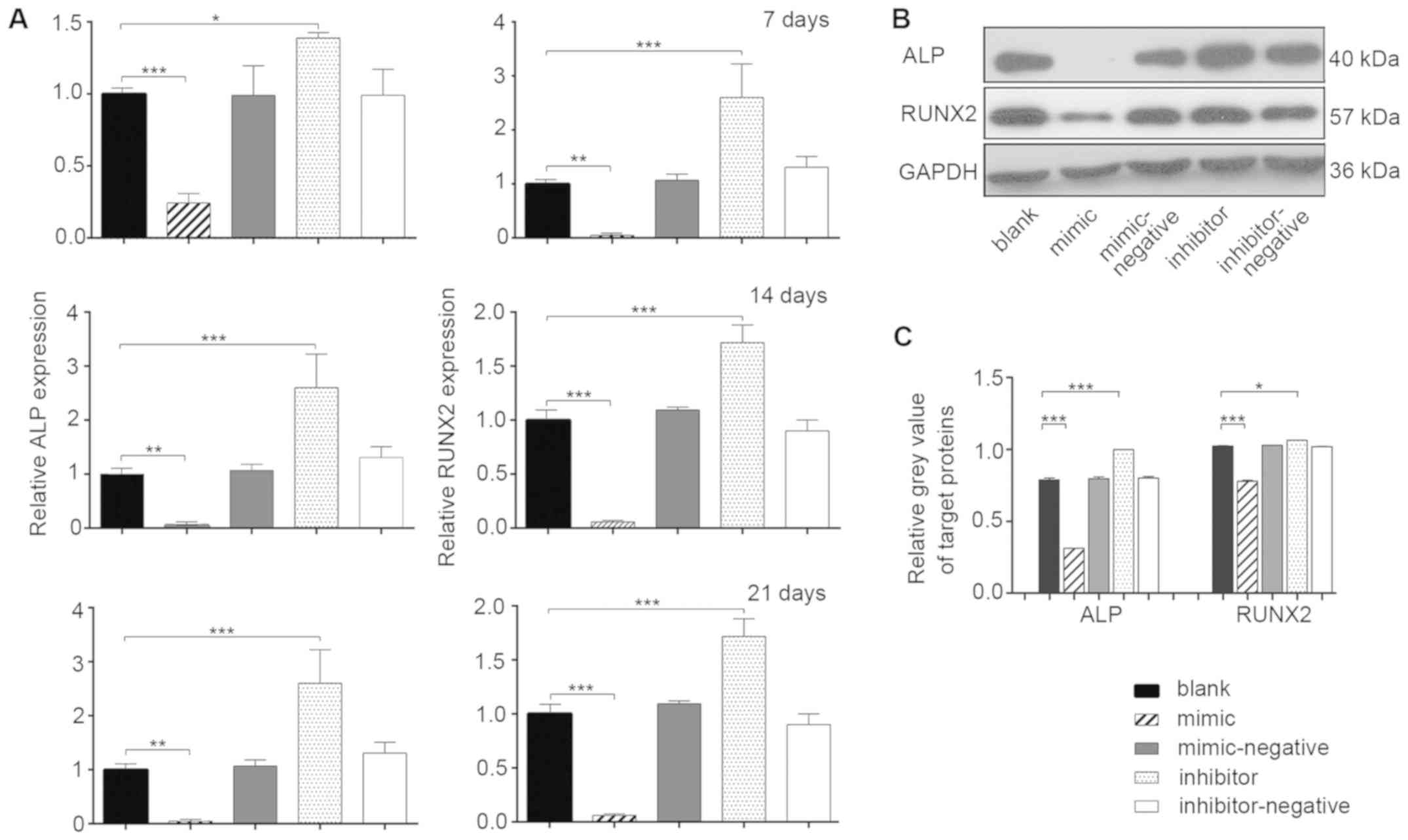
Osteoblast differentiation of hBMSCs
in vitro
mRNA expression levels of mineralization-related
genes were measured during osteoblast differentiation by RT-qPCR at
7, 14 and 21 days post-transfection (Fig. 3A). At all three time points, the
relative expression of ALP in the mimic group was ~1-fold lower
than the blank group (P<0.05). The inhibitor group exhibited a
higher expression level of ALP compared with the blank group
(P<0.05). The changes in the relative expression of RUNX2 in the
various groups were even more significant (P<0.01). The results
indicated that miR-10a-5p had a negative regulatory effect on the
osteogenic processes of hBMSCs (P<0.05). To further investigate
the effects of miR-10a-5p on post-transcription expression, the
protein expression levels of ALP and RUNX2 were evaluated in the
experimental groups using western blotting 14 days
post-transfection (Fig. 3B).
Compared with the blank group, the protein levels of RUNX2 and ALP
were significantly decreased in the mimic group, whereas the levels
were significantly increased in the inhibitor group (P<0.05)
(Fig. 3B and C). No significant
differences were found among the blank, mimic-negative and
inhibitor-negative groups. The protein expression of RUNX2 and ALP
confirmed that miR-10a-5p was a potential negative regulator of
osteoblast differentiation in vitro.
