Introduction
Primary hepatic cancer is the third leading cause of
cancer-associated mortality worldwide, and ~90% of primary hepatic
cases are hepatocellular carcinoma (1). Surgical resection and liver
transplantation are the first-line treatments used for liver cancer
therapy. However, only 10–20% of patients with liver cancer are
surgical candidates. Although liver-directed locoregional therapy,
systemic chemotherapy or molecular therapy, such as sorafenib
administration, are used for liver cancer therapy, these treatment
strategies provide limited success due to high cancer cell
heterogeneity (2). Cancer stem
cells (CSCs) are a small population of cancer cells exhibiting
self-renewal and differentiation abilities (3). CSCs are involved in tumor initiation,
tumor metastasis and recurrence, and drug resistance (4). Liver CSCs can be identified by
specific surface markers, such as epithelial cell adhesion
molecule, CD133, CD44, CD90, CD13 and CD24 (5–7).
However, the exact biological function of liver CSCs remains
unclear. The investigation of the molecular mechanisms critical to
the generation of liver CSC populations may provide novel
therapeutic strategies for liver cancer.
Since the development of the Human Genome Project,
long non-coding RNAs (lncRNAs), which are non-protein coding RNA
transcripts >200 nucleotides, have attracted considerable
attention. Although lncRNAs were initially believed to be
transcriptional noise, subsequent studies have demonstrated that
they play functional roles in various physiological and
pathological processes, including carcinogenesis, tumor metastasis
and drug resistance (8,9). However, the mechanism by which
lncRNAs modulate liver CSC features remains largely unknown.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is
a ubiquitously expressed and highly conserved lncRNA, which has
been demonstrated to be markedly overexpressed in several human
malignancies (10). The expression
of MALAT1 has been reported to be associated with clinical
parameters and cancer prognosis (10). MALAT1 may also be involved in tumor
growth, metastasis and drug resistance of liver cancer (11,12),
and has been reported to be associated with the CSC features of
pancreatic cancer (13) and glioma
(14). These findings suggest that
MALAT1 can modulate stemness in liver cancer cells.
In the present study, MALAT1 was demonstrated to be
a regulator of liver CSCs by effecting the expression levels of
Yes-associated protein 1 (YAP1), a major transcriptional effector
of the Hippo pathway. MALAT1 performed these functions by acting as
a competitive endogenous RNA (ceRNA) for microRNA (miRNA/miR)-375.
The MALAT1/miR-375/YAP1 axis may provide a potential target for
liver cancer therapy and could be used to develop novel therapeutic
drugs against liver CSCs.
Materials and methods
Patients and clinical tissues
Fresh liver tumor specimens and matched non-tumor
liver samples were obtained from 20 patients (15 male patients and
5 female patients; age, 35–71 years) with hepatocellular carcinoma
(without other malignant tumors) during hepatic resection at The
Second Affiliated Hospital, Zhejiang University (Hangzhou, China)
between January 2017 and October 2019. The present study was
approved by the Hospital Ethics Committee (ethics no. 2019-099) and
all patients signed informed consent for their participation in the
study. The samples were immediately snap-frozen in liquid
nitrogen.
Cell culture
The human liver cancer cell lines Huh7, Hep3B, HepG2
and 293T cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.), and were
maintained at 37°C in a humidified atmosphere containing 5%
CO2. Cell line authentication was performed by genetic
profiling using polymorphic short tandem repeat loci.
MALAT1 expression analysis by reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was prepared from liver tumor tissues and
cancer cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). A PrimeScript™ RT Reagent kit with
genomic DNA Eraser (Takara Bio, Inc.) was used to reverse
transcribe RNA into cDNA at 42°C for 1 h according to the
manufacturer's instructions. Subsequently, qPCR was performed using
an ABI Prism 7900 kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were used
for qPCR: Initial denaturation at 95°C for 30 sec; followed by 40
cycles at 95°C for 5 sec and at 60°C for 30 sec. The following
primers were used for qPCR: MALAT1 forward,
5′-ATACCTAACCAGGCATAACA-3′ and reverse, 5′-AGTAGACCAACTAAGCGAAT-3′;
and GAPDH forward, 5′-GACCTGACCTGCCGTCTAG-3′ and reverse,
5′-AGGAGTGGGTGTCGCTGT-3′. The 2−ΔΔCq method was used to
determine relative MALAT1 expression levels (15), which were normalized to the
internal reference gene GAPDH.
Sphere formation assay
The cells (1×103 cells/ml) were seeded on
ultralow attachment culture dishes (Corning Inc.) in DMEM/F12
serum-free medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10 ng/ml fibroblast growth factor-2 (Peprotech,
Inc.), 20 ng/ml epithelial growth factor (Peprotech, Inc.), N2
(Thermo Fisher Scientific, Inc.), B27 (Thermo Fisher Scientific,
Inc.), 1% sodium pyruvate (Thermo Fisher Scientific, Inc.), 2 mM
L-glutamine (Thermo Fisher Scientific, Inc.) and
penicillin/streptomycin. After 2 weeks of incubation at 37°C, the
spheres with a size >100 µm were counted and images were
obtained using a light stereomicroscope (Olympus Corporation).
Plasmid construction
The fragments of wild-type MALAT1 and of the 3′-UTR
of wild-type YAP1 containing the putative binding site of miR-375,
as well as their corresponding mutant sequences, which were
generated using a Site-Directed Mutagenesis Kit (cat. no. E0554S;
New England Biolabs, Inc.), were synthetized and cloned into a
pmirGLO vector (Promega Corporation) using the XhoI and
NotI sites in order to generate wild-type MALAT1 reporter
(MALAT1-WT), mutant MALAT1 reporter (MALAT1-MUT), wild-type YAP1
reporter (YAP1-WT) or mutant YAP1 reporter (YAP1-MUT)
dual-luciferase reporter plasmids. To assess the function of YAP1,
the full-length sequence of YAP1 was ligated into the pcDNA 3.1
plasmid (Thermo Fisher Scientific, Inc.) to construct a YAP1
overexpression plasmid (oeYAP1). An empty pcDNA3.1 plasmid was used
as a control (oeCtrl). All constructs were established using the
In-Fusion® Clone kit (Clontech Laboratories, Inc.).
