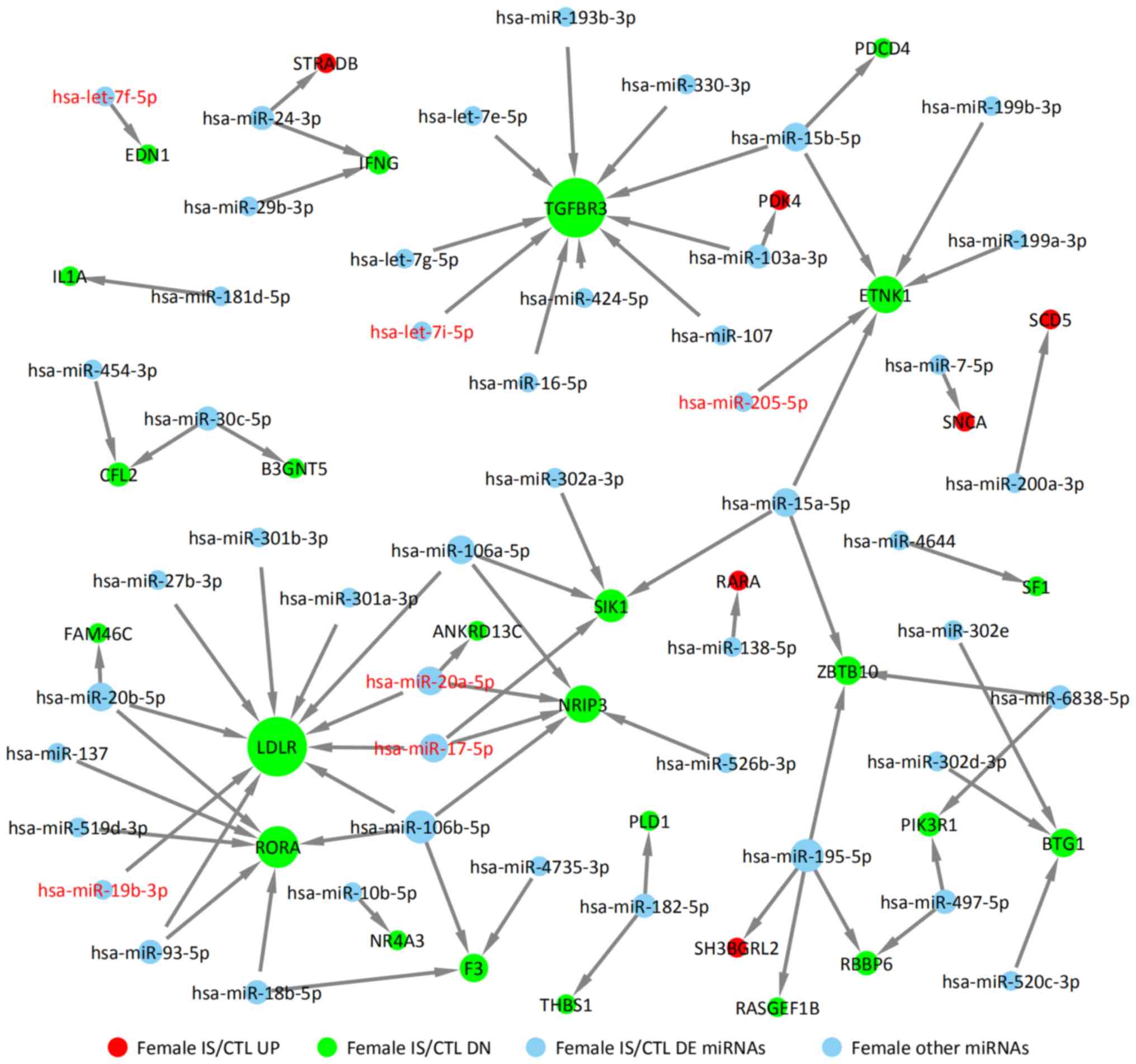
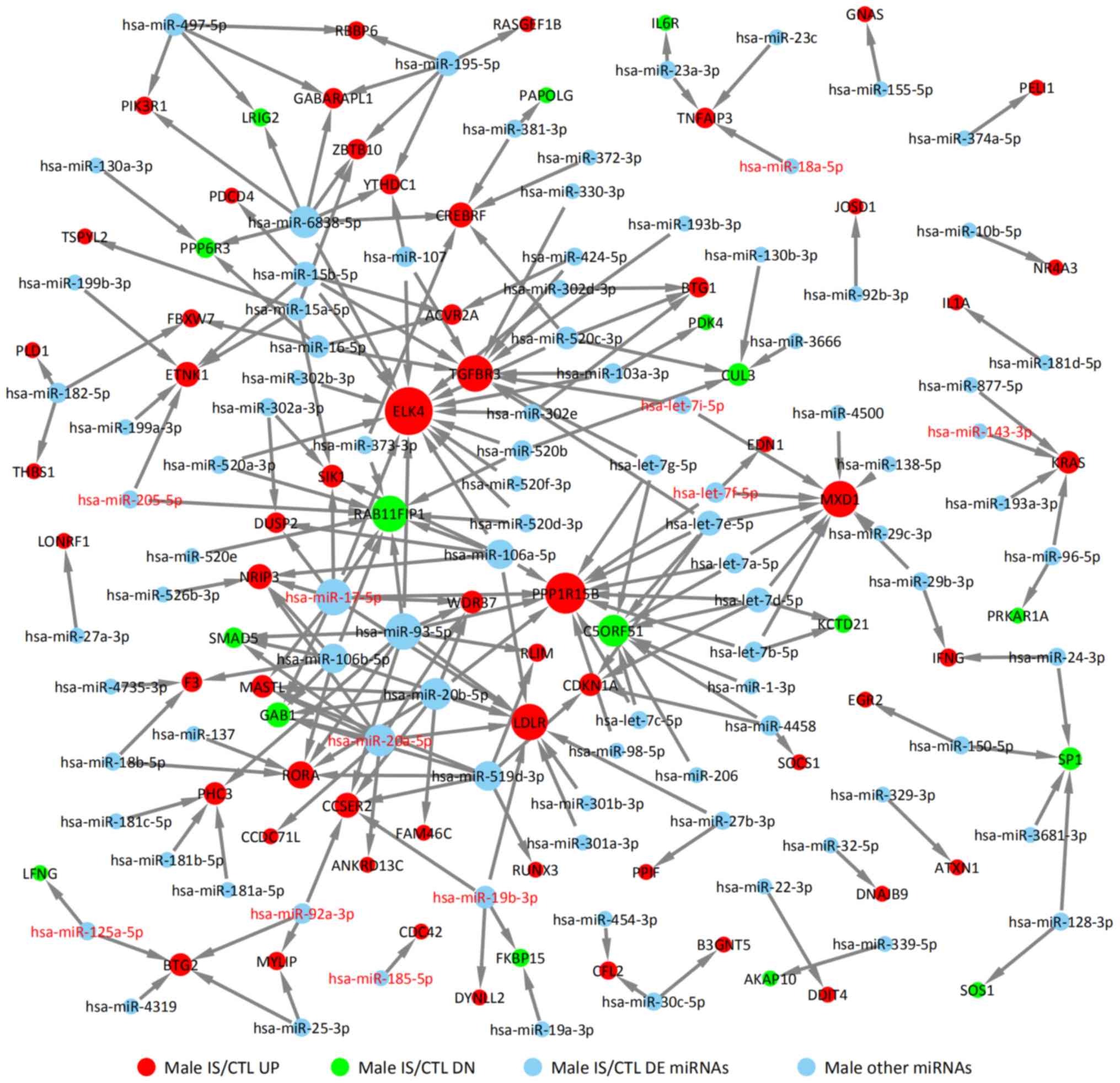
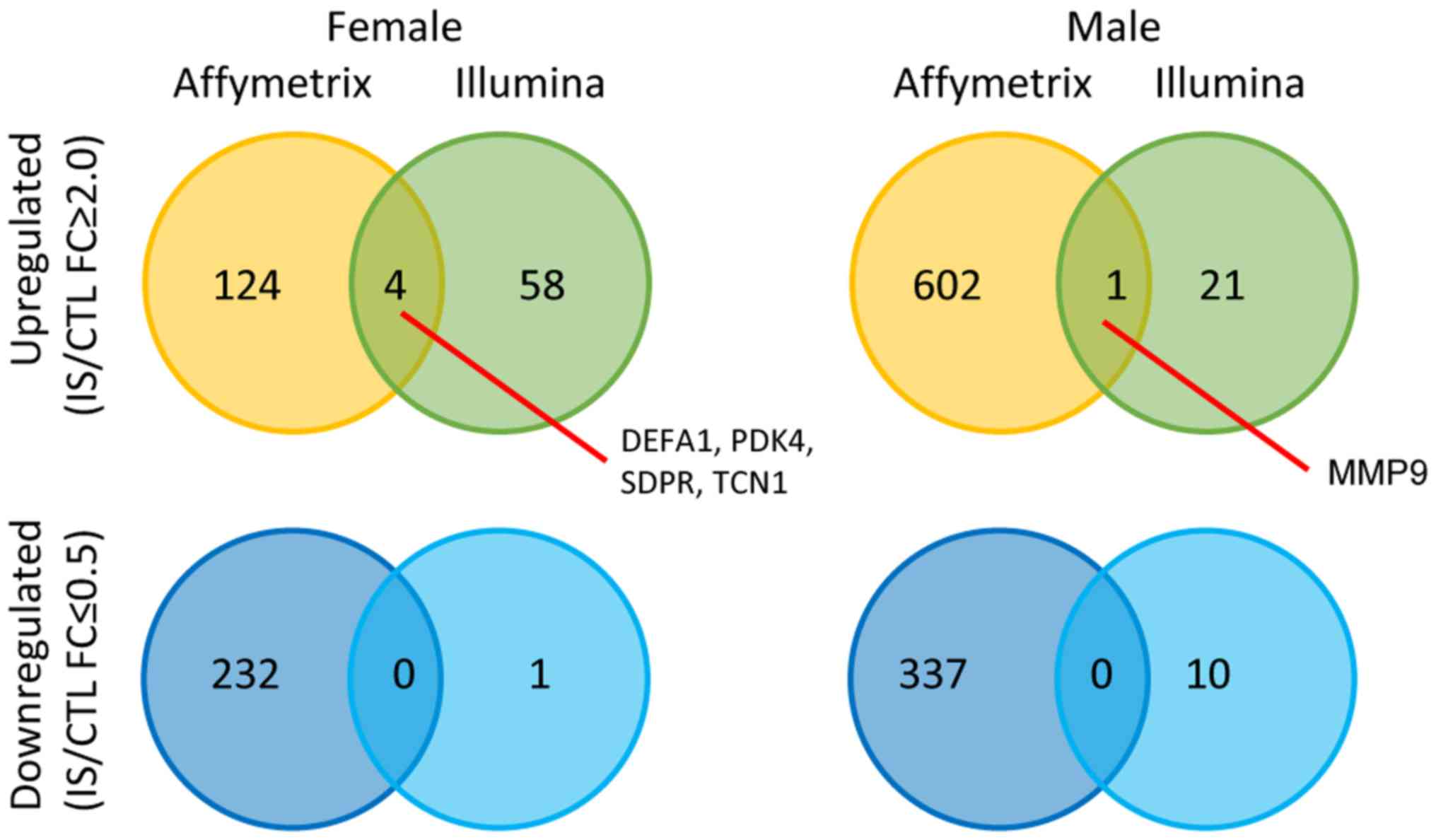
|
1
|
Ishak B, Abdul-Jabbar A, Singla A, Yilmaz
E, von Glinski A, Ramey WL, Blecher R, Tymchak Z, Oskouian R and
Chapman JR: Intraoperative ischemic stroke in elective spine
surgery: A retrospective study of incidence and risk. Spine (Phila
Pa 1976). 45:109–115. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weiss HR, Mellender SJ, Kiss GK, Liu X and
Chi OZ: Improvement in microregional oxygen supply/consumption
balance and infarct size after cerebral ischemia-reperfusion with
inhibition of p70 ribosomal S6 kinase (S6K1). J Stroke Cerebrovasc
Dis. 28:1042762019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weisberg LA: ‘Stroke and struck’:
Protecting the brain from cerebrovascular disease. South Med J.
96:3312003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Singh RB, Suh IL, Singh VP, Chaithiraphan
S, Laothavorn P, Sy RG, Babilonia NA, Rahman AR, Sheikh S,
Tomlinson B and Sarraf-Zadigan N: Hypertension and stroke in Asia:
Prevalence, control and strategies in developing countries for
prevention. J Hum Hypertens. 14:749–763. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
West R, Hill K, Hewison J, Knapp P and
House A: Psychological disorders after stroke are an important
influence on functional outcomes: A prospective cohort study.