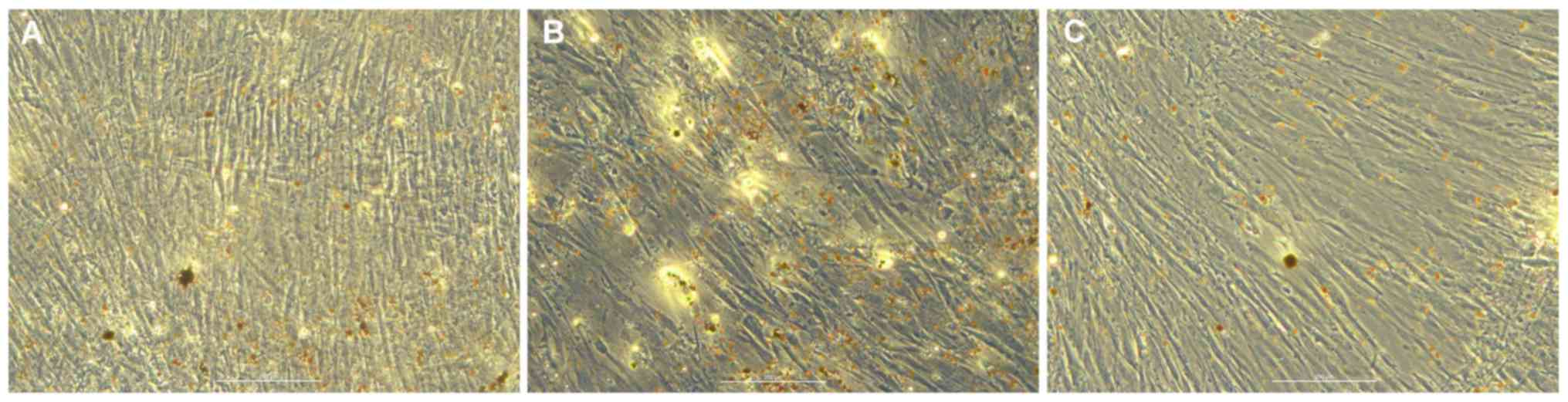
Adipogenic differentiation of hBMSCs
in vitro
Oil red O staining of transfected hBMSCs was
performed 7 days after induction in the blank (Fig. 4A), mimic (Fig. 4B) and inhibitor (Fig. 4C) groups. Compared with the blank
group, the number of lipid droplets in the miR-10a-5p group was
increased, whereas the number of lipid droplets in the inhibitor
group was reduced. Therefore, it was hypothesized that miR-10a-5p
may have a positive regulatory role in hBMSCs adipogenic
differentiations.
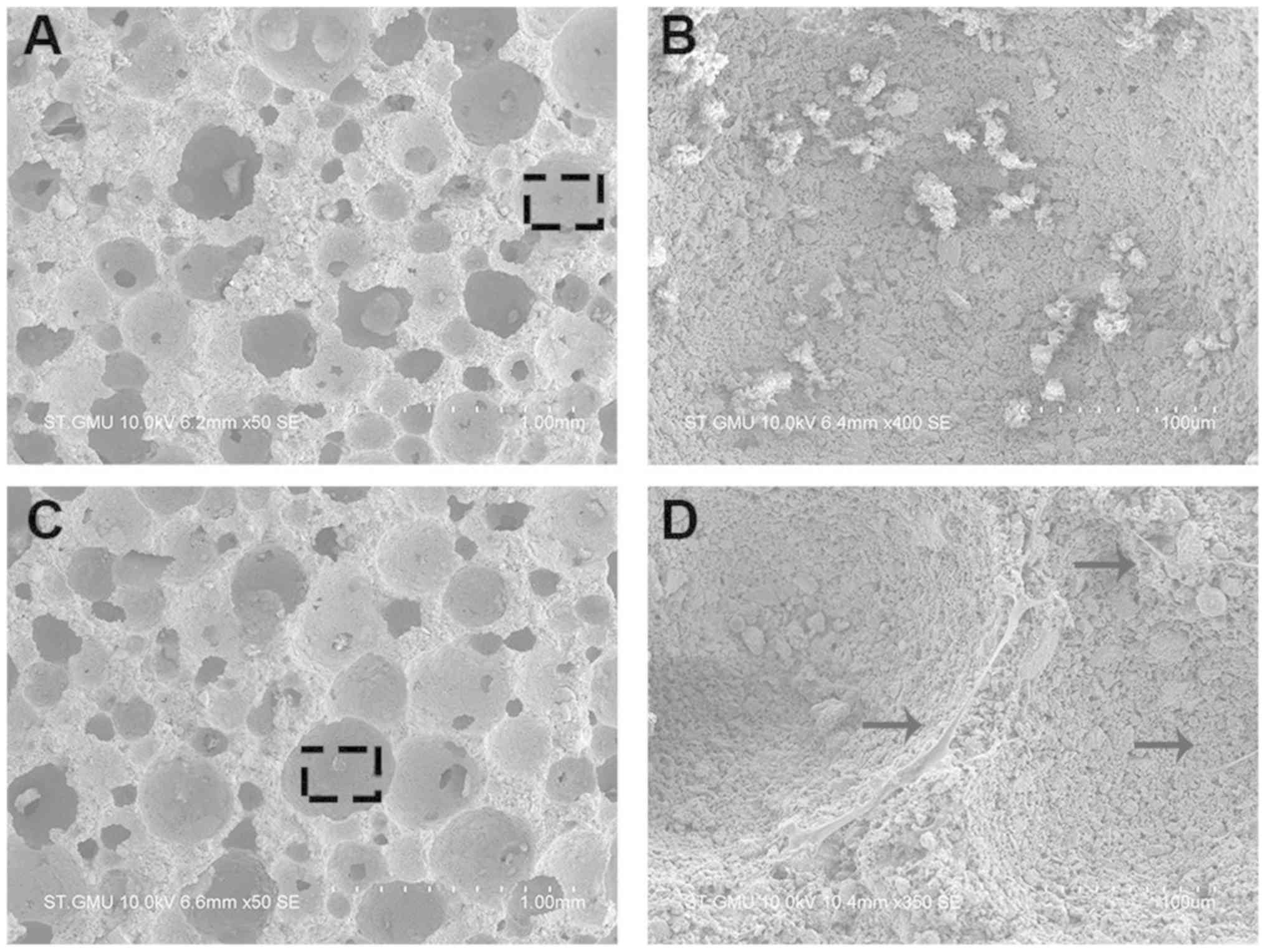
Culturing hBMSCs in the scaffold
To explore whether the regulation of miR-10a-5p
expression in hBMSCs influenced bone formation in vivo, the
transfected hBMSCs were transferred into scaffolds and cultured for
4 days. In an effort to promote ectopic bone formation, hBMSCs were
loaded into the HA/TCP scaffolds, incubated for 24 h and
subcutaneously implanted in BALB/c mice. The hBMSCs morphology and
attachment to the porous surface of the HA/TCP scaffold (Fig. 5A), in comparison with non-cellular
scaffolds used as a control group, was observed using SEM (Fig. 5B). hBMSCs adhered to the surface of
the scaffold and extended their pseudopodia confirming a conducive
environment for growth conditions (Fig. 5C and D).
miR-10a-5p suppresses ectopic bone
formation in vivo
Specimens stained with Goldner's trichrome
illustrated different stages of bone mineralization under the
microscope. Fig. 6A displays a
general overview of histological sections. Bone and bone-like
tissues are presented as a substructure visualized in green and
light purple and the HA/TCP scaffold was presented as a dark green
positive control. The connective tissue is displayed as a
structural network of cells and collagen fibers in orange and pink
(Fig. 6A-F). Inside the pores of
the scaffold, less new bone formation was noted in the
scaffold-only groups in comparison with the scaffolds containing