Transfection of small interfering RNA
(siRNA), miRNA and plasmids
siRNA sequences targeting MALAT1 expression
(si-MALAT1; cat. no. siB170613115806-1-5) and control siRNA
sequences (si-Ctrl; siN0000002-1-5), miR-375 mimic (sense,
5′-UUUGUUCGUUCGGCUCGCGUGA-3′ and antisense,
5′-UCACGCGAGCCGAACGAACAAA-3′), miRNA mimic negative control
(miR-NC, sense, 5′-UUUGUACUACACAAAAGUACUG-3′ and antisense,
5′-CAGUACUUUUGUGUAGUACAAA-3′), miR-375 inhibitor
(5′-UCACGCGAGCCGAACGAACAAA-3′), and miRNA inhibitor negative
control (miR-NC, 5′-CAGUACUUUUGUGUAGUACAAA-3′) were purchased from
Guangzhou RiboBio Co., Ltd. Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for siRNA and
plasmid transfection, and Lipofectamine™ RNAiMAX (Invitrogen;
Thermo Fisher Scientific, Inc.) was used for miRNA transfection. At
75% confluence, cells were transfected with siRNA (100 nm), miRNA
(50 nM) or plasmid (1 µg/ml) at 37°C. At 24 h post-transfection,
MALAT1 expression in siRNA-transfected cells and miR-375 expression
in miRNA mimic- or miRNA inhibitor-transfected cells were assessed
using RT-qPCR analysis, and YAP1 expression in plasmid-transfected
cells was assessed using Western blotting (Fig. S1).
Dual-luciferase assay
MiRcode (version 11; www.mircode.org) and TargetScan (version 7.2;
www.targetscan.org/vert_72) were used
for predicting the potential target sequences between MALAT1 and
miR-375 and between miR-375 and YAP1, respectively. The
dual-luciferase reporter plasmids MALAT1-WT, MALAT1-MUT, YAP1-WT or
YAP1-MUT (1 µg/ml), and miR-375 mimics or miRNA mimic negative
control (50 nM) were co-transfected into 293T cells
(5×104 cells/well) using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C. A total of 48
h post-transfection, the Renilla and Firefly luciferase
activities were measured using a Dual-Luciferase Reporter Assay
system (Promega Corporation) according to manufacturer's protocol.
The results were quantified by the ratio of the activity of
Renilla luciferase to that of Firefly luciferase.
RNA pull-down assay
Biotinylated miR-375 was labeled using the RNA 3′
End Desthiobiotinylation Kit (Pierce; Thermo Fisher Scientific,
Inc.) according to manufacturer's protocol. RNA pull-down assays
were performed following transfection of biotin-labeled-miR-375
into liver cancer cells (1×106 cells/well). Three
sequences were used as follows: 3′-biotin-labeled miR-375
(bio-miR-375-WT), 3′-biotin-labeled miR-375 with a mutation at the
binding site between MALAT1 and miR-375 (bio-miR-375-MUT), and
3′-biotin-labeled miR-NC (bio-miR-NC). The cells were harvested
after 48 h of transfection at 37°C and subsequently lysed in
RNase-free cell lysis solution (1 Mm HEPES, 200 mM NaCl, 1% Triton
X-100, 10 mM MgCl2, 200 U/ml RNase Inhibitor) at 4°C.
Streptavidin agarose beads (Pierce; Thermo Fisher Scientific, Inc.)
were incubated with the cell lysis overnight at 4°C for pulling
down the biotin-labeled miRNA and associated RNAs from the
solution. After washing and centrifugation (1,500 × g; 10 min,
4°C), the pellet was lysed with TRIzol®. Subsequently,
the expression levels of MALAT1 in the pull-down samples were
measured by RT-qPCR.
Verteporfin treatment
Verteporfin (Sigma-Aldrich; Merck KGaA) was
dissolved in DMSO and added to the cell medium (1×103
cells/ml) for a final concentration of 10 µg/ml for 24 h at 37°C.
Equal concentration of DMSO was used as control.
Western blot analysis
A RIPA peptide lysis buffer (Beyotime Institute of
Biotechnology) containing 1% protease inhibitor (Pierce; Thermo
Fisher Scientific, Inc.) was used for tissue and cell lysis. Total
protein was quantified using a bicinchoninic acid assay kit (Thermo
Fisher Scientific, Inc.). Proteins (30 µg/lane) were separated via
10% SDS-PAGE and transferred to a 0.22-µm PVDF membrane. After
blocking with 5% non-fat milk for 1 h at room temperature, the
membranes were incubated overnight at 4°C with the following
primary antibodies: anti-YAP1 (1:1,000; cat. no. ab52771; Abcam),
anti- phosphorylated (p)-YAP1 (1:1,000; cat. no. ab172374; Abcam),
anti-c-Myc (1:1,000; cat. no. ab32072; Abcam), anti-Oct4 (1:1,000;
cat. no. ab181557; Abcam), anti-Sox2 (1:1,000; cat. no. ab92494;
Abcam) and anti-GAPDH (1:3,000; cat. no. 5174S; Cell Signaling
Technology). Membranes were incubated with primary antibodies at
room temperature for 3 h. A horseradish peroxidase-conjugated goat
anti-rabbit IgG H&L (1:3,000; cat. no. ab7090; Abcam) was used
as the secondary antibody and was used to incubate the membranes at
room temperature for 1 h. The protein bands were visualized using
an Enhanced Chemiluminescence system (PerkinElmer Inc.) and
detected using the ChemiScope Western Blot Imaging system (Clinx
Science Instruments Co., Ltd.). ImageJ software (version 1.46;
National Institutes of Health) was used for semi-quantification of
the results.
Statistical analysis
Each experiment was repeated at least three times.
The differences between two groups were compared using the
independent samples t-test. One-way ANOVA followed by Bonferroni
post-hoc test was used to determine significant differences between
multiple groups. The correlation between YAP1 and MALAT1 was
determined using Spearman correlation tests. The data were
presented as mean ± SD. P<0.05 was considered to indicate a
statistically significant difference.
Results
MALAT1 is overexpressed in liver
cancer
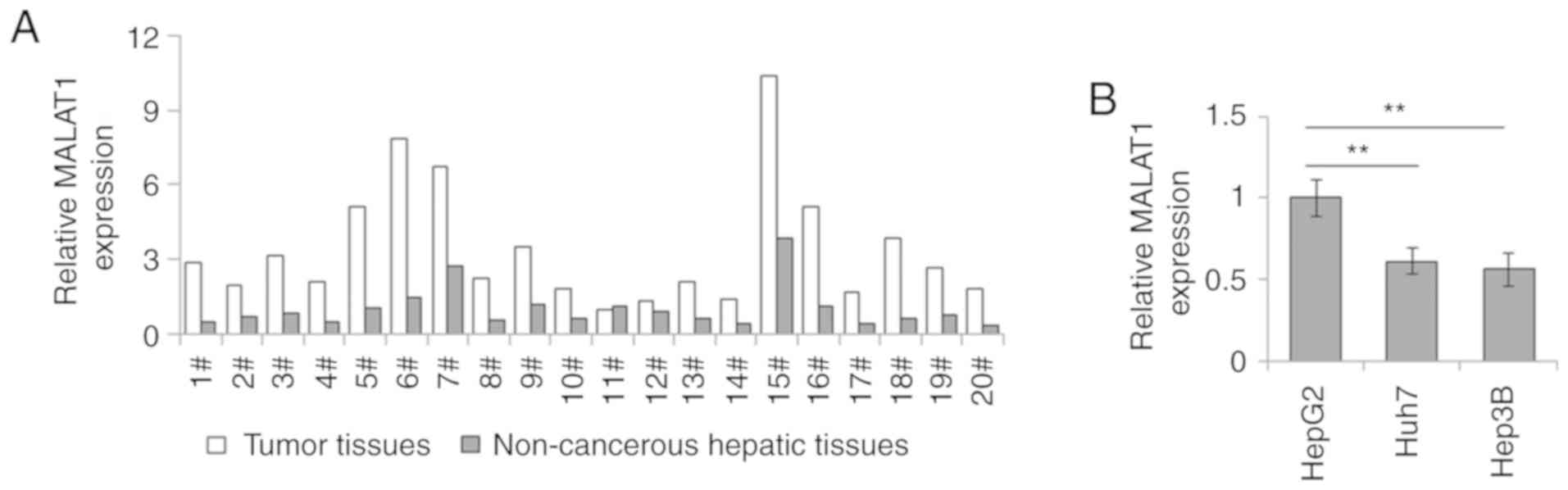
The RT-qPCR results indicated that MALAT1 expression
was markedly higher in liver tumor tissues than that in the
corresponding non-cancerous hepatic tissues (Fig. 1A). Moreover, the expression levels
of lncRNA MALAT1 in the hepatoblastoma cell line HepG2 were higher
compared with those in the common hepatoma cell lines Huh7 and
Hep3B (Fig. 1B). These data
indicated that MALAT1 expression was upregulated in liver cancer,
especially in hepatoblastoma cells.
