Introduction
Advanced gastric cancer often recurs and
metastasizes subsequent to surgery, and eventually the metastatic
cancer cells develop resistance to the chemotherapeutic drugs
(1–3). Multidrug resistance (MDR) accounts
for poor prognosis in gastric cancer (4), the development of multidrug
resistance is a key issue for tumor recurrence and metastasis,
leading to treatment failure of gastric cancer. Cancer cells may
become unresponsive to chemotherapeutics via multidrug resistance,
which interrupts apoptosis signaling. Multidrug resistance involves
the overexpression of energy-dependent ATP-binding cassette
transporter protein, which detoxifies cancer cells and lowers
intracellular concentrations under the therapeutic threshold by
pumping drugs out at the expense of ATP hydrolysis (5,6).
Therefore, it is necessary to identify multidrug resistant
molecules in gastric cancer cells, and to develop more effective
diagnostic and therapeutic clinical strategies in order to treat
advanced gastric cancer.
Siva-1 exists in a wide variety of tissues and
cells, and serves as a proapoptotic protein (7). Siva-1 was elucidated by Prasad et
al (8) from a HeLa cell
library using yeast two-hybrid screening with a tumor necrosis
factor receptor. Although numerous studies (9–11)
have demonstrated that Siva-1 functions in the cytoplasm, studies
have also determined that Siva-1 can relocate into the nucleus
(12,13). The human Siva gene is located on
chromosome 14 (14), a region
which is often targeted for chromosomal translocation. Previous
studies have demonstrated that Siva-1 arrests apoptosis and
facilitates cancer development in osteosarcoma, hepatocellular
carcinoma and non-small cell lung cancer (15–18).
However, the molecular function of Siva-1 regulating multidrug
resistant in gastric cancer currently remains uncertain as previous
studies have presented confusing and ambiguous results (16), which has prompted further
investigation into both its function and associated signaling
pathway. Whether Siva-1 acts as a determinant for gastric cancer
development therefore requires further examination.
With the aim of gaining further knowledge regarding
the specific mechanisms of Siva-1 in gastric cancer and elucidating
molecular determinants for multidrug resistance, the current study
overexpressed Siva-1 in gastric cancer cells using a lentiviral
vector. Subsequently, effects on chemotherapeutic compound 50%
inhibitory concentration (IC50) values, apoptosis,
colony formation, metastasis and invasion were observed.
Preliminary experiments revealed that drug-sensitive (native)
gastric cancer cells were sensitive to chemotherapeutic drugs at
very low doses. Thus, it is appropriate to choose a
chemotherapeutic drug resistance gastric cancer cell line as a
research tool. The five commonly used chemotherapeutic drugs,
including vincristine (VCR), doxorubicin (DOX), platinum drugs,
5-fluorouracil (5-FU) and paclitaxel (PTX), in gastric cancer is of
great clinical interest (19).
Therefore, the vincristine (VCR)-resistant KATO III/VCR gastric
cancer cell line was selected for further experimentation. DOX is a
common substrate for P-glycoprotein (P-gp), which is one of the
major energy-dependent efflux transporters that contribute to MDR.
The ability to pump DOX was analyzed by flow cytometry to reveal
potential molecular determinants for multidrug resistance.
Additionally, the possible underlying mechanisms of multidrug
resistance were also investigated in the current study.
Materials and methods
Reagents
VCR, trypsin, penicillin and streptomycin were
obtained from Sigma-Aldrich (Merck KGaA). DOX (0.4 µg/ml) was
purchased from Sigma-Aldrich (Merck KGaA). Cells were cultured in
RPMI-1640 medium, which was purchased from Invitrogen (Thermo
Fisher Scientific, Inc.) along with fetal bovine serum (FBS).
Siva-1 (cat. no. 12532), NF-κB (cat. no. 8242), multidrug
resistance 1 (MDR1; cat. no. 13342), multidrug resistance protein
(MRP1; cat. no. 72202), Lamin B1 (cat. no. 13435) and GAPDH (cat.
no. 5174) antibodies were purchased from Cell Signaling Technology,
Inc. Horseradish peroxide (HRP)-conjugated goat anti-rabbit IgG
H&L (cat. no. ab205718) was purchased from Abcam. VCR (1.8
µg/ml), and 5-FU (20 µM) were purchased from SHRbio.
Cell culture
KATO III gastric cancer cells (obtained from the
Experimental Center of the People's Hospital of Guangxi Zhuang
Autonomous Region) and 293T cells (obtained from the Xiangya
Central Laboratory at Central South University, Changsha Hunan,
China) were cultured in RPMI-1640 supplemented with 10% FBS and
antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin), and
incubated at 37°C in a fully-humidified atmosphere containing 5%
CO2. KATO III/VCR cells were maintained culture medium
was supplemented with 0.6 µg/ml VCR to maintain drug-resistant
phenotypes.