cells. The mineralization area seen with the blank group was only
slightly more than that seen in the negative controls. As
hypothesized, the inhibitor group produced more new bone tissues
than the blank group. Conversely, the mimic-only group had small
and barely visible purple staining, which confirmed limited amounts
of early stage bone formation (Fig.
6G-L). These results suggested that miR-10a-5p also had a
negative regulation in vivo. Regular breeding mice were
sacrificed at 8 weeks and the scaffold transplants were collected
with some margins of soft tissue. The bone volume values of the
samples were 101.9±3.16 (control), 113.4±3.38 (blank), 107.9±3.21
(mimic), 103.8±8.40 (mimic-negative), 108.3±3.54 (inhibitor) and
104.4±1.84 mm3 (inhibitor-negative) (P>0.05) with no
significant difference noted among the groups. Therefore, with the
volume of the scaffolds ~equal, this would not have an overall
effect on the final results (Fig.
6M).
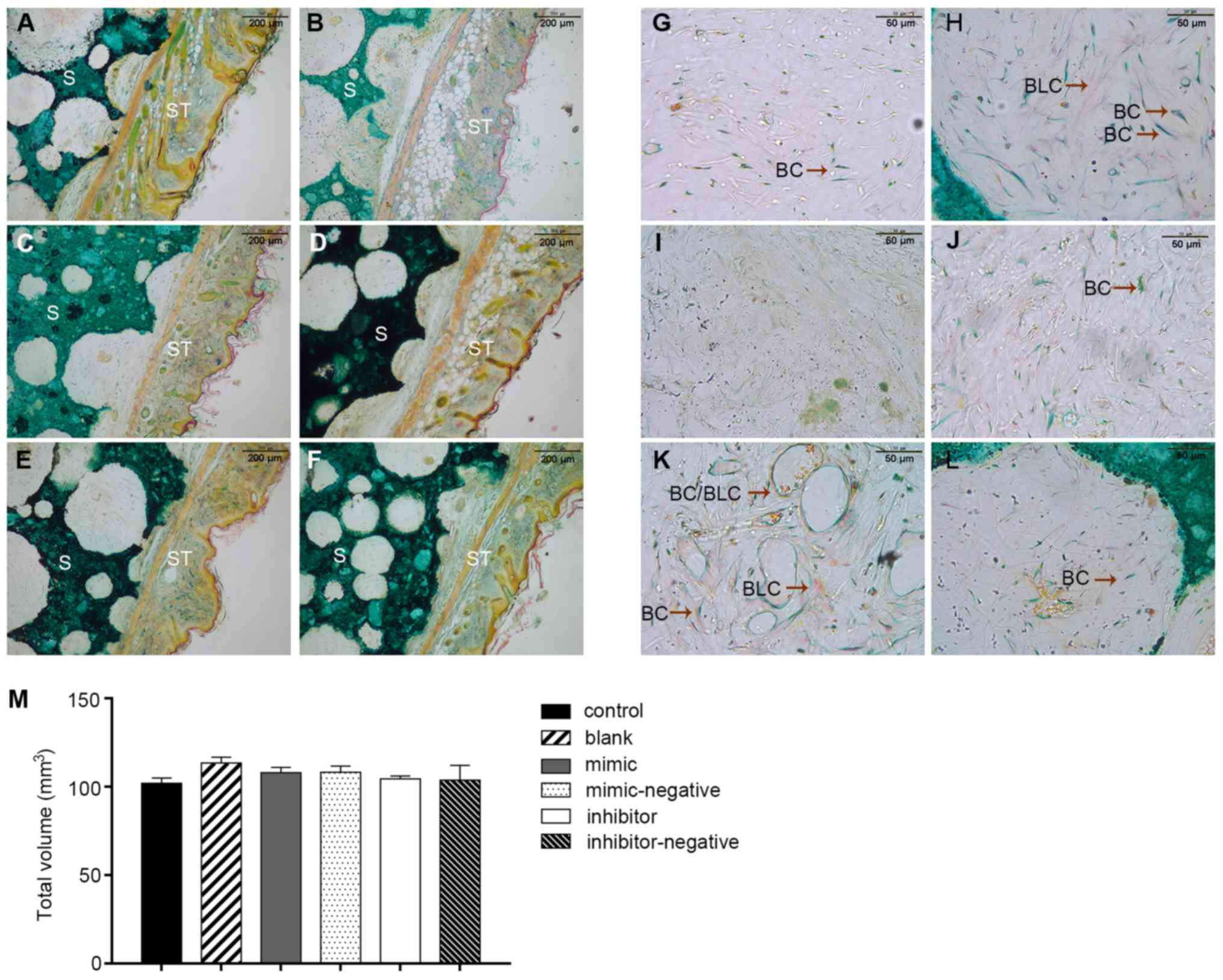 | Figure 6.Two transplants per treatment were
engrafted into mice and three sections of each transplant were
stained and investigated. Images from (A) control, (B) blank, (C)
mimic, (D) mimic-negative, (E) inhibitor and (F) inhibitor-negative
group. Embedded S and ST were stained with Goldner's trichrome. The
cuticular, dermal, subcutaneous tissue and HA/TCP were easily
observed (magnification, ×10). (G-L) Inside the pores of scaffolds,
BC were green and BLC were purple (magnification, ×40). The
scaffold transplants from mice were collected with some margins of
soft tissue. (M) Bone volume values of samples were 101.9±3.16,
113.4±3.38, 107.9±3.21, 103.8±8.40, 108.3±8.4 and 104.4±1.84
mm3 in the groups, with no significant difference noted
among the groups. BC, bone cells; BLC, bone-like cells; HA/TCP,
hydroxyapatite/tricalcium-phosphate; S, scaffolds; ST, soft
tissues. |
Discussion
hBMSCs have a great potential for differentiation