MALAT1 modulates CSC features of liver
cancer cells
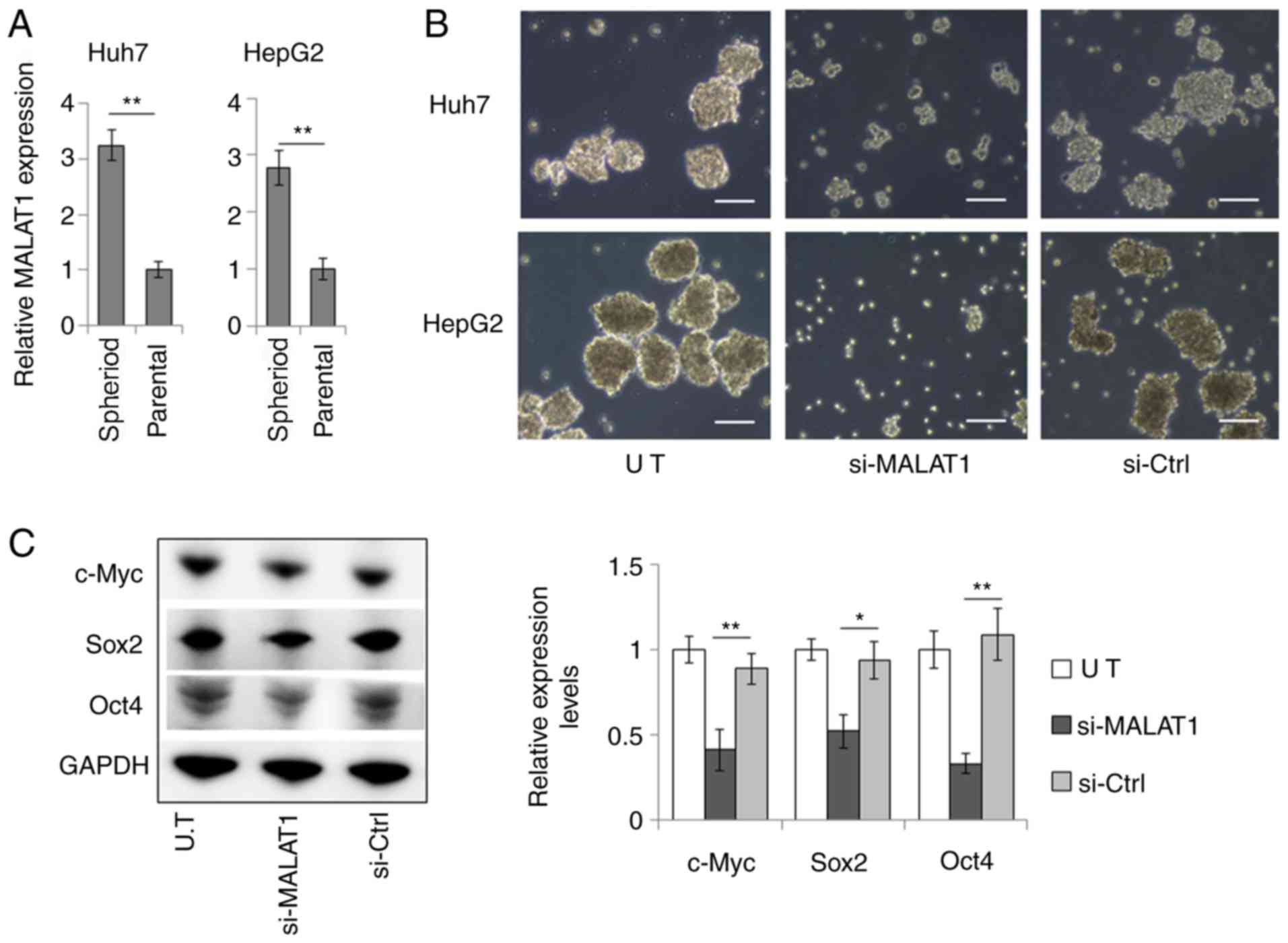
The role of MALAT1 in modulating the stemness of
liver CSCs was examined. MALAT1 was highly expressed in spheroids
derived from Huh7 and HepG2 cells (Fig. 2A). siRNA-induced downregulation of
MALAT1 expression levels (Fig.
S1) resulted in considerably suppressed sphere formation and
reduced sphere size of the Huh7 and HepG2 liver cancer cell lines
(Fig. 2B). In HepG2 cells, MALAT1
suppression significantly reduced the expression levels of c-Myc,
Sox2 and Oct4 (Fig. 2C), which are
key transcription factors that modulate cancer cell stemness
(16–18). These results indicated that MALAT1
could promote the stem-like properties of liver cancer cells.