Gene transfection
The DNA sequence of SIVA-1 was obtained from the
Gene Bank (ID No. NM_6427) and the cDNA, which included the entire
coding sequence (CDS) of SIVA-1 was obtained from Shanghai Cancer
Institute. pGV358-GFP (Shanghai Cancer Institute), which express
green fluorescent protein, were used to construct the
Siva-1-overexpression lentivirus. The Siva-1-overexpression
lentiviral vector (pGV358-GFP-SIVA-1) and negative control vector
(pGV358-GFP) was stored in the laboratory of Guangxi People's
Hospital. Lentiviruses were generated by co-transfecting
pGV358-GFP-SIVA-1 with pHelper1.0 and pHelper2.0 plasmids (Shanghai
Genechem Co., Ltd) into 293T cells according to the manufacturer's
protocol (20). Recombinant
pGV358-GFP-SIVA-1 plasmid was transfected into 293T cells to
determine LV titers using the end-point dilution method, which
involved counting the number of infected green cells under
fluorescence microscopy (magnification, ×100). The lentiviral titer
was calculated by the formula: lentiviral titer (TU/ml) = number of
positive cells × dilution times/volume of lentivirus used. KATO
III/VCR cells were plated at a low density (5×104
cells/well) in 6-well plates. Following incubation for 24 h, KATO
III/VCR cells were infected with polybrene (2 mg/ml; Shanghai
Genechem Co., Ltd) combined with recombinant lentiviruses at a
multiplicity of infection (MOI) value of 12 PFU/cell (MOI, 12), as
previously described (21).
Transfected cells were subsequently cultured in the presence of 600
mg/ml G418 (Invitrogen; Thermo Fisher Scientific) for 4 weeks,
after which stably overexpressed cell lines were generated. Cells
were then divided into three groups: i) KATO III/VCR + LV-Siva-1;
ii) KATO III/VCR + LV-negative control (NC); and iii) KATO
III/VCR.
Cytotoxicity assay
Cells (5×104 cells/ml) were cultured in
96-well tissue microplates (100 µl/well) and exposed to VCR (1.8
µg/ml). Following incubation for 48 h at 37°C, the cytotoxicity of
KATO III/VCR + LV-Siva-1, KATO III/VCR + LV-NC and KATO III/VCR
cells was determined via an MTT assay (Biological Industries) in
accordance with the manufacturer's protocol. MTT (0.1 mg/well) was
added for 4 h at 37°C before harvesting, and then DMSO (150
µl/well) was added to dissolve all the precipitation. Absorbance
values were measured at 450 nm using a microplate reader (PR 3100
TSC; Bio-Rad Laboratories, Inc.). Relative drug resistance folds
were analyzed and compared with IC50 values.
Measurement of DOX pump rate via flow
cytometry
The fluorescence intensity of intracellular
doxorubicin was determined using flow cytometry. KATO III gastric
cancer cells were seeded into 6-well plates, after which
doxorubicin was added to each well to a final concentration of 4
mg/ml. Samples were then cultured at 37°C for 30 min. Cells were
subsequently washed twice with fresh culture medium and incubated
at 37°C for 1 h. Doxorubicin levels were subsequently determined by
measuring the fluorescence intensity of doxorubicin in cells with
an excitation wavelength of 488 nm and an emission wavelength of
575 nm (21,22). The cells were analyzed using an
EPICS XL-MCL flow cytometry system (Beckman Coulter) and the data
was analyzed using MultiCycle Software for Windows (version 3.0,
Phoenix Flow Systems). The procedure was performed in triplicate
and an average value was obtained to calculate the pump rate of
doxorubicin using the following formula: Releasing index =
(accumulation value - retention value)/accumulation value.
Quantification of apoptosis via flow
cytometry
KATO III/VCR + LV-Siva-1, KATO III/VCR + LV-NC and
KATO III/VCR cells were harvested using 0.25% trypsin. Cells
(1×106 cells/ml) were subsequently washed twice with
ice-cold (4°C) PBS, treated with trypsin and fixed with 70% ethanol
at 4°C for 30 min. The cell suspension was incubated with Annexin
V-PE (2 µl/ml, BD Biosciences) and 7-amino-actinomycin D (7-AAD; 2
µl/ml) apoptosis detection kits (BD Biosciences) at room
temperature for 15 min according to the manufacturer's protocols.
The percentage of apoptotic cells were analyzed using flow
cytometry with an EPICS XL-MCL FACSCanto II cytometer (BD
Biosciences). The percentage of apoptotic cells (early and late) in
each quadrant was calculated using MultiCycle Software for Windows
(Beckman Instruments, Inc.) with the following equation: Apoptotic
index = the rate of Early apoptotic cells in lower right quadrant +
the rate of late apoptosis or necrosis in the upper right
quadrant.