into multiple lineages and serve as the ideal source for remodeling
bone tissue to recover body functions. Their clinical potential for
gene-based therapy applications is rapidly being recognized
(21,22). Several miRNAs are currently being
studied for their involvement in the inhibition of osteoblast
differentiation and may become important for regulating bone
remodeling. The present study explored the role and regulation of
miR-10a-5p in osteogenesis, including in vitro and in
vivo experiments. The influence of miR-10a-5p on the adipogenic
process was also investigated.
RNAi, a conserved evolutionary mechanism widely
found in nature, can induce gene silencing by prohibiting the
translation of the target Mrna (23). RNAi affects the
post-transcriptional expression levels of several genes. miRNAs
pair with mRNA regions and target them for degradation (24). The first step in this process is
ensuring the RNA can base pair with a region of its target gene;
once base pairing is completed, the target proteins, including
drosha, dicer and argonaute, direct the mRNA to nuclease
destruction (25). miRNA mimics
are small, chemically modified dsRNAs, which mimic endogenous
miRNAs and make functional analysis of miRNAs possible by
upregulating miRNA activity. miRNA inhibitors are small, chemically
modified ssRNA molecules that specifically bind and inhibit
endogenous miRNA molecules, thereby enabling functional analysis of
miRNAs by downregulating miRNA activity. Both are recommended for
in vivo and in vitro applications.
In the present study, the role of miR-10a-5p in the
process of bone formation in vitro was studied to provide
evidence for bone formation in vivo. Two classic target
genes are associated with bone formation in the transcriptional and
post-transcriptional stages. RUNX2 belongs to the runt-associated
transcription factor family (26)
and is closely associated with the osteoblast phenotype. The human
RUNX2 gene has been identified and located on chromosome 6p21.