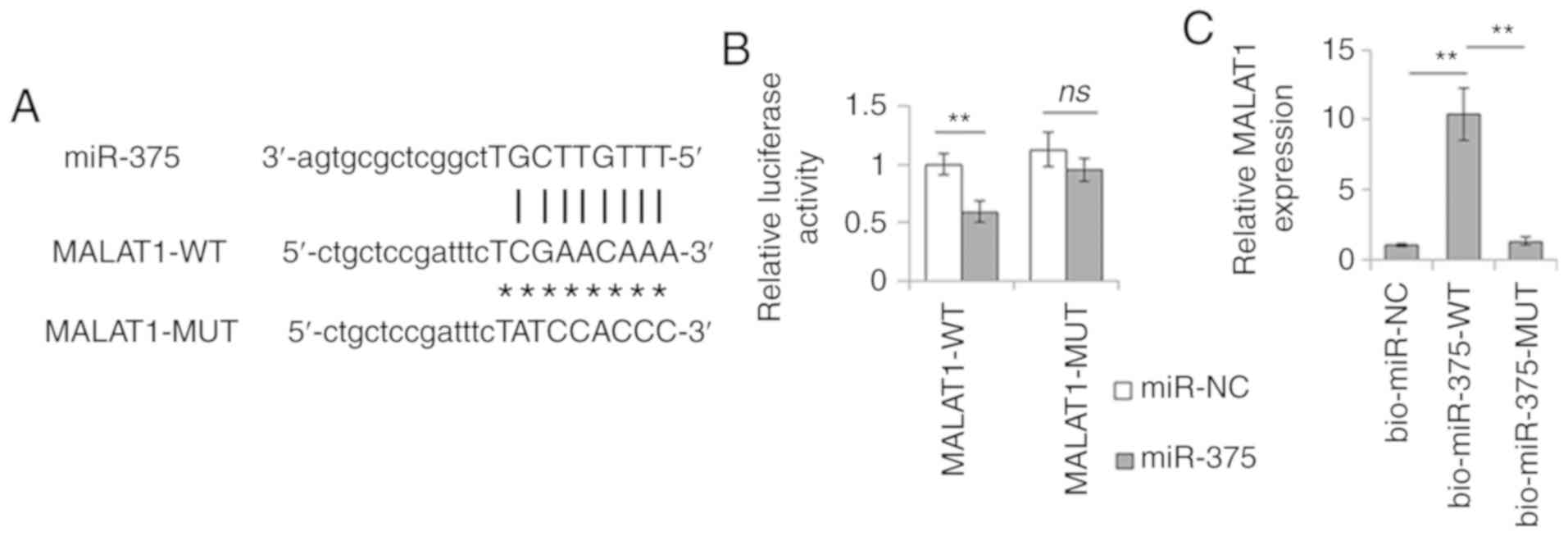
MALAT1 interacts with miR-375
lncRNAs can act as miRNA ceRNAs. To confirm the role
of MALAT1 in modulating CSC features of liver cancer cells as a
ceRNA molecule, bioinformatics analysis was performed. The data led
to the identification of several miRNA-binding sites on MALAT1.
Among these miRNAs, miR-375 has previously been shown to be
associated with the CSC phenotype (19). Therefore, the present study focused
on miR-375 and further investigated the interaction between MALAT1
and miR-375 by dual-luciferase assay. miR-375 mimics significantly
reduced the luciferase activity of MALAT1-WT compared with that of
miR-NC, whereas no change was observed in the luciferase activity
of MALAT1-MUT, which had a mutation in the miR-375 binding site
(Fig. 3A and B). Subsequently, the
biotin-streptavidin RNA pull-down assay was used to further
determine the interaction of MALAT1 with miR-375. As expected,
miR-375 could successfully pull down MALAT1. However, the mutation
in the binding site of miR-375 to MALAT1 disturbed the pull-down of
MALAT1 by this miRNA (Fig. 3C).
These data confirmed the interaction between MALAT1 and miR-375,
and suggested that MALAT1 may regulate liver CSC features by
sponging miR-375.
YAP1 is a direct target of
miR-375
As miRNAs are known to regulate gene expression at a
post-transcriptional level by interacting with the 3′-UTR of target
genes, TargetScan was employed to predict the targets of miR-375.
YAP1 is a major transcriptional effector for regulating liver CSC
self-renewal capacity (20). In
the present study, YAP1 was identified as a candidate target of
miR-375 (Fig. 4A).
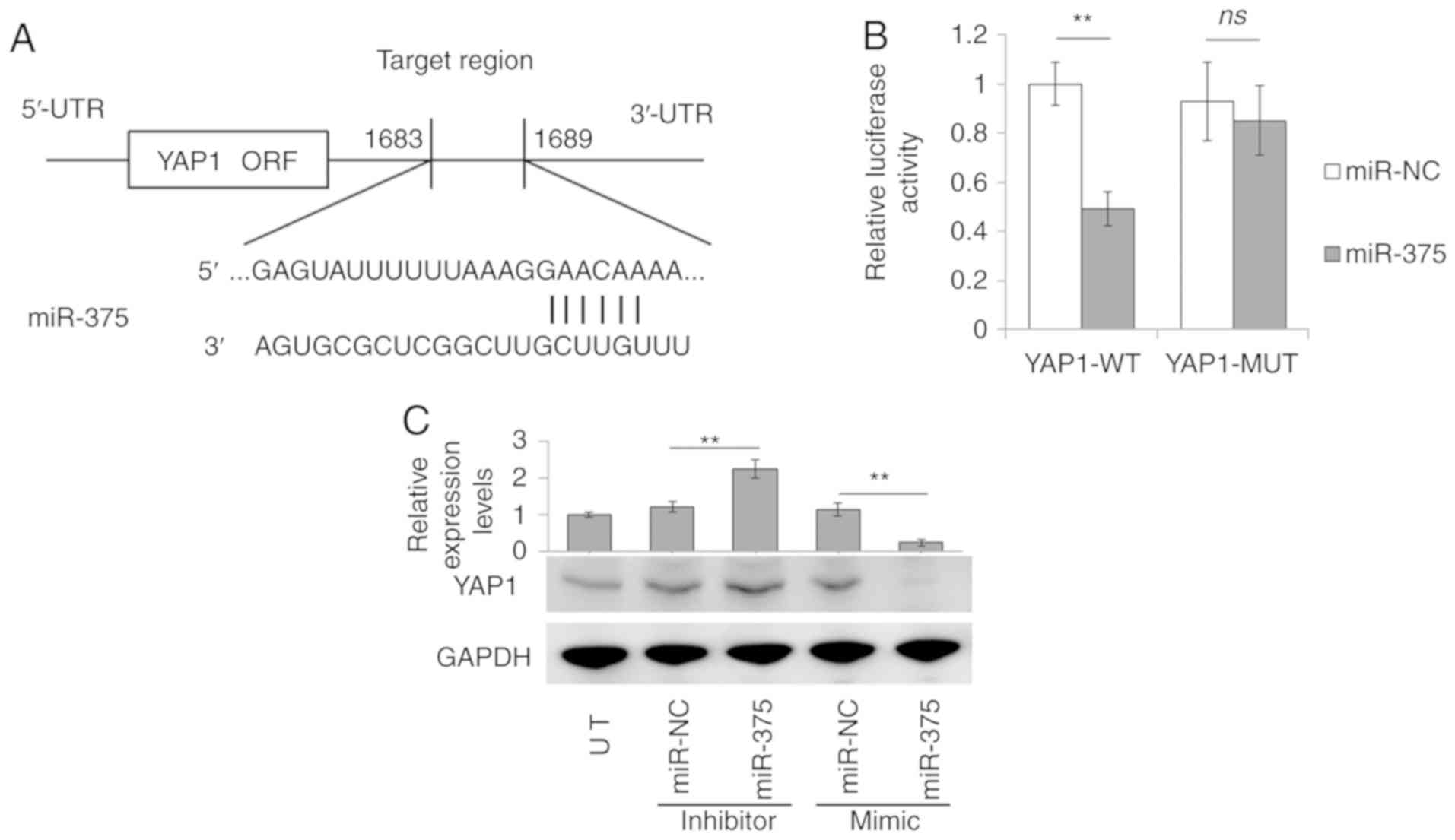 | Figure 4.miR-375 directly targets YAP1. (A)
Target site of miR-375 in YAP1 3′-UTR. (B) Dual-luciferase activity
of the WT and MUT YAP1 3′-UTR reporters in the presence of miR-375
or miR-NC mimics in 293T cells. (C) Western blot analysis and gray
value assay of YAP1 expression in Huh7 cells transfected with
miR-375 mimics, miR-375 inhibitors, and their corresponding
controls (miR-NC). Data are presented as the mean ± SD.
