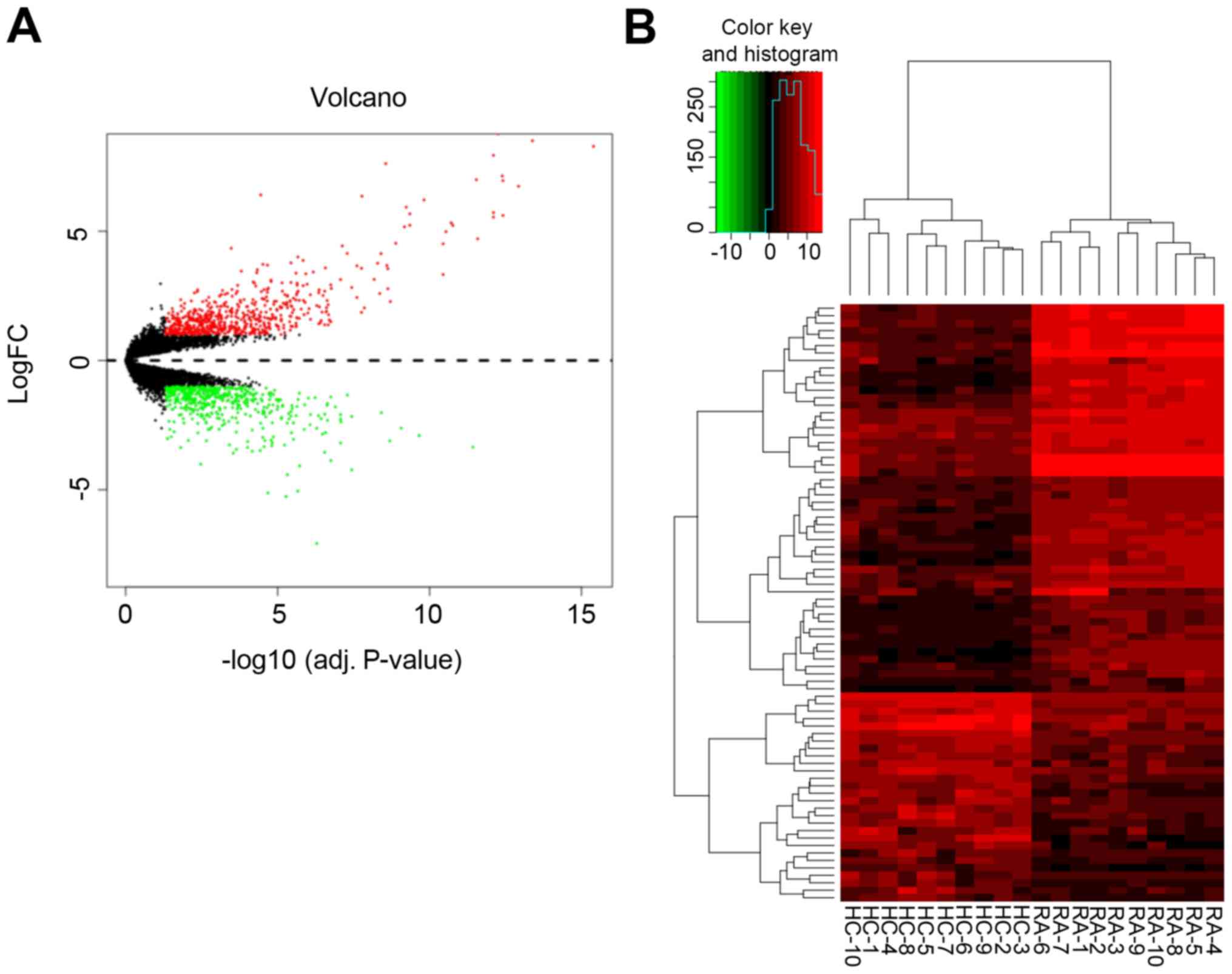
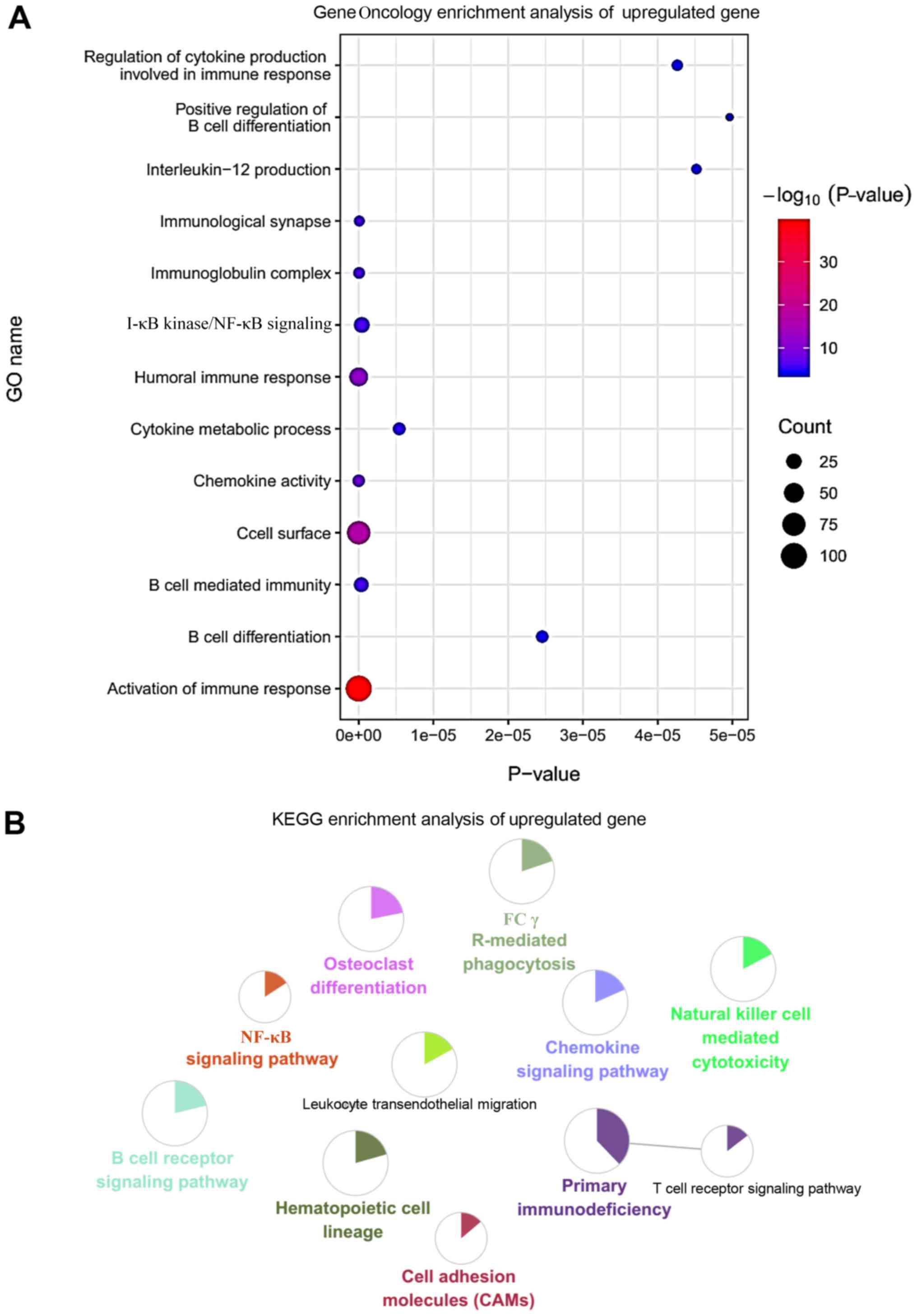
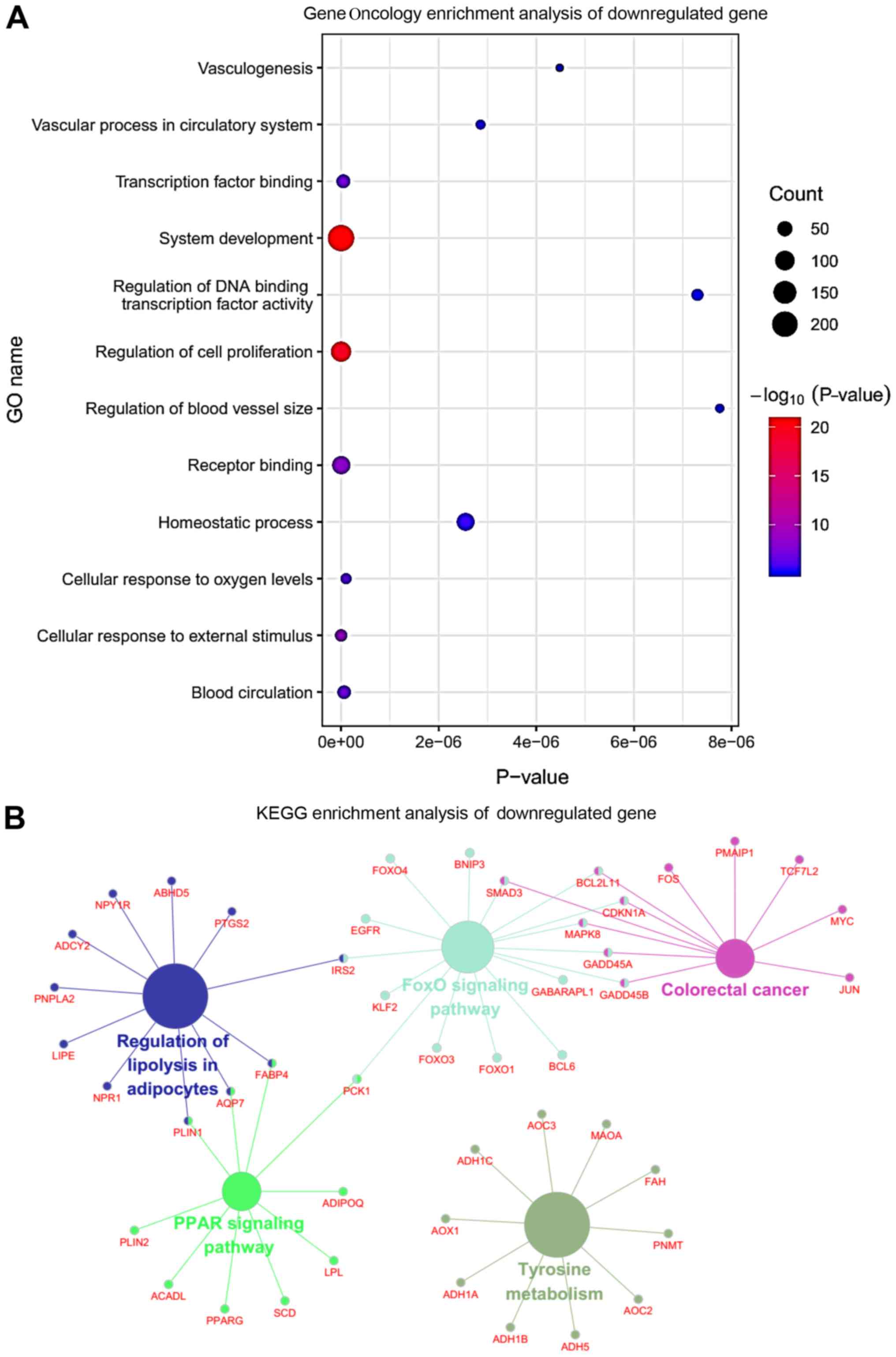
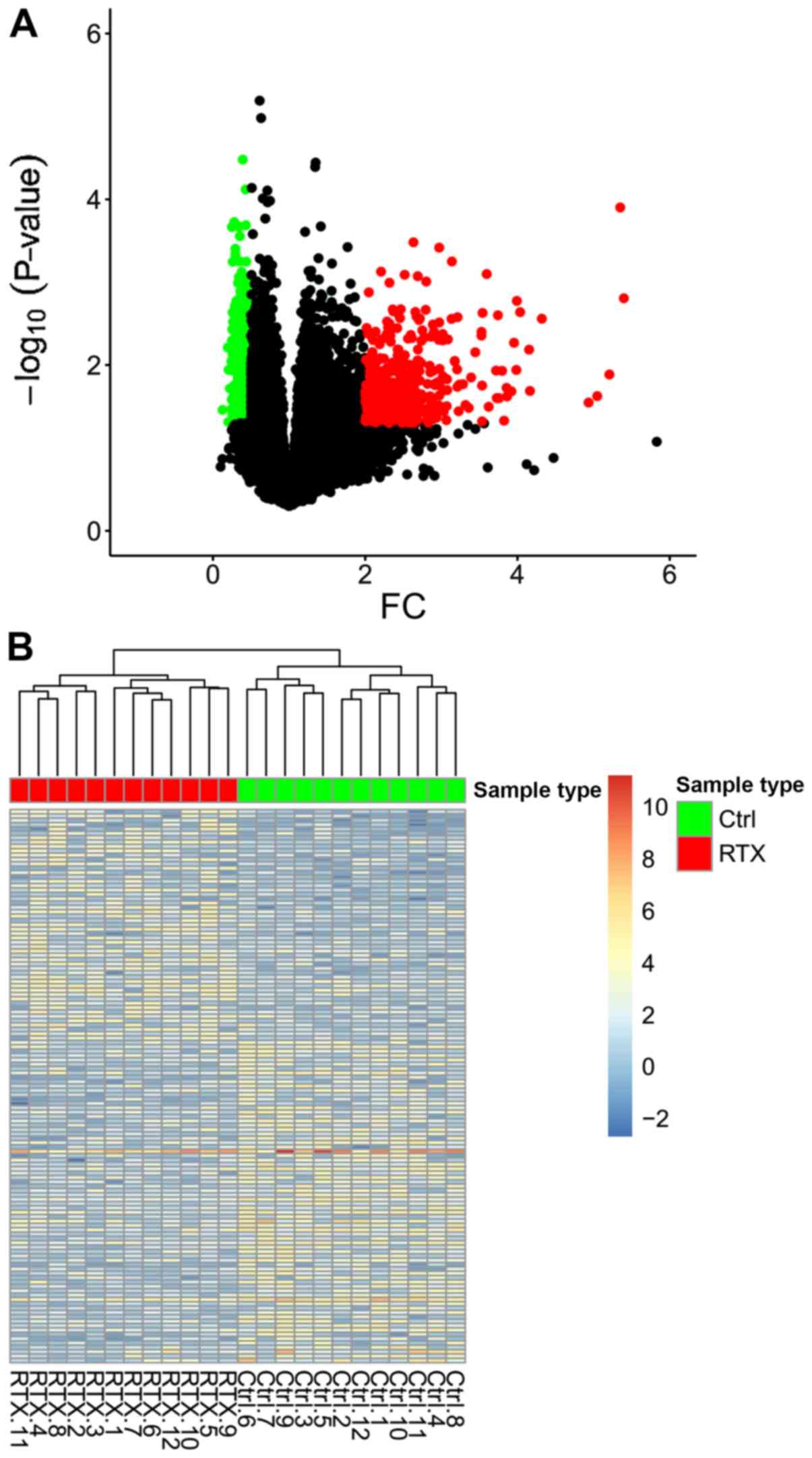
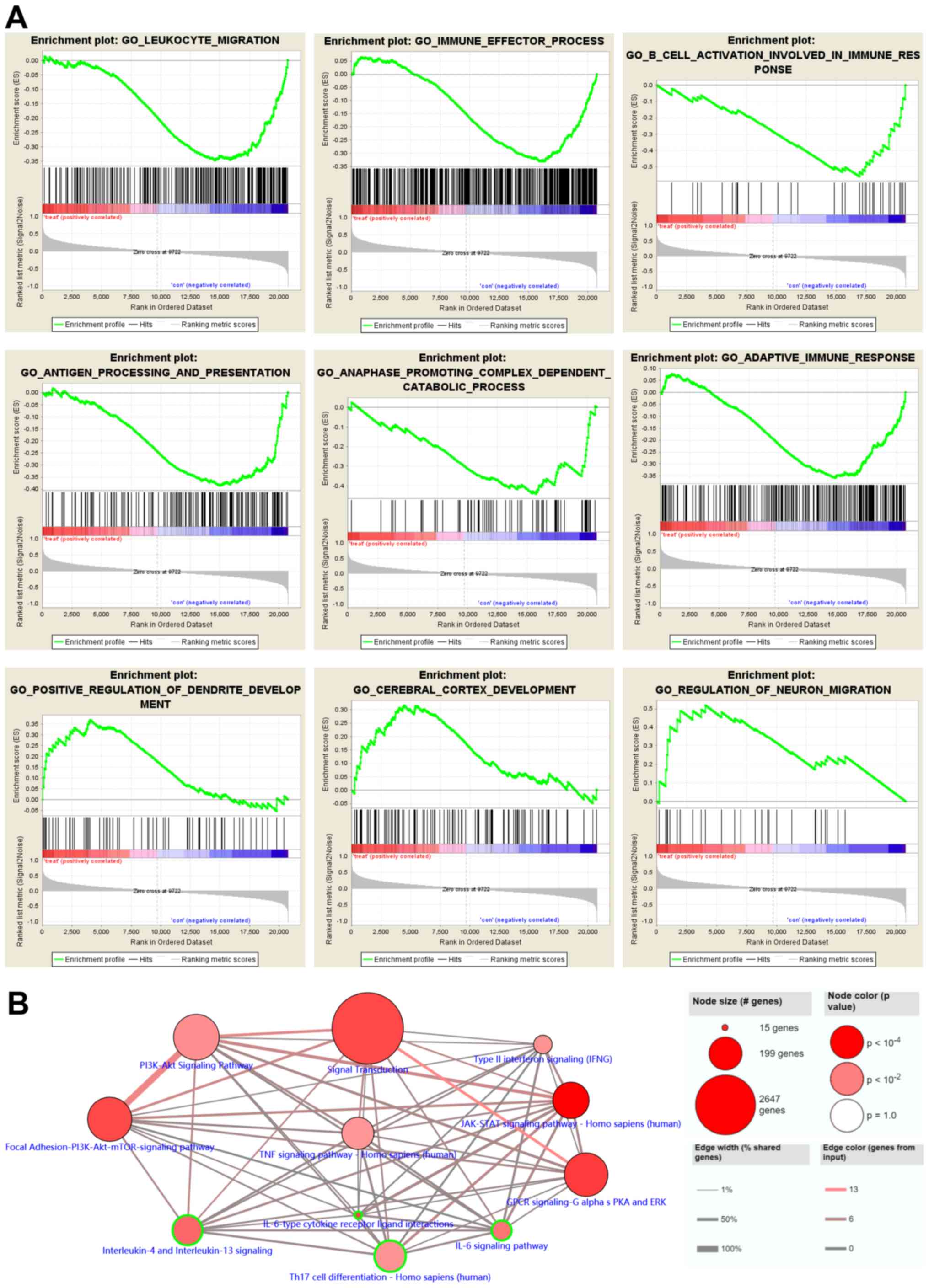
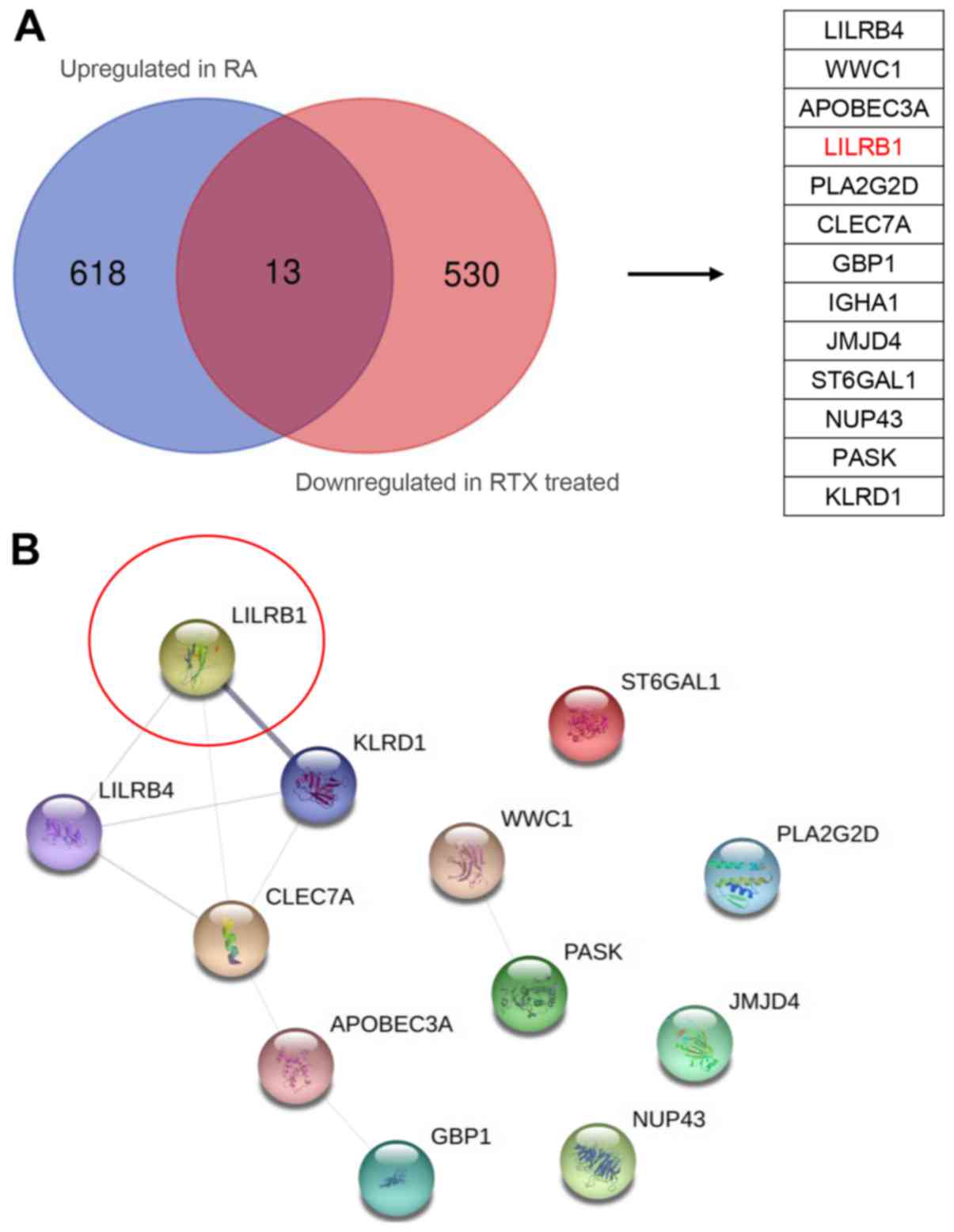