Stroke. 41:1723–1727. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ickenstein GW, Horn M, Schenkel J,
Vatankhah B, Bogdahn U, Haberl R and Audebert HJ: The use of
telemedicine in combination with a new stroke-code-box
significantly increases t-PA use in rural communities. Neurocrit
Care. 3:27–32. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Keller L, Hobohm C, Zeynalova S, Classen J
and Baum P: Does treatment with t-PA increase the risk of
developing epilepsy after stroke? J Neurol. 262:2364–2372. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rudnick SI, Swaminathan J, Sumaroka M,
Liebhaber S and Gewirtz AM: Effects of local mRNA structure on
posttranscriptional gene silencing. Proc Natl Acad Sci USA.
105:13787–13792. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park JH, Ahn JH, Song M, Kim H, Park CW,
Park YE, Lee TK, Lee JC, Kim DW, Lee CH, et al: A 2-min transient
ischemia confers cerebral ischemic tolerance in non-obese gerbils,
but results in neuronal death in obese gerbils by increasing
abnormal mTOR activation-mediated oxidative stress and
neuroinflammation. Cell. 8:11262019. View Article : Google Scholar
|
|
10
|
Liu S, Jin R, Xiao AY, Zhong W and Li G:
Inhibition of CD147 improves oligodendrogenesis and promotes white
matter integrity and functional recovery in mice after ischemic
stroke. Brain Behav Immun. 82:13–24. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feschotte C, Jiang N and Wessler SR: Plant
transposable elements: Where genetics meets genomics. Nat Rev
Genet. 3:329–341. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sunkar R and Zhu JK: Novel and
stress-regulated microRNAs and other small RNAs from Arabidopsis.
Plant Cell. 16:2001–2019. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Crippa S, Cassano M and Sampaolesi M: Role
of miRNAs in muscle stem cell biology: Proliferation,
differentiation and death. Curr Pharm Des. 18:1718–1729. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumar P, Wu H, McBride JL, Jung KE, Kim
MH, Davidson BL, Lee SK, Shankar P and Manjunath N: Transvascular
delivery of small interfering RNA to the central nervous system.
Nature. 448:39–43. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mehler MF and Mattick JS: Non-coding RNAs
in the nervous system. J Physiol. 575:333–341. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jung HL, Choi J, Hwang YK, Kim DH, Yoo KH,
Sung KW and Koo HH: Inhibition of survivin expression by small
interfering RNA (siRNA) in brain tumor cell lines. Cancer Res.
65:2005.
|
|
18
|
Pantano L, Friedländer MR, Escaramís G,
Lizano E, Pallarès-Albanell J, Ferrer I, Estivill X and Martí E:
Specific small-RNA signatures in the amygdala at premotor and motor
stages of Parkinson's disease revealed by deep sequencing analysis.
Bioinformatics. 32:673–681. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roy J, Sarkar A, Parida S, Ghosh Z and
Mallick B: Small RNA sequencing revealed dysregulated piRNAs in
Alzheimer's disease and their probable role in pathogenesis. Mol
Biosyst. 13:565–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bam M, Yang X, Sen S, Zumbrun EE, Dennis
L, Zhang J, Nagarkatti PS and Nagarkatti M: Characterization of
dysregulated miRNA in peripheral blood mononuclear cells from
ischemic stroke patients. Mol Neurobiol. 55:1419–1429. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma Q, Zhao H, Tao Z, Wang R, Liu P, Han Z,
Ma S, Luo Y and Jia J: MicroRNA-181c exacerbates brain injury in
acute ischemic stroke. Aging Dis. 7:705–714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Watson SJ and Akil H: Gene chips and
arrays revealed: A primer on their power and their uses. Biol
Psychiatry. 45:533–543. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Johnston M: Gene chips: Array of hope for
understanding gene regulation. Curr Biol. 8:R171–R174. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hochreiter S, Clevert DA and Obermayer K:
A new summarization method for Affymetrix probe level data.
Bioinformatics. 22:943–949. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Teng L, Wang K, Liu Y, Ma Y, Chen W and Bi
L: Based on integrated bioinformatics analysis identification of
biomarkers in hepatocellular carcinoma patients from different
regions. Biomed Res Int. 2019:17423412019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gelaw YA, Williams G, Assefa Y, Asressie M
and Soares Magalhães RJ: Sociodemographic profiling of tuberculosis
hotspots in Ethiopia, 2014–2017. Trans R Soc Trop Med Hyg.