**P<0.01, n=3. UT, untreated control cells; ns, not significant;
YAP1, Yes-associated protein 1; WT, wild-type; MUT, mutant; miR,
microRNA; UTR, untranslated region; NC, negative control; ORF, open
reading frame. |
To verify whether YAP1 was an actual target of
miR-375, the 3′-UTR fraction of YAP1 containing the predicted
miR-375 binding site was cloned into a pmirGLO vector for YAP1-WT
reporter construction. The dual-luciferase report assay indicated
that miR-375 significantly reduced the luciferase activity of
YAP1-WT compared with that of miR-NC. However, mutation of the
miR-375 binding site of YAP1 3′-UTR (YAP1-MUT) abrogated the
inhibitory effects of miR-375 (Fig.
4B).
The miR-375-mediated regulation of YAP1 expression
was further examined in liver cancer cells. miR-375 mimic
transfection significantly reduced YAP1 protein levels, whereas
miR-375 inhibitor transfection increased YAP1 expression in Huh7
cells (Fig. 4C). Together, these
data strongly suggested that YAP1 was a direct target of
miR-375.
MALAT1 regulates YAP1 expression by
sponging miR-375
To further support the ceRNA hypothesis and
investigate the regulatory mechanism of MALAT1 on YAP1 expression,
YAP1 levels were measured in si-MALAT1-transfected Huh7 cells by
Western blot analysis. As shown in Fig. 5A, the YAP1 expression levels were
notably decreased in Huh7 cells transfected with si-MALAT1, which
is similar to the finding in cells transfected with miR-375 mimic
(Fig. 4C). In addition, si-MALAT1
caused a reduction in the expression levels of YAP1 that could be
partially abolished by transfection with the miR-375 inhibitor
(Fig. 5B). The association between
the expression levels of YAP1 and MALAT1 was further determined in
liver cancer cell lines and tumor samples. As shown in Fig. 5C, the highest YAP1 expression was
detected in HepG2 cells, which also exhibited the highest level of
MALAT1 expression compared with the Huh7 and Hep3B cell lines
(Fig. 1B). Moreover, YAP1
expression, alongside MALAT1, was markedly increased in liver tumor
tissues compared with in the corresponding non-cancerous hepatic
tissues (Fig. 5E and F). A
Spearman correlation analysis also determined that there was a
positive correlation between YAP1 expression and MALAT1 expression
in liver tumor tissues (Fig. 5D).
These results indicated that MALAT1 could regulate YAP1 expression
by sponging miR-375.
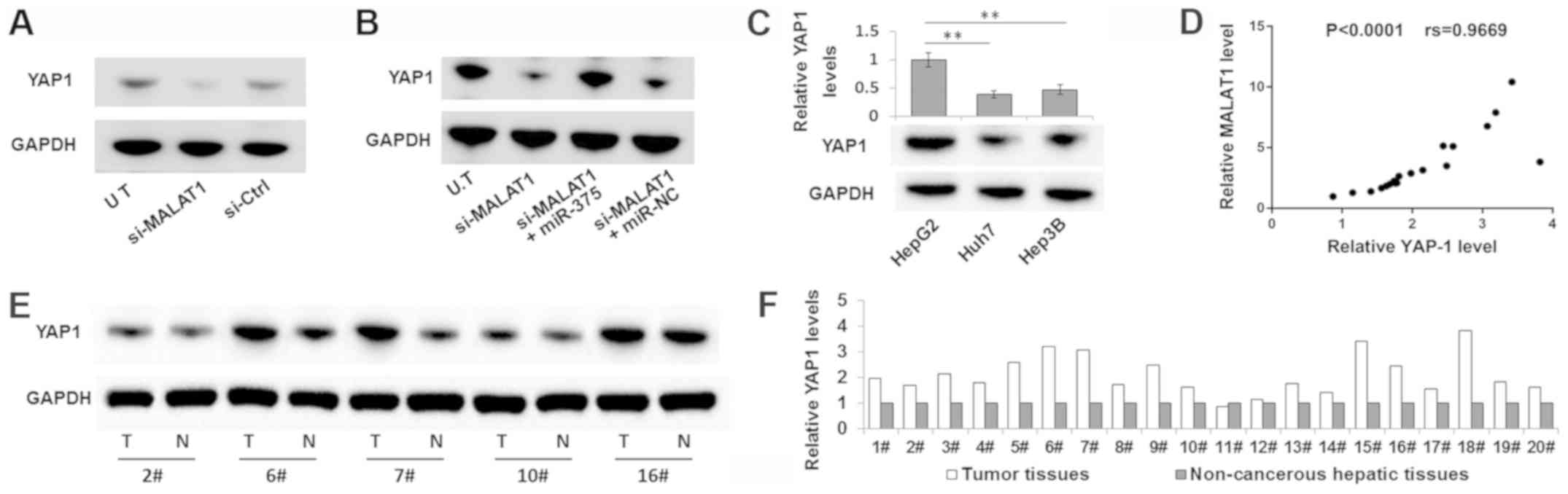 | Figure 5.MALAT1 can regulate YAP1 expression
by sponging miR-375. Western blot analysis of (A) YAP1 expression
in the Huh7 cells transfected with si-MALAT1 or si-Ctrl, and (B) in
the Huh7 cells co-transfected with si-MALAT1 and miR-375 inhibitor
or si-MALAT1 and control inhibitor (miR-NC). (C) Western blot
analysis of YAP1 expression in three liver cancer cell lines. (D)
Spearman correlation analysis of MALAT1 and miR-375 expression
levels in tumor samples. YAP1 expression levels in the T and paired
N samples were detected by (E) Western blot analysis and
subsequently (F) semi-quantified, relative to GAPDH. Data are
presented as the mean ± SD. **P<0.01, n=3. MALAT1,
metastasis-associated lung adenocarcinoma transcript 1; YAP1,
Yes-associated protein 1; miR, microRNA; si-, small interfering
RNA; NC, negative control; T, hepatic tumor tissues; N,
non-cancerous hepatic tissues; UT, untreated control cells. |
MALAT1-mediated upregulation of YAP1
affects the CSC features of liver cancer cells
YAP1 is ubiquitously activated in human malignancies
and its expression is associated with the CSC features of various
tumors (21). Therefore, the
present study further explored whether the modulatory effects of
MALAT1 on CSC features were mediated by YAP1. As expected, YAP1
overexpression in non-sphere cells markedly promoted sphere
formation (Fig. 6A), and increased
the expression levels of c-Myc, Sox2 and Oct4 (Fig. 6B). By contrast, verteporfin, a
specific inhibitor for YAP1 expression and YAP1 phosphorylation
inhibition (22), considerably
impaired the sphere formation capacity of tumor cells (Fig. 6A), and reduced the expression
levels of the corresponding stemness genes (Fig. 6C).