Colony formation assay
Cells were seeded in 6-well plates (200 cells/well)
and incubated in the presence of VCR (0.6 g/ml) in a humidified
atmosphere of 95% air, 5% CO2 at 37°C for 14 days. Cells
were then washed twice with PBS, fixed with glutaraldehyde (6.0%
v/v) and stained with crystal violet (0.5% w/v). Colony numbers
were counted manually under an Olympus CKX53 inverted microscope
(Olympus Corporation) at ×40 magnification.
Transwell invasion assay
Cell invasion was assessed using an 8-µm Matrigel
invasion chamber (BD Bioscience) with 24-well plates. Cells in the
upper chamber (5×104) were suspended in 100 µl RPMI-1640
containing Matrigel (BD Biosciences) without serum, and the lower
chamber was seeded with 700 µl RPMI-1640 containing 10% FBS.
Following 48 h incubation, cells that had invaded through the
membranes were fixed using 4% polyoxymethylene for 5 min and
stained with Giemsa dye for 20 min at room temperature. The number
of visible cells was counted in five random fields of view under a
light microscope (magnification, ×200).
Wound healing assay
A total of ~3×106 cells were seeded in
6-well plates. When cell confluence reached between 90–100%, a
straight central linear wound was created in confluent cells using
a 200-µl sterile pipette tip. Subsequently, cells were rinsed twice
with PBS to remove any debris prior to culture in serum-free growth
medium. Wound healing was observed at different time points (0, 24,
48 and 72 h), and the wound size was imaged under an Olympus CKX53
inverted light microscope (magnification, ×40; Olympus
Corporation). The Digimizer software system (version no. 5.3.4;
MedCalc Software) was used to measure the distance between the two
edges of the scratch.
Western blotting
Cytoplasmic proteins were extracted using a cell
lysate extraction kit (Beijing Solarbio Science & Technology
Co., Ltd.) following the manufacturer's protocol. Nuclear proteins
were extracted using a EpiQuik™ Nuclear Extraction kit (cat. no.
OP-0002; EpiGentek Group, Inc.), 1×107 cells were
transferred to a 1.5-ml microcentrifuge tube and centrifuged at 500
× g at 4°C for 3 min to harvest the cell pellet. Ice-cold
cytoplasmic extraction reagent was added, and the microcentrifuge
tubes were left to incubate on ice for 1 min. Following incubation,
the tubes were centrifuged at 4°C for 5 min in a microcentrifuge
(16,000 × g at 4°C) to get the insoluble fraction, and subsequently
suspended in ice-cold nuclear extraction reagent. Then, the tubes
were centrifuged at 16,000 × g at 4°C for 10 min to get nuclear
extract (the supernatant). The concentration of extracted protein
was measured using a bicinchoninic acid protein assay kit (Thermo
Fisher Scientific, Inc.). A total of 2 µg of protein was loaded per
lane and separated via 12% SDS-PAGE, transferred to PVDF membranes
and blocked with 5% skimmed milk at 37°C for 60 min. Membranes were
subsequently incubated with the following primary antibodies
overnight at 4°C: Anti-Siva-1 (1:100), anti-MDR1 (1:1,500),
anti-MRP1 (1:1,000), anti-NF-κB (1:1,500), anti-Lamin B1 (1:1,000)
and anti-GAPDH (1:1,000). After three washes in TBS with 0.1%[v/v]
Tween-20 (Thermo Fisher Scientific, Inc.), membranes were incubated
with the HRP-conjugated goat anti-rabbit IgG H&L secondary
antibody (1:1,000) for 1 h at room temperature. The Odyssey Fc
Imaging System (LI-COR Biosciences) was used to analyze the optical
density of samples. The semi-quantitative analysis was performed
using ImageJ software (v 1.8.0; National Institutes of Health).
GAPDH and Lamin B1 served as loading controls.