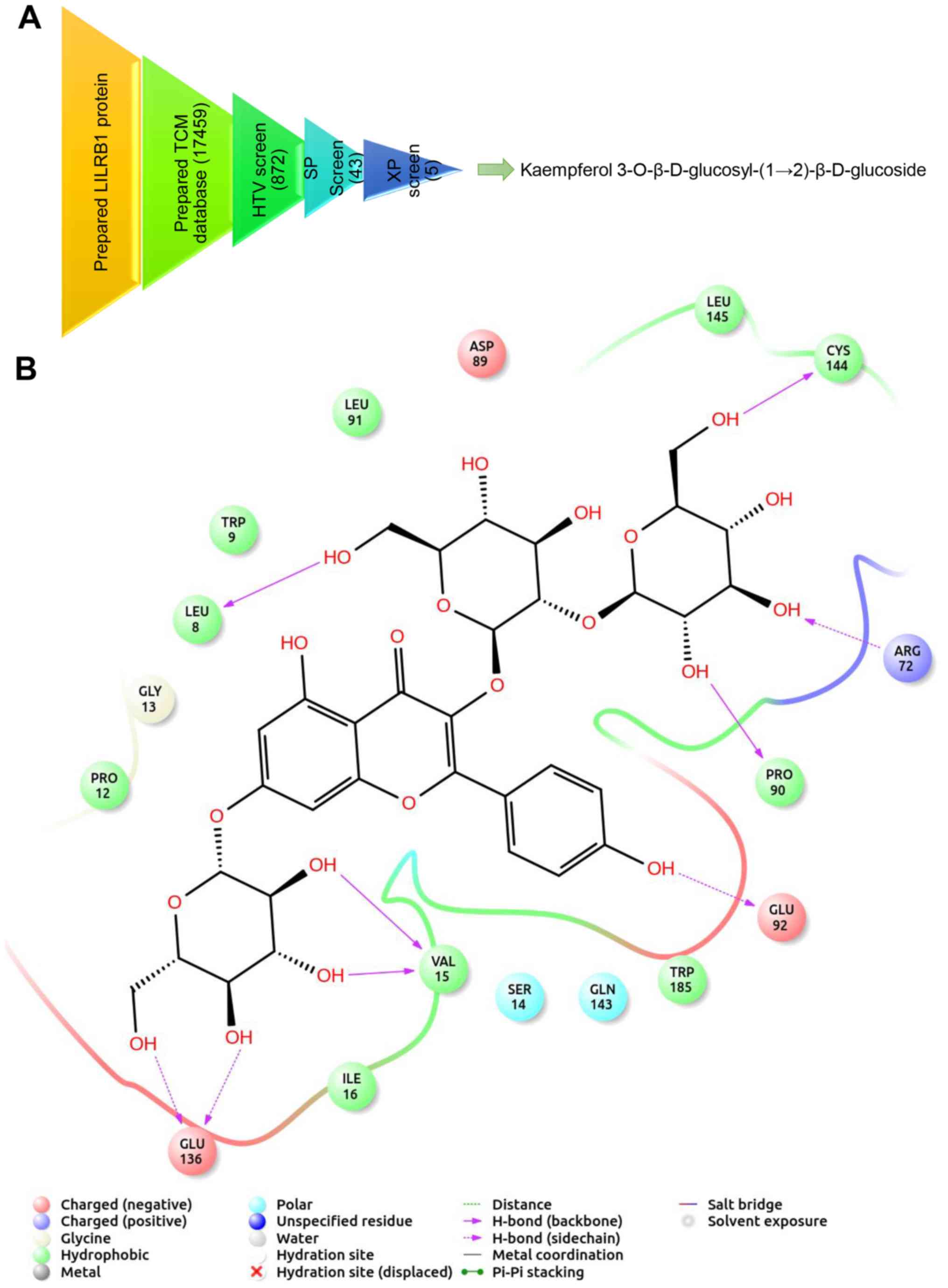
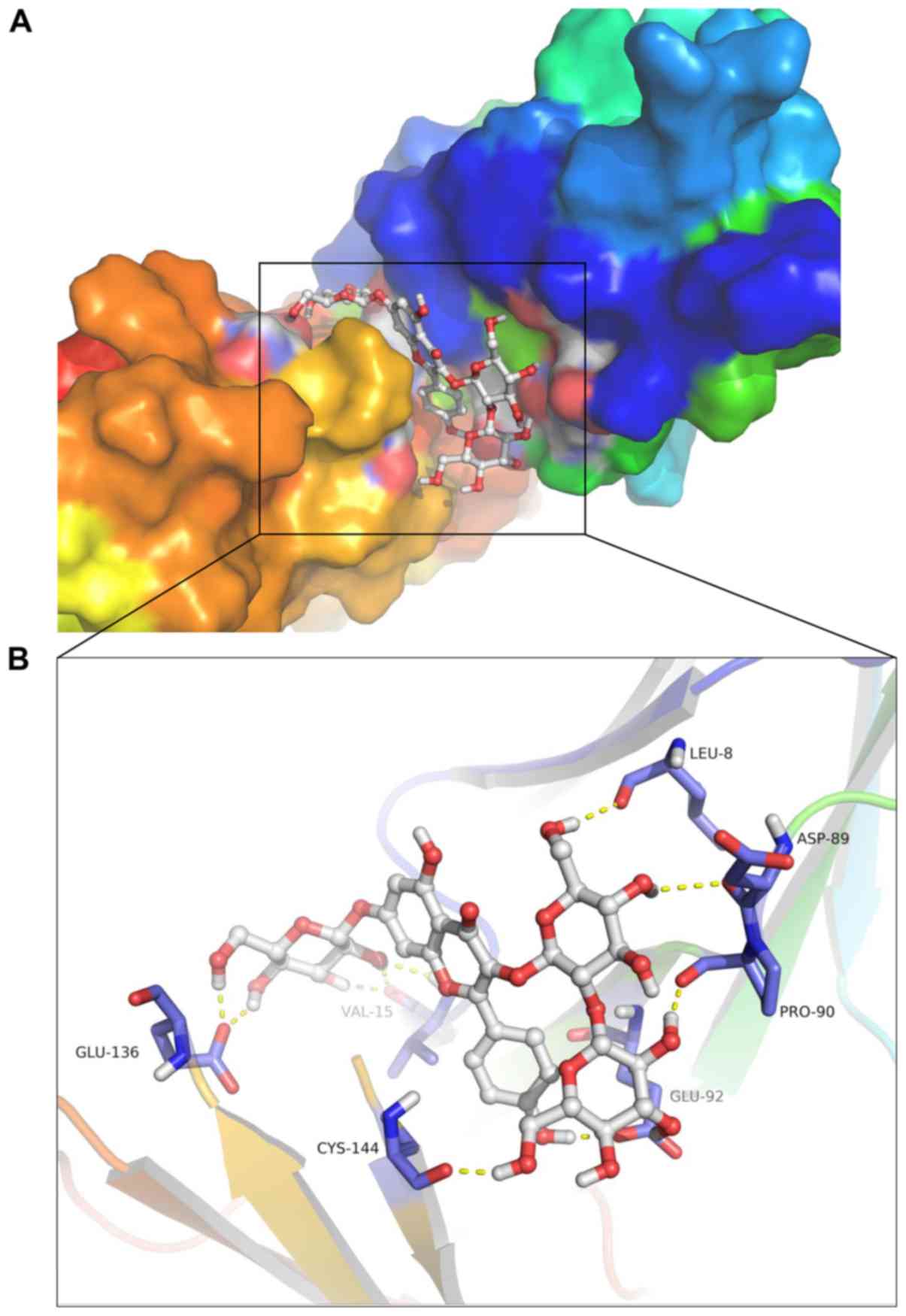
|
1
|
Falconer J, Murphy AN, Young SP, Clark AR,
Tiziani S, Guma M and Buckley CD: Review: Synovial cell metabolism
and chronic inflammation in rheumatoid arthritis. Arthritis
Rheumatol. 70:984–999. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nakajima A, Aoki Y, Sonobe M, Takahashi H,
Saito M, Terayama K and Nakagawa K: Radiographic progression of
large joint damage in patients with rheumatoid arthritis treated
with biological disease-modifying anti-rheumatic drugs. Mod
Rheumatol. 26:517–521. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Firestein GS and McInnes IB:
Immunopathogenesis of rheumatoid arthritis. Immunity. 46:183–196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arend WP and Firestein GS: Pre-rheumatoid
arthritis: Predisposition and transition to clinical synovitis. Nat
Rev Rheumatol. 8:573–586. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Firestein GS: Evolving concepts of
rheumatoid arthritis. Nature. 423:356–361. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singh JA, Saag KG, Bridges SL Jr, Akl EA,
Bannuru RR, Sullivan MC, Vaysbrot E, McNaughton C, Osani M,
Shmerling RH, et al: 2015 American college of rheumatology
guideline for the treatment of rheumatoid arthritis. Arthritis
Rheumatol. 68:1–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McInnes IB and Schett G: Pathogenetic
insights from the treatment of rheumatoid arthritis. Lancet.