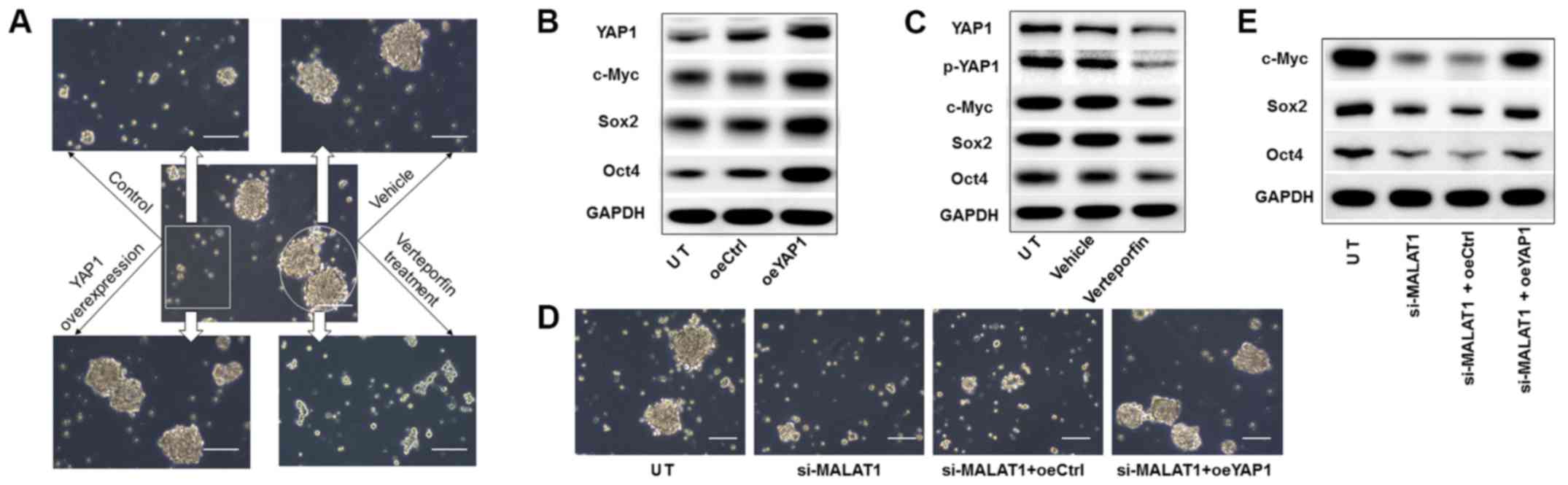 | Figure 6.YAP1 mediates the regulation of
MALAT1 on CSC features of liver cancer cells. (A) YAP1
overexpression in non-spherical Huh7 cells (box) or YAP1
suppression in spherical Huh7 cells (circle), followed by sphere
formation assay (arrows). (B) At 2 weeks after sphere formation,
UT, oeYAP1 and oeCtrl spheres of Huh7 cells were collected for
Western blot analysis of expression of the stemness markers. (C) UT
spheres, verteportin-treated spheres and vehicle-treated spheres
from Huh7 cells were collected for Western blot analysis at 2 weeks
after sphere formation. (D) Sphere formation assay and (E) Western
blot analysis of expression of stemness factors on Huh7 cells
transfected with si-MALAT1, or co-transfected with si-MALAT1 and
oeYAP1 or oeCtrl. Scale bar, 100 µm. UT, untreated control cells.
YAP1, Yes-associated protein 1; MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; CSC, cancer stem cell; oe,
overexpression plasmid; Ctrl, control; si-, small interfering RNA;
p-, phosphorylated. |
Subsequently, the role of YAP1 was investigated in
MALAT1-modulated CSC properties. YAP1 overexpression rescued
si-MALAT1-reduced sphere formation (Fig. 6D) and recovered the stemness factor
expression levels in Huh7 cells (Fig.
6E). Therefore, these data indicated that MALAT1-mediated YAP1
signaling was required for the CSC features of liver cancer
cells.
Discussion
Liver CSCs are one of the major factors contributing
to the refractory nature of liver cancer to conventional treatment,
and are closely associated with tumor growth, relapse, metastasis
and chemoresistance (4,23). Therefore, the successful
development of potential therapeutic approaches that target liver
CSCs is promising in improving the outcomes of patients with liver
cancer. In the present study, it was revealed that the lncRNA
MALAT1 was significantly increased in liver CSCs and that this
lncRNA could maintain the stemness of liver CSCs by acting as a
ceRNA for miR-375. This in turn regulated the expression of YAP1,
which was functionally required for CSC attributes. The findings
may contribute to liver cancer therapy by providing a new target
for liver CSCs.
MALAT1 was one of the first lncRNAs shown to be
involved in cancer progression, and a series of studies established
the importance of MALAT1 as a cancer marker (24). MALAT1 upregulation has been
observed in various cancer types, including lung, gastric,
colorectal, prostate, melanoma, cervical, breast and liver cancer
(25). In addition, MALAT1
upregulation may facilitate cell proliferation, migration, invasion
and multidrug resistance of tumor cells, and predict the malignant
progression and poor prognosis of liver cancer (11,12,26).
Recently, the role of MALAT1 in regulating the expression of
stemness markers and CSC properties of cancer cells has attracted
the attention of various researchers. In pancreatic cancer cells,
MALAT1 knockdown reduced the formation of spheres and the
expression of self-renewal related factors, such as Sox2,
indicating that MALAT-1 may enhance stemness phenotypes of
pancreatic CSCs by upregulating Sox2 expression (13). The MALAT1-mediated induction of
sphere formation and the upregulation of stemness biomarkers were
also observed in osteosarcoma and glioma stem cells (14,27).