113:379–391. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thodberg M, Thieffry A, Vitting-Seerup K,
Andersson R and Sandelin A: CAGEfightR: Analysis of 5′-end data
using R/Bioconductor. 20:4872019.PubMed/NCBI
|
|
28
|
Postma M and Goedhart J: PlotsOfData-A web
app for visualizing data together with their summaries. PLoS Biol.
17:e30002022019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doncheva NT, Morris JH, Gorodkin J and
Jensen LJ: Cytoscape StringApp: Network analysis and visualization
of proteomics data. J Proteome Res. 18:623–632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Scardoni G, Tosadori G, Faizan M, Spoto F,
Fabbri F and Laudanna C: Biological network analysis with
CentiScaPe: Centralities and experimental dataset integration.
F1000Res. 3:1392014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kalhori MR, Arefian E, Fallah Atanaki F,
Kavousi K and Soleimani M: miR-548× and miR-4698 controlled cell
proliferation by affecting the PI3K/AKT signaling pathway in
Glioblastoma cell lines. Sci Rep. 10:15582020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang D, Zhao X, Zhang L and Wang C:
Comprehensive analysis of pseudogene HSPB1P1 and its potential
roles in hepatocellular carcinoma. J Cell Physiol. Jan
27–2020.(Epub ahead of print). View Article : Google Scholar
|
|
33
|
Jiang W, Zheng L, Yan Q, Chen L and Wang
X: miR-532-3p inhibits metastasis and proliferation of non-small
cell lung cancer by targeting FOXP3. J BUON. 24:2287–2293.
2019.PubMed/NCBI
|
|
34
|
Sticht C, De La Torre C, Parveen A and
Gretz N: miRWalk: An online resource for prediction of microRNA
binding sites. PLoS One. 13:e02062392018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Monbailliu T, Goossens J and
Hachimi-Idrissi S: Blood protein biomarkers as diagnostic tool for
ischemic stroke: A systematic review. Biomark Med. 11:503–512.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li P, Teng F, Gao F, Zhang M, Wu J and
Zhang C: Identification of circulating microRNAs as potential
biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiol.
35:433–447. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dolmans LS, Rutten FH, Koenen NCT,
Bartelink MEL, Reitsma JB, Kappelle LJ and Hoes AW: Candidate
biomarkers for the diagnosis of transient ischemic attack: A
systematic review. Cerebrovasc Dis. 47:207–216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McColl BW, Rothwell NJ and Allan SM:
Systemic inflammatory stimulus potentiates the acute phase and CXC
chemokine responses to experimental stroke and exacerbates brain
damage via interleukin-1- and neutrophil-dependent mechanisms. J
Neurosci. 27:4403–4412. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miller MC and Mayo KH: Chemokines from a
structural perspective. Int J Mol Sci. 18(pii): E20882017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mirabelli-Badenier M, Braunersreuther V,
Viviani GL, Dallegri F, Quercioli A, Veneselli E, Mach F and
Montecucco F: CC and CXC chemokines are pivotal mediators of
cerebral injury in ischaemic stroke. Thromb Haemost. 105:409–420.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Griffith JW, Sokol CL and Luster AD:
Chemokines and chemokine receptors: Positioning cells for host
defense and immunity. Annu Rev Immunol. 32:659–702. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brait VH, Rivera J, Broughton BR, Lee S,
Drummond GR and Sobey CG: Chemokine-related gene expression in the
brain following ischemic stroke: No role for CXCR2 in outcome.
Brain Res. 1372:169–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Villa P, Triulzi S, Cavalieri B, Di
Bitondo R, Bertini R, Barbera S, Bigini P, Mennini T, Gelosa P,
Tremoli E, et al: The interleukin-8 (IL-8/CXCL8) receptor inhibitor
reparixin improves neurological deficits and reduces long-term
inflammation in permanent and transient cerebral ischemia in rats.
Mol Med. 13:125–133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Harris RA, Bowker-Kinley MM, Huang B and
Wu PJ: Regulation of the activity of the pyruvate dehydrogenase
complex. Adv Enzyme Regul. 42:249–259. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Holness MJ and Sugden MC: Regulation of
pyruvate dehydrogenase complex activity by reversible
phosphorylation. Biochem Soc Trans. 31:1143–1151. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cohoon KP, Criqui MH, Budoff MJ, Lima JA,
Blaha MJ, Decker PA, Durazo R, Liu K and Kramer H: Relationship of
aortic wall distensibility to mitral and aortic valve
calcification: The multi-ethnic study of atherosclerosis.