RUNX2, also known as Cbfa1, is an early major transcription factor
that initiates the process of osteoblastic lineage transcription
(27,28). Izu et al (29) demonstrated that the expression of
RUNX2 was increased in early-stage osteoblast development when
compared with wild type cells. Several studies regarding miRNAs
that regulate RUNX2 expression have used MC3T3-E1 and ATDC5 cells,
which originate from human mesenchymal stem cells (30,31).
A previous study noted that miR-30c, miR-133a, miR-135 and miR-338
strongly inhibited RUNX2 protein expression (32). Another study confirmed that C-X-C
motif chemokine ligand 13 mediated upregulation of RUNX2 by
inhibiting miR-23a (33). These
studies all support the results from the present study, which
indicated that RUNX2 expression was inhibited by miR-10a-5p.
However, overexpression of RUNX2 induced the transcription of
miR-10a/10b in breast cancer cell lines, and also impaired cell
motility. Whereas, inhibition of RUNX2 decreased miR-10a/10b
expression and impaired cell migration and invasion ability
(34). ALP is a type of
extracellular membrane protein that is downregulated during bone
formation (35). A previous study
on miR-214 suggested that ALP mRNA expression levels were
continuously decreased in MC3T3-E1 cells for 33 days, starting from
day 6 with an interval of 3 days (36), and the trend observed in the
present study produced similar results. Conversely, overexpression
of miR-210 promoted the osteoblast differentiation of hBMSCs via
the PI3K/AKT pathway by increasing the expression of ALP and
osterix (37). miRNAs modulate
their target genes through different signaling pathways, which
might have opposite outcomes (38–40).
RUNX2 has been reported to directly regulate the transcription of
target genes, including ALP and osteocalcin (41). This suggests why the high
expression of RUNX2 is easily found, whereas there is a slight
discrepancy in the role of ALP expression in the late stage of
osteogenesis.
Only a few studies have examined the role of miRNAs
in bone formation or osteoblast phenotypic development in
vivo (42–45). However, the present study performed
an in vivo investigation and confirmed that new bone
formation resulted from hBMSCs and not host mouse cells. The
chemical composition and crystal structure of HA are very similar
to those of natural bone tissue, and it has valuable
biocompatibility and osteoinductive effects. However, the
absorption of HA is very low (46). In contrast, TCP is an absorbable,
biocompatible bone substitute with improved degradability and bone
regeneration ability compared with HA. Since the degradation of HA
and TCP are complementary, specific proportions must be used in
order for the bone substitute material to have the ideal properties
(optimized dissolution and resorption) for the bone remodeling
process to be effective (47). The
combination of HA and TCP can improve bone regeneration and extend
the medical applications of these artificial materials (48). It has previously been shown that
bidirectional calcium phosphate created by mixing HA and TCP at a
ratio of 1:3 had improved absorbability than that observed with TCP
alone (49).
BALB/c mice lack a thymus, leading to a deficiency
in cellular immunity; however, their humoral immunity is normal. In
this way, rejection and inflammatory reactions are not seen as
serious following surgery, thereby creating a more stable
microenvironment, which is beneficial to transfected hBMSCs
proliferation and differentiation. By choice, the hard tissue
slicing embedment did not require decalcification of the samples.
Therefore, the present study avoided the normal mineralized tissue
being damaged by acid or EDTA, and more clearly observed the
difference between experimental and control groups. Goldner's
trichrome staining contains Weigert's iron hematoxylin, ponceau
acid fuchsin, phosphomolybdic acid-orange G and light green
solutions. This staining procedure is a commonly used method in
bone histology and allows for tissue identification by colorimetric
and morphological differences. In addition, the staining can also
allow for a clear separation of connective tissue and bone matrix
(50). The results of the present
study indicated that miR-10a-5p downregulated bone formation
post-transfection. It has previously been reported that miR-206,
miR-124 and miR-138 inhibited osteoblastogenesis in vivo, by
targeting different signal molecules (17,51,52).
In order to apply these findings to the clinic, further studies are
required to elucidate the mechanism involved.
In the present study, the specific mechanism by
which miR-10a-5p affects the expression of RUNX2 and ALP, and
ultimately affects the process of osteogenic differentiation was
not studied in detail. Liu et al (53) demonstrated that inhibiting the
expression of miR-29b, upregulating DNA methyltransferase 1 and
downregulating Notch1 and Notch intracellular domain, ultimately
upregulated the expression of RUNX2 and ALP via the Notch signaling
pathway. In high-fat diet-fed mice, the Notch1 expression levels
were increased when compared with normal diet-fed mice, whereas the
expression levels of RUNX2 and ALP were relatively low in the
high-fat diet group (54). Zou
et al (55) also reported
that a forward genetic screen yielded the microtubule-associated
protein, DCAMKL1, which suppressed osteoblast activation by
antagonizing RUNX2. DCAMKL1 is associated with several miRNAs,
including miR-200a, let-7a and miR-144, in different types of
tissue (56–58). Notably, marrow adipocytes have been
reported to exhibit osteogenic and adipogenic characteristics, and
are particularly responsive to parathyroid hormone (PTH) and
secrete receptor activator of nuclear factor-κΒ ligand (RANKL)
(59). IL-17A may also mediate the
bone catabolic activity of continuous PTH by upregulating the
production of RANKL to induce bone loss via
Gαs/cAMP/Ca2+ signaling (60). Therefore, there remains much work
to be performed in order to understand the mechanism underlying
miR-10a-5p-regulated osteogenesis.