In the present study, MALAT1 was required for the CSC features of
liver cancer cells, as determined by reduced sphere formation and
sphere size, as well as decreased CSC-related factor expression,
following siRNA-induced knockdown of MALAT1 expression levels.
lncRNAs act through various modes, including
cotranscriptional regulation, gene expression modulation, nuclear
or cytoplasmic complex scaffolding, and pairing with other RNAs
(28). In addition, lncRNAs can
exert oncogenic functions by mediating alternative splicing, and
transcriptional and post-transcriptional regulation (29). One of the main mechanisms of
MALAT1-mediated post-transcriptional regulation is its action as a
ceRNA (30,31). This mechanism suggests that lncRNAs
act as molecular sponges for other RNA transcripts through the
lncRNA-miRNA binding sites (also termed miRNA response elements) by
competing for pool miRNAs (30,31).
MALAT1 has been shown to promote the growth, migration and invasion
of liver cancer cells via the MALAT1/miR-195/epidermal growth
factor receptor (11),
MALAT1/miR-30a-5p/vimentin (32)
and MALAT1/miR-143-3p/zinc finger E-box binding homeobox 1 axes
(33), respectively. However, to
the best of our knowledge, the exact role of MALAT1 in liver CSCs
remains unknown. The present study further suggested the ceRNA role
of MALAT1 in modulating the stemness of liver CSCs through the
post-transcriptional regulation of YAP1 expression caused by
sponging miR-375.
miR-375 was initially considered a pancreatic
islet-specific miRNA that regulates insulin secretion, but was
subsequently identified as a multifunctional miRNA with roles in
pancreatic islet development, mucosal immunity, glucose homeostasis
and even tumorigenesis (34).
Previously, the significant reduction of miR-375 has been observed
in multiple cancer types, including liver cancer (35,36).
Low miR-375 expression levels have been reported to be correlated
with poor disease-free survival and may be a potential prognostic
biomarker of disease progression in hepatocellular carcinoma
(36). Further studies identified
that miR-375 could suppress liver cancer cell proliferation and
migration, and overcome chemoresistance by targeting several
important oncogenes, such as JAK2, receptor tyrosine-protein kinase
erbB-2, AEG-1 and autophagy-related 7 (37–40).
Nanoparticle-mediated miR-375 delivery enhanced chemosensitivity
and overcame multiple drug resistance in hepatocellular carcinoma,
thus suggesting that miR-375 could be a potential target in liver
cancer therapy (41). In the
present study, a novel pathway was identified that explained the
antitumor activity of miR-375 in liver cancer. MALAT1 knockdown may
reverse sponging of miR-375, release miR-375 and boost its
regulatory effects on oncogene expression at the
post-transcriptional level.
In the current study, YAP1 was identified as a
target of miR-375. YAP1 has an important role in the Hippo
signaling pathway, which is an essential pathway in cell
proliferation, apoptosis and organ growth regulation (42). YAP1 has also been reported to be
involved in cancer development, since it can promote cell
proliferation, suppress cell apoptosis, and induce the
epithelial-mesenchymal transition and drug resistance of cancer
cells (43). An increasing number
of studies have determined the role of YAP1 in modulating cellular
stemness. YAP1 overexpression may induce CSC properties, such as
self-renewal, sphere-formation, invasiveness and drug resistance
(21,44). Further studies showed that these
YAP1-related CSC biological features were mediated by regulating
embryonic stem cell factors, such as Oct4, Sox2 and Nanog (45,46).
The results of the present study were consistent with the
observations that YAP1 overexpression promoted sphere formation,
and upregulated Sox2 and Oct4 expression levels. MALAT1
knockdown-induced suppression of sphere formation and the
corresponding reduction in expression of stemness markers could
also be rescued by YAP1 overexpression, suggesting that MALAT1
contributed to CSC properties via YAP1. These observations provide
information regarding the contribution of MALAT1 to CSC-associated
biological features via the competitive regulation of
miR-375-mediated YAP1 expression.
In summary, the present study demonstrated that
MALAT1 was highly expressed in liver CSCs and that it promoted the
CSC properties in cell culture by upregulating YAP1 expression via
sponging miR-375. These findings elucidated the potential role of
the MALAT1/miR-375/YAP1 axis in modulating the stemness of liver
cancer cells and improved our understanding of the molecular
characteristics of liver CSCs. The current study lays the
foundation for the development of novel therapeutic strategies to
eradicate CSCs that are potentially harbored in residual cancerous
liver lesions.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the general research
project of education of Zhejiang province (grant no. Y201738069),
the National Natural Science Fund of China (grant no. 81870428),
and the Key R&D Projects of Zhejiang Province (grant no.
2018C03019).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ and GL performed the research and drafted the
manuscript. AL participated in in vitro studies and
performed the statistical analysis. YL conceived the study, and
participated in its design and coordination. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Second
Affiliated Hospital, Zhejiang University Hospital Ethics Committee
(ethics no. 2019-099), and all patients signed informed consent for
their participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Prager BC, Xie Q, Bao S and Rich JN:
Cancer stem cells: The architects of the tumor ecosystem. Cell Stem
Cell. 24:41–53. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Najafi M, Farhood B and Mortezaee K:
Cancer stem cells (CSCs) in cancer progression and therapy. J Cell
Physiol. 234:8381–8395. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiang Y, Yang T, Pang BY, Zhu Y and Liu
YN: The progress and prospects of putative biomarkers for liver
cancer stem cells in hepatocellular carcinoma. Stem Cells Int.
2016:76149712016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang N, Wang S, Li MY, Hu BG, Liu LP, Yang
SL, Yang S, Gong Z, Lai PBS and Chen GG: Cancer stem cells in
hepatocellular carcinoma: An overview and promising therapeutic
strategies. Ther Adv Med Oncol. 10:17588359188162872018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamashita T, Honda M, Nakamoto Y, Baba M,
Nio K, Hara Y, Zeng SS, Hayashi T, Kondo M, Takatori H, et al:
Discrete nature of EpCAM+ and CD90+ cancer
stem cells in human hepatocellular carcinoma. Hepatology.
57:1484–1497. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sanchez Calle A, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Hamblin MH and Yin KJ: The long
noncoding RNA Malat1: Its physiological and pathophysiological
functions. RNA Biol. 14:1705–1714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu D, Zhu Y, Pang J, Weng X, Feng X and
Guo Y: Knockdown of long non-coding RNA MALAT1 inhibits growth and
motility of human hepatoma cells via modulation of miR-195. J Cell
Biochem. 119:1368–1380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan P, Cao W, Zang Q, Li G, Guo X and Fan
J: The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance
of hepatocellular carcinoma cells via modulating autophagy. Biochem
Biophys Res Commun. 478:1067–1073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiao F, Hu H, Han T, Yuan C and Wang L,
Jin Z, Guo Z and Wang L: Long noncoding RNA MALAT-1 enhances stem
cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci.