389:2328–2337. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lopez-Olivo MA, Amezaga Urruela M, McGahan
L, Pollono EN and Suarez-Almazor ME: Rituximab for rheumatoid
arthritis. Cochrane Database Syst Rev. 1:CD0073562015.PubMed/NCBI
|
|
9
|
Smolen JS and Aletaha D: Rheumatoid
arthritis therapy reappraisal: Strategies, opportunities and
challenges. Nat Rev Rheumatol. 11:276–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Young NT, Waller EC, Patel R, Roghanian A,
Austyn JM and Trowsdale J: The inhibitory receptor LILRB1 modulates
the differentiation and regulatory potential of human dendritic
cells. Blood. 111:3090–3096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Xu Z, Ren X, Chen X, Wei J, Lin W,
Li Z, Ou C, Gong Z and Yan Y: Function of low ADARB1 expression in
lung adenocarcinoma. PLoS One. 14:e02222982019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan Y, Xu Z, Hu X, Qian L, Li Z, Zhou Y,
Dai S, Zeng S and Gong Z: SNCA is a functionally low-expressed gene
in lung adenocarcinoma. Genes (Basel). 9:E162018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Woetzel D, Huber R, Kupfer P, Pohlers D,
Pfaff M, Driesch D, Häupl T, Koczan D, Stiehl P, Guthke R and Kinne
RW: Identification of rheumatoid arthritis and osteoarthritis
patients by transcriptome-based rule set generation. Arthritis Res
Ther. 16:R842014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lauwerys BR, Hernández-Lobato D, Gramme P,
Ducreux J, Dessy A, Focant I, Ambroise J, Bearzatto B, Nzeusseu
Toukap A, Van den Eynde BJ, et al: Heterogeneity of synovial
molecular patterns in patients with arthritis. PLoS One.
10:e01221042015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Perry M: Heatmaps: Flexible Heatmaps for
Functional Genomics and Sequence Features. R package version
1.12.0. 2020.10.18129/B9.bioc.heatmaps.
|
|
17
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bindea G, Galon J and Mlecnik B: CluePedia
Cytoscape plugin: Pathway insights using integrated experimental
and in silico data. Bioinformatics. 29:661–663. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Kuehn H, Gould J, Tamayo P
and Mesirov JP: GSEA-P: A desktop application for gene set
enrichment analysis. Bioinformatics. 23:3251–3253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Herwig R, Hardt C, Lienhard M and Kamburov
A: Analyzing and interpreting genome data at the network level with
ConsensusPathDB. Nat Protoc. 11:1889–1907. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kamburov A, Wierling C, Lehrach H and
Herwig R: ConsensusPathDB-a database for integrating human
functional interaction networks. Nucleic Acids Res. 37:D623–628.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Snel B, Lehmann G, Bork P and Huynen MA:
STRING: A web-server to retrieve and display the repeatedly
occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442–3444.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sanderson K: Databases aim to bridge the
East-West divide of drug discovery. Nat Med. 17:15312011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Halgren TA, Murphy RB, Friesner RA, Beard
HS, Frye LL, Pollard WT and Banks JL: Glide: A new approach for
rapid, accurate docking and scoring. 2. Enrichment factors in
database screening. J Med Chem. 47:1750–1759. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Friesner RA, Banks JL, Murphy RB, Halgren
TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK,
et al: Glide: A new approach for rapid, accurate docking and
scoring. 1. Method and assessment of docking accuracy. J Med Chem.
47:1739–1749. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nair SK and Burley SK: X-ray structures of
Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation
by proto-oncogenic transcription factors. Cell. 112:193–205. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gupta S and Bajaj AV: Extra precision
glide docking, free energy calculation and molecular dynamics
studies of 1,2-diarylethane derivatives as potent urease
inhibitors. J Mol Model. 24:2612018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Friesner RA, Murphy RB, Repasky MP, Frye
LL, Greenwood JR, Halgren TA, Sanschagrin PC and Mainz DT: Extra
precision glide: Docking and scoring incorporating a model of
hydrophobic enclosure for protein-ligand complexes. J Med Chem.
49:6177–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhong W, Liu P, Zhang Q, Li D and Lin J:
Structure-based QSAR, molecule design and bioassays of
protease-activated receptor 1 inhibitors. J Biomol Struct Dyn.
35:2853–2867. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan Y, Su W, Zeng S, Qian L, Chen X, Wei
J, Chen N, Gong Z and Xu Z: Effect and mechanism of tanshinone I on
the radiosensitivity of lung cancer cells. Mol Pharm. 15:4843–4853.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Janossy G, Panayi G, Duke O, Bofill M,
Poulter LW and Goldstein G: Rheumatoid arthritis: A disease of
T-lymphocyte/macrophage immunoregulation. Lancet. 2:839–842. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Catrina AI, Joshua V, Klareskog L and
Malmström V: Mechanisms involved in triggering rheumatoid
arthritis. Immunol Rev. 269:162–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barrett T, Suzek TO, Troup DB, Wilhite SE,
Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W and Edgar R: NCBI
GEO: Mining millions of expression profiles-database and tools.
Nucleic Acids Res. 33:D562–566. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kashyap D, Sharma A, Tuli HS, Sak K, Punia
S and Mukherjee TK: Kaempferol-A dietary anticancer molecule with
multiple mechanisms of action: Recent trends and advancements. J
Funct Foods. 30:203–219. 2017. View Article : Google Scholar : PubMed/NCBI
|