In conclusion, miR-10a-5p could be utilized as a
negative regulator of osteoblastic differentiation of hBMSCs and
may have a positive effect on adipogenesis. Notably, the results of
the present study demonstrated that the functional inhibition of
miR-10a-5p expedited osteogenic differentiation of hBMSCs and led
to promotion of bone formation in vivo. Furthermore, the
present study suggested that use of miR-10a-5p-targeting treatments
could promote bone formation, and ultimately may be used for the
treatment of pathological bone loss, cardiovascular disorders, bone
and cartilage regeneration, and neuronal damage.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Planning Project of Guangdong Province, China (grant no.
509197952061), the Scientific and Technological Planning Project of
Guangzhou, China (grant no. 201704020118), the Southern Medical
University (youth cultivation project, grant no. PY2017N036), the
Medical Scientific Research Foundation of Guangdong Province, China
(grant nos. C2017061, C2018076 and A2018358), and the Natural
Science Foundation of Guangdong Province, China (grant no.
2018A030313759).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ and ZL conceived and designed the experiments.
LZ, ZZ, FR and LC performed the experiments. YZ, LZ and ZL analyzed
the data and wrote the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was reviewed and approved as
appropriate and humane by the Institutional Animal Care and Use
Committee of Guangzhou Medical University. All research was
performed in accordance with the regulations as outlined by the
National Institutes of Health. Animals were anesthetized and
sacrificed in accordance with the council directive of the European
Community of 24 November 1986 (86/609/EEC), the Care and Use of
Animal Testing Procedures and local laws and regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pichler K, Loreto C, Leonardi R, Reuber T,
Weinberg AM and Musumeci G: Rankl is downregulated in bone cells by
physical activity (treadmill and vibration stimulation training) in
rat with glucocorticoid-induced osteoporosis. Histol Histopathol.
28:1185–1196. 2013.PubMed/NCBI
|
|
2
|
Castrogiovanni P, Trovato FM, Szychlinska
MA, Nsir H, Imbesi R and Musumeci G: The importance of physical
activity in osteoporosis. From the molecular pathways to the
clinical evidence. Histol Histopathol. 31:1183–1194.
2016.PubMed/NCBI
|
|
3
|
Cardinale M and Bosco C: The use of
vibration as an exercise intervention. Exerc Sport Sci Rev. 31:3–7.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Srinivasaiah S, Musumeci G, Mohan T,
Castrogiovanni P, Absenger-Novak M, Zefferer U, Mostofi S, Bonyadi
Rad E, Grün NG, Weinberg AM and Schäfer U: A 300 µm organotypic
bone slice culture model for temporal investigation of endochondral
osteogenesis. Tissue Eng Part C Methods. 25:197–212. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Szychlinska MA, Castrogiovanni P, Nsir H,
Di Rosa MD, Guglielmino C, Parenti R, Calabrese G, Pricoco E,
Salvatorelli L, Magro G, et al: Engineered cartilage regeneration
from adipose tissue derived-mesenchymal stem cells: A
morphomolecular study on osteoblast, chondrocyte and apoptosis
evaluation. Exp Cell Res. 357:222–235. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamada Y, Ueda M, Hibi H and Nagasaka T:
Translational research for injectable tissue-engineered bone
regeneration using mesenchymal stem cells and platelet-rich plasma:
From basic research to clinical case study. Cell Transplant.
13:343–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yamada Y, Nakamura S, Ito K, Kohgo T, Hibi
H, Nagasaka T and Ueda M: Injectable tissue-engineered bone using
autogenous bone marrow-derived stromal cells for maxillary sinus
augmentation: Clinical application report from a 2-6-year
follow-up. Tissue Eng Part A. 14:1699–1707. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rana TM: Illuminating the silence:
Understanding the structure and function of small RNAs. Nat Rev Mol
Cell Biol. 8:23–36. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanzer A, Amemiya CT, Kim CB and Stadler
PF: Evolution of microRNAs located within hox gene clusters. J Exp
Zool B Mol Dev Evol. 304:75–85. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Woltering JM and Durston AJ: Mir-10
represses hoxb1a and hoxb3a in zebrafish. PLoS One. 3:e13962008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mu N, Gu J, Huang T, Zhang C, Shu Z, Li M,
Hao Q, Li W, Zhang W, Zhao J, et al: A novel NF-κB/YY1/MicroRNA-10a
regulatory circuit in fibroblast-like synoviocytes regulates
inflammation in rheumatoid arthritis. Sci Rep. 6:200592016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang D, Zhen L, Yuan T, Huang J, Deng F,
Wuyahan, Zhang H, Pan L, Liu Y, The E, et al: miR-10a regulates
proliferation of human cardiomyocyte progenitor cells by targeting
GATA6. PLoS One. 9:e1030972014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiong G, Huang H, Feng M, Yang G, Zheng S,
You L, Zheng L, Hu Y, Zhang T and Zhao Y: miR-10a-5p targets TFAP2C
to promote gemcitabine resistance in pancreatic ductal
adenocarcinoma. J Exp Clin Cancer Res. 37:762018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and mesenchymal
progenitor cell differentiation. Stem Cells. 28:357–364.