16:6677–6693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han Y, Zhou L, Wu T, Huang Y, Cheng Z, Li
X, Sun T, Zhou Y and Du Z: Downregulation of lncRNA-MALAT1 affects
proliferation and the expression of stemness markers in glioma stem
cell line SHG139S. Cell Mol Neurobiol. 36:1097–1107. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Akita H, Marquardt JU, Durkin ME, Kitade
M, Seo D, Conner EA, Andersen JB, Factor VM and Thorgeirsson SS:
MYC activates stem-like cell potential in hepatocarcinoma by a
p53-dependent mechanism. Cancer Res. 74:5903–5913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leis O, Eguiara A, Lopez-Arribillaga E,
Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R
and Martin AG: Sox2 expression in breast tumours and activation in
breast cancer stem cells. Oncogene. 31:1354–1365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kuo KK, Lee KT, Chen KK, Yang YH, Lin YC,
Tsai MH, Wuputra K, Lee YL, Ku CC, Miyoshi H, et al: Positive
feedback loop of OCT4 and c-JUN expedites cancer stemness in liver
cancer. Stem Cells. 34:2613–2624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu H, Fu L, Xie C, Zuo WS, Liu YS, Zheng
MZ and Yu JM: miR-375 inhibits cancer stem cell phenotype and
tamoxifen resistance by degrading HOXB3 in human ER-positive breast
cancer. Oncol Rep. 37:1093–1099. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L, et al: LncBRM initiates YAP1 signalling
activation to drive self-renewal of liver cancer stem cells. Nat
Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zanconato F, Cordenonsi M and Piccolo S:
YAP/TAZ at the Roots of Cancer. Cancer Cell. 29:783–803. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kandasamy S, Adhikary G, Rorke EA,
Friedberg JS, Mickle MB, Alexander HR and Eckert RL: The YAP1
signaling inhibitors, verteporfin and CA3, suppress the
mesothelioma cancer stem cell phenotype. Mol Cancer Res.
18:343–351. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nio K, Yamashita T and Kaneko S: The
evolving concept of liver cancer stem cells. Mol Cancer. 16:42017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G and
Zhu YS: MALAT1: A potential biomarker in cancer. Cancer Manag Res.
10:6757–6768. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshimoto R, Mayeda A, Yoshida M and
Nakagawa S: MALAT1 long non-coding RNA in cancer. Biochim Biophys
Acta. 1859:192–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Konishi H, Ichikawa D, Yamamoto Y, Arita
T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, et
al: Plasma level of metastasis-associated lung adenocarcinoma
transcript 1 is associated with liver damage and predicts
development of hepatocellular carcinoma. Cancer Sci. 107:149–154.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Huang W, Sun W, Zheng B, Wang C,
Luo Z, Wang J and Yan W: LncRNA MALAT1 promotes cancer metastasis
in osteosarcoma via activation of the PI3K-Akt signaling pathway.
Cell Physiol Biochem. 51:1313–1326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kondo Y, Shinjo K and Katsushima K: Long
non-coding RNAs as an epigenetic regulator in human cancers. Cancer
Sci. 108:1927–1933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang WT, Ye H, Wei PP, Han BW, He B, Chen
ZH and Chen YQ: LncRNAs H19 and HULC, activated by oxidative
stress, promote cell migration and invasion in cholangiocarcinoma
through a ceRNA manner. J Hematol Oncol. 9:1172016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan Y, Tong S, Cui R, Fan J, Liu C, Lin Y,
Tang J, Xie H, Lin P, Zheng T, et al: Long non-coding MALAT1
functions as a competing endogenous RNA to regulate vimentin
expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell
Physiol Biochem. 50:108–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen L, Yao H, Wang K and Liu X: Long
non-coding RNA MALAT1 regulates ZEB1 expression by sponging
miR-143-3p and promotes hepatocellular carcinoma progression. J
Cell Biochem. 118:4836–4843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li X: MiR-375, a microRNA related to
diabetes. Gene. 533:1–4. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yan JW, Lin JS and He XX: The emerging
role of miR-375 in cancer. Int J Cancer. 135:1011–1018. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou N, Wu J, Wang X, Sun Z, Han Q and
Zhao L: Low-level expression of microRNA-375 predicts poor
prognosis in hepatocellular carcinoma. Tumour Biol. 37:2145–2152.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cao S, Wang G, Wang J, Li C and Zhang L:
Hsa_circ_101280 promotes hepatocellular carcinoma by regulating
miR-375/JAK2. Immunol Cell Biol. 97:218–228. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li L, Jia L and Ding Y: Upregulation of
miR-375 inhibits human liver cancer cell growth by modulating cell
proliferation and apoptosis via targeting ErbB2. Oncol Lett.
16:3319–3326. 2018.PubMed/NCBI
|
|
39
|
He XX, Chang Y, Meng FY, Wang MY, Xie QH,
Tang F, Li PY, Song YH and Lin JS: MicroRNA-375 targets AEG-1 in
hepatocellular carcinoma and suppresses liver cancer cell growth in
vitro and in vivo. Oncogene. 31:3357–3369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chang Y, Yan W, He X, Zhang L, Li C, Huang
H, Nace G, Geller DA, Lin J and Tsung A: miR-375 inhibits autophagy
and reduces viability of hepatocellular carcinoma cells under
hypoxic conditions. Gastroenterology. 143:177–87.e8. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao P, Wu S, Cheng Y, You J, Chen Y, Li
M, He C, Zhang X, Yang T, Lu Y, et al: MiR-375 delivered by
lipid-coated doxorubicin-calcium carbonate nanoparticles overcomes
chemoresistance in hepatocellular carcinoma. Nanomedicine (Lond).
13:2507–2516. 2017. View Article : Google Scholar
|
|
42
|
Hansen CG, Moroishi T and Guan KL: YAP and
TAZ: A nexus for Hippo signaling and beyond. Trends Cell Biol.
25:499–513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shibata M, Ham K and Hoque MO: A time for
YAP1: Tumorigenesis, immunosuppression and targeted therapy. Int J
Cancer. 143:2133–2144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lu T, Li Z, Yang Y, Ji W, Yu Y, Niu X,
Zeng Q, Xia W and Lu S: The Hippo/YAP1 pathway interacts with FGFR1
signaling to maintain stemness in lung cancer. Cancer Lett.
423:36–46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bora-Singhal N, Nguyen J, Schaal C,
Perumal D, Singh S, Coppola D and Chellappan S: YAP1 regulates OCT4
activity and SOX2 expression to facilitate self-renewal and
vascular mimicry of stem-like cells. Stem Cells. 33:1705–1718.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Strnadel J, Choi S, Fujimura K, Wang H,
Zhang W, Wyse M, Wright T, Gross E, Peinado C, Park HW, et al:
eIF5A-PEAK1 signaling regulates YAP1/TAZ protein expression and
pancreatic cancer cell growth. Cancer Res. 77:1997–2007. 2017.
View Article : Google Scholar : PubMed/NCBI
|