Angiology. 69:443–448. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang R, Zhu Y, Wang Y, Ma W, Han X, Wang X
and Liu N: HIF-1α/PDK4/autophagy pathway protects against advanced
glycation end-products induced vascular smooth muscle cell
calcification. Biochem Biophys Res Commun. 517:470–476. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sugden MC and Holness MJ: Recent advances
in mechanisms regulating glucose oxidation at the level of the
pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol
Metab. 284:E855–E862. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tobin MK, Bonds JA, Minshall RD,
Pelligrino DA, Testai FD and Lazarov O: Neurogenesis and
inflammation after ischemic stroke: What is known and where we go
from here. J Cereb Blood Flow Metab. 34:1573–1584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cao Q, Wu J, Wang X and Song C: Noncoding
RNAs in vascular aging. Oxid Med Cell Longev. 2020:79149572020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gupta MK, Halley C, Duan ZH, Lappe J,
Viterna J, Jana S, Augoff K, Mohan ML, Vasudevan NT, Na J, et al:
miRNA-548c: A specific signature in circulating PBMCs from dilated
cardiomyopathy patients. J Mol Cell Cardiol. 62:131–141. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cho EY, Lee SJ, Nam KW, Shin J, Oh KB, Kim
KH and Mar W: Amelioration of oxygen and glucose
deprivation-induced neuronal death by chloroform fraction of bay
leaves (Laurus nobilis). Biosci Biotechnol Biochem. 74:2029–2035.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He W, Chen S, Chen X, Li S and Chen W:
Bioinformatic analysis of potential microRNAs in ischemic stroke. J
Stroke Cerebrovasc Dis. 25:1753–1759. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sørensen SS, Nygaard AB, Nielsen MY,
Jensen K and Christensen T: miRNA expression profiles in
cerebrospinal fluid and blood of patients with acute ischemic
stroke. Transl Stroke Res. 5:711–718. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huang D, Chen Z, Wang J, Chen Y, Liu D and
Lin K: MicroRNA-221 is a potential biomarker of myocardial
hypertrophy and fibrosis in hypertrophic obstructive
cardiomyopathy. Biosci Rep. 40(pii): BSR201912342020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Di Martino MT, Arbitrio M, Fonsi M,
Erratico CA, Scionti F, Caracciolo D, Tagliaferri P and Tassone P:
Allometric scaling approaches for predicting human pharmacokinetic
of a locked nucleic acid oligonucleotide targeting
cancer-associated miR-221. Cancers (Basel). 12(pii): E272019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wei YS, Xiang Y, Liao PH, Wang JL and Peng
YF: An rs4705342 T>C polymorphism in the promoter of miR-143/145
is associated with a decreased risk of ischemic stroke. Sci Rep.
6:346202016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liang Y, Xu J, Wang Y, Tang JY, Yang SL,
Xiang HG, Wu SX and Li XJ: Inhibition of miRNA-125b decreases
cerebral ischemia/reperfusion injury by targeting CK2α/NADPH
oxidase signaling. Cell Physiol Biochem. 45:1818–1826. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li JQ, Hu SY, Wang ZY, Lin J, Jian S, Dong
YC, Wu XF, Lan D and Cao LJ: MicroRNA-125-5p targeted CXCL13: A
potential biomarker associated with immune thrombocytopenia. Am J
Transl Res. 7:772–780. 2015.PubMed/NCBI
|
|
60
|
Chen T, Huang Z, Wang L, Wang Y, Wu F,
Meng S and Wang C: MicroRNA-125a-5p partly regulates the
inflammatory response, lipid uptake, and ORP9 expression in
oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 83:131–139.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shi C, Na N, Zhu X and Xu J: Estrogenic
effect of ginsenoside Rg1 on APP processing in post-menopausal
platelets. Platelets. 24:51–62. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Giustino G, Overbey J, Taylor D, Ailawadi
G, Kirkwood K, DeRose J, Gillinov MA, Dagenais F, Mayer ML,
Moskowitz A, et al: Sex-based differences in outcomes after mitral
valve surgery for severe ischemic mitral regurgitation: From the
cardiothoracic surgical trials network. JACC Heart Fail. 7:481–490.
2019. View Article : Google Scholar : PubMed/NCBI
|