2010.PubMed/NCBI
|
|
17
|
Inose H, Ochi H, Kimura A, Fujita K, Xu R,
Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, et al: A
microRNA regulatory mechanism of osteoblast differentiation. Proc
Natl Acad Sci USA. 106:20794–20799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative pcr and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
National Research Council, . Guide for the
Care and Use of Laboratory Animals: Eighth Edition. The National
Academies Press. (Washington, DC). 2011.
|
|
20
|
Kanczler JM, Ginty PJ, Barry JJ, Clarke
NM, Howdle SM, Shakesheff KM and Oreffo RO: The effect of
mesenchymal populations and vascular endothelial growth factor
delivered from biodegradable polymer scaffolds on bone formation.
Biomaterials. 29:1892–1900. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reiser J, Zhang XY, Hemenway CS, Mondal D,
Pradhan L and La Russa VF: Potential of mesenchymal stem cells in
gene therapy approaches for inherited and acquired diseases. Expert
Opin Biol Ther. 5:1571–1584. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen L, Xu Y, Zhao J, Zhang Z, Yang R, Xie
J, Liu X and Qi S: Conditioned medium from hypoxic bone
marrow-derived mesenchymal stem cells enhances wound healing in
mice. PLoS One. 9:e961612014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bernstein E, Caudy AA, Hammond SM and
Hannon GJ: Role for a bidentateribonuclease in the initiation step
of RNA interference. Nature. 409:363–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Morris KV and Mattick JS: The rise of
regulatory RNA. Nat Rev Genet. 15:423–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Van Wijnen AJ, Stein GS, Gergen JP, Groner
Y, Hiebert SW, Ito Y, Liu P, Neil JC, Ohki M and Speck N:
Nomenclature for Runt-related (RUNX) proteins. Oncogene.
23:4209–4210. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rosen CJ: Bone remodeling, energy
metabolism, and the molecular clock. Cell Metab. 7:7–10. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Musumeci G, Mobasheri A, Trovato FM,
Szychlinska MA, Graziano AC, Lo Furno D, Avola R, Mangano S,
Giuffrida R and Cardile V: Biosynthesis of collagen I, II, RUNX2
and lubricin at different time points of chondrogenic
differentiation in a 3D in vitro model of human mesenchymal stem
cells derived from adipose tissue. Acta Histochem. 116:1407–1417.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Izu Y, Sun M, Zwolanek D, Veit G, Williams
V, Cha B, Jepsen KJ, Koch M and Birk DE: Type XII collagen
regulates osteoblast polarity and communication during bone
formation. J Cell Biol. 193:1115–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Y, Xu F, Pei HX, Zhu X, Lin X, Song
CY, Liang QH, Liao EY and Yuan LQ: Vaspin regulates the osteogenic
differentiation of MC3T3-E1 through the Pi3K-Akt/miR-34c loop. Sci
Rep. 6:255782016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bonyadi Rad E, Musumeci G, Pichler K,
Heidary M, Szychlinska MA, Castrogiovanni P, Marth E, Böhm C,
Srinivasaiah S, Krönke G, et al: Runx2 mediated induction of novel
targets ST2 and Runx3 leads to cooperative regulation of
hypertrophic differentiation in ATDC5 chondrocytes. Sci Rep.
7:179472017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y, Xie RL, Croce CM, Stein JL, Lian
JB, Van Wijnen AJ and Stein GS: A program of microRNAs controls
osteogenic lineage progression by targeting transcription factor
Runx2. Proc Natl Acad Sci USA. 108:9863–9868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian F, Ji XL, Xiao WA, Wang B and Wang F:
CXCL13 promotes osteogenic differentiation of mesenchymal stem
cells by inhibiting miR-23a expression. Stem Cells Int.
2015:6323052015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang CH, Fan TC, Yu JC, Liao GS, Lin YC,
Shih AC, Li WH and Yu AL: The prognostic significance of RUNX2 and
miR-10a/10b and their inter-relationship in breast cancer. J Transl
Med. 12:2572014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Saltiel AR: Structural and functional
roles of glycosylphosphoinositides. Subcell Biochem. 26:65–185.
1996.
|
|
36
|
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang
A, Li D, Hou Z, Lv K, Kan G, et al: miR-214 targets ATF4 to inhibit
bone formation. Nat Med. 19:93–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu XD, Cai F, Liu L, Zhang Y and Yang AL:
MicroRNA-210 is involved in the regulation of postmenopausal
osteoporosis through promotion of VEGF expression and osteoblast
differentiation. Biol Chem. 396:339–347. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen E, Diao X, Wei C, Wu Z, Zhang L and
Hu B: MicroRNAs target gene and signaling pathway by bioinformatics
analysis in the cardiac hypertrophy. Biochem Biophys Res Commun.
397:380–385. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao Q, Li YY, He WF, Zhang ZZ, Zhou Q, Liu
X, Shen Y and Huang TT: Interplay between microRNAs and the STAT3
signaling pathway in human cancers. Physiol Genomics. 45:1206–1214.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang W, Xie Y, Xu L, Wang Y, Zhu X, Wang
R, Zhang Y, Muleke EM and Liu L: Identification of microRNAs and
their target genes explores miRNA-mediated regulatory network of
cytoplasmic male sterility occurrence during anther development in
radish (Raphanus sativus L.). Front Plant Sci.
7:10542016.PubMed/NCBI
|
|
41
|
Otto F, Lübbert M and Stock M: Upstream
and downstream targets of RUNX proteins. J Cell Biochem. 89:9–18.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park J, Wada S, Ushida T and Akimoto T:
The microRNA-23a has limited roles in bone formation and
homeostasis in vivo. Physiol Res. 64:711–719. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Taipaleenmä H, Eskildsen T, Stenvang J,
Abdallah MB, Ditzel N, Säämäne AM, Kauppine S and Kasse M: Mir-138
is a novel regulator of in vivo bone formation. Bone. 47 (Suppl
1):S462010. View Article : Google Scholar
|
|
44
|
Chen L, Holmstrøm K, Qiu W, Ditzel N, Shi
K, Hokland L and Kassem M: MicroRNA-34a inhibits osteoblast
differentiation and in vivo bone formation of human stromal stem
cells. Stem Cells. 32:902–912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu L, Liu M, Li R, Liu H, Du L, Chen H,
Zhang Y, Zhang S and Liu D: MicroRNA-503-5p inhibits
stretch-induced osteogenic differentiation and bone formation. Cell
Biol Int. 41:112–123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hattori S: Structural features of ectopic
bone-like tissue in porous hydroxyapatite blocks. Kokubyo Gakkai
Zasshi. 75:120–137. 2008.(In Japanese). View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mayr H, Schlüfter S, Detsch R and Ziegler
G: Influence of phase composition on degradation and resorption of
biphasic calcium phosphate ceramics. Key Eng Mater 361-363.
1043–1046. 2008.
|
|
48
|
Daculsi G, Laboux O, Malard O and Weiss P:
Current state of the art of biphasic calcium phosphate bioceramics.
J Mater Sci Mater Med. 14:195–200. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yamada S, Heymann D, Bouler JM and Daculsi
G: Osteoclasticresorption of calcium phosphate ceramics with
different hydroxyapatite/beta-tricalcium phosphate ratios.
Biomaterials. 18:1037–1041. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rentsch C, Schneiders W, Manthey S,
Rentsch B and Rammelt S: Comprehensive histological evaluation of
bone implants. Biomatter. 4:e279932014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Eskildsen T, Taipaleenmäki H, Stenvang J,
Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S and Kassem M:
MicroRNA-138 regulates osteogenic differentiation of human stromal
(mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA.
108:6139–6144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qadir AS, Um S, Lee H, Baek K, Seo BM, Lee
G, Kim GS, Woo KM, Ryoo HM and Baek JH: miR-124 negatively
regulates osteogenic differentiation and in vivo bone formation of
mesenchymal stem cells. J Cell Biochem. 116:730–742. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu S, Liu D, Chen C, Hamamura K,
Moshaverinia A, Yang R, Liu Y, Jin Y and Shi S: MSC transplantation
improves osteopenia via epigenetic regulation of notch signaling in
lupus. Cell Metab. 22:606–618. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Luo Y, Chen GL, Hannemann N, Ipseiz N,
Krönke G, Bäuerle T, Munos L, Wirtz S, Schett G and Bozec A:
Microbiota from obese mice regulate hematopoietic stem cell
differentiation by altering the bone niche. Cell Metab. 22:886–894.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zou W, Greenblatt MB, Brady N, Lotinun S,
Zhai B, de Rivera H, Singh A, Sun J, Gygi SP, Baron R, et al: The
microtubule-associated protein DCAMKL1 regulates osteoblast
function via repression of Runx2. J Exp Med. 210:1793–1806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sureban SM, May R, Lightfoot SA, Hoskins
AB, Lerner M, Brackett DJ, Postier RG, Ramanujam R, Mohammed A, Rao
CV, et al: DCAMKL-1 regulates epithelial-mesenchymal transition in
human pancreatic cells through a miR-200a-dependent mechanism.
Cancer Res. 71:2328–2338. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sureban SM, May R, Ramalingam S,
Subramaniam D, Natarajan G, Anant S and Houchen CW: Selective
blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a
MicroRNA-dependent mechanism. Gastroenterology. 137:649–659. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cao T, Li H, Hu Y, Ma D and Cai X: miR-144
suppresses the proliferation and metastasis of hepatocellular
carcinoma by targeting E2F3. Tumour Biol. 35:10759–10764. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fan Y, Hanai JI, Le PT, Bi R, Maridas D,
DeMambro V, Figueroa CA, Kir S, Zhou X, Mannstadt M, et al:
Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell
Metab. 25:661–672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li JY, D'Amelio P, Robinson J, Walker LD,
Vaccaro C, Luo T, Tyagi AM, Yu M, Reott M, Sassi F, et al: IL-17A
is increased in humans with primary hyperparathyroidism and
mediates PTH-induced bone loss in mice. Cell Metab. 22:799–810.
2015. View Article : Google Scholar : PubMed/NCBI
|