Statistical analysis
All statistical analyses were performed using SPSS
version 13.0 (SPSS, Inc.), using Student's t-test, χ2
test or one-way ANOVA. Bonferroni post hoc analysis was employed to
perform multiple comparison tests. Data are presented as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Construction and identification of
LV-Siva-1-GFP lentiviral vectors
DNA sequence analysis demonstrated that the RNA
coding frames and frame sequences, as well as the recombinant
pGV-Siva-1-GFP and pGV-NC-GFP plasmids were constructed
successfully. Siva-1 and NC lentiviral vectors, LV-Siva-1-GFP and
LV-NC-GFP, were produced following co-transfection with a packaging
vector (pHelper 1.0) and a vesicular stomatitis virus glycoprotein
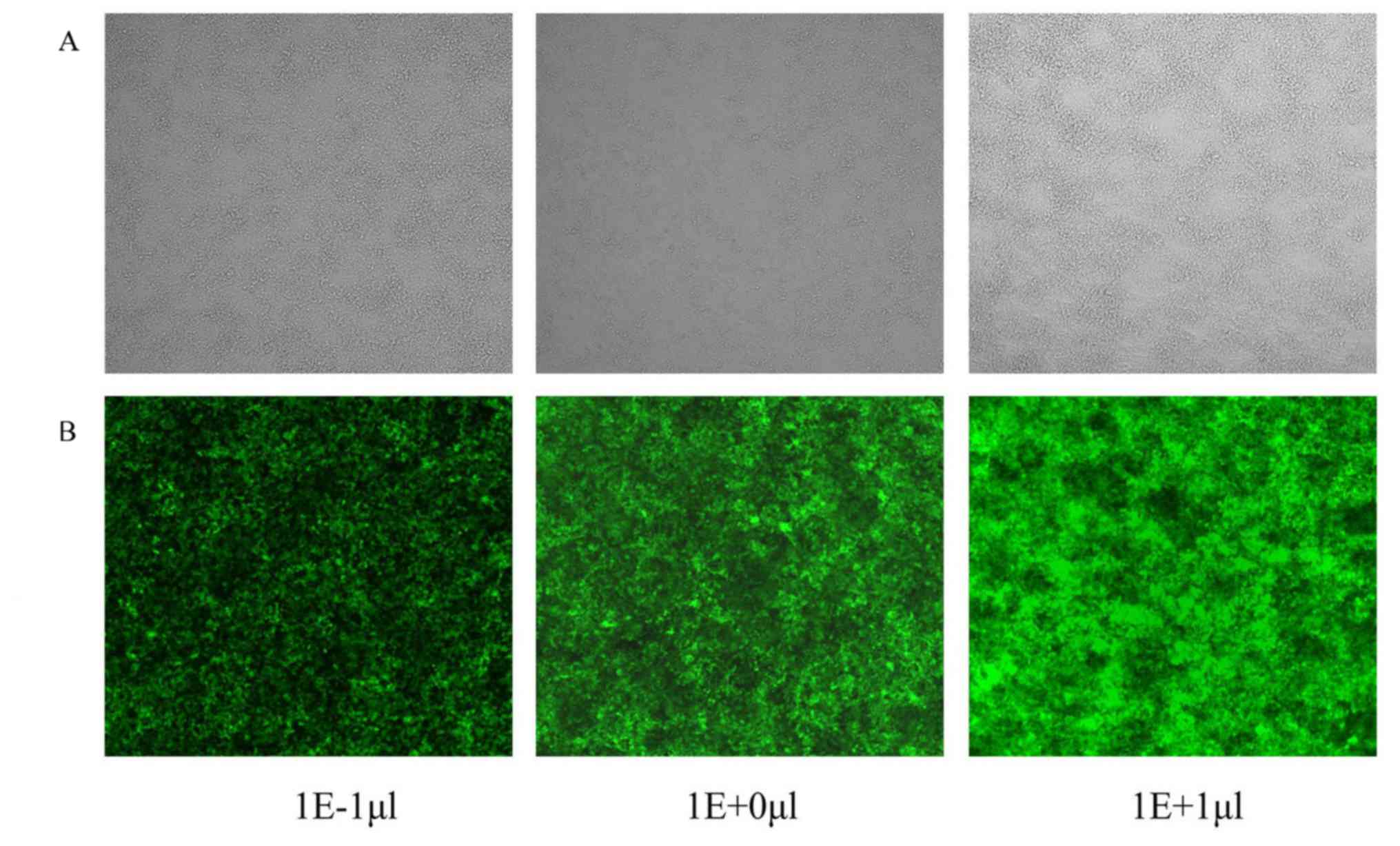
expression plasmid (pHelper 2.0) in 293T cells. As presented in
Fig. 1, GFP fluorescence indicated
that the lentiviral vector was successfully generated for use in
the present study. The viral titer was 5×108 TU/ml
medium.
Overexpression of Siva-1 in gastric
cancer cells with recombinant lentivirus
To determine the effects of Siva-1 overexpression in
gastric cancer cells transfected with recombinant lentivirus,
proteins were extracted from KATO III/VCR + LV-Siva-1, KATO III/VCR
+ LV-NC and KATO III/VCR cells, after which Siva-1 expression was
assessed by western blotting. The results indicated significantly
increased (1.6-fold) Siva-1 protein expression in KATO III/VCR +
LV-Siva-1 cells compared with KATO III/VCR + LV-NC and KATO III/VCR
cells (P<0.05; Fig. 2 and
Table I). No significant
differences were identified between the KATO III/VCR + LV-NC and
KATO III/VCR groups. The results indicated that KATO III/VCR cells
transfected with LV-Siva-1-GFP effectively translated more Siva-1
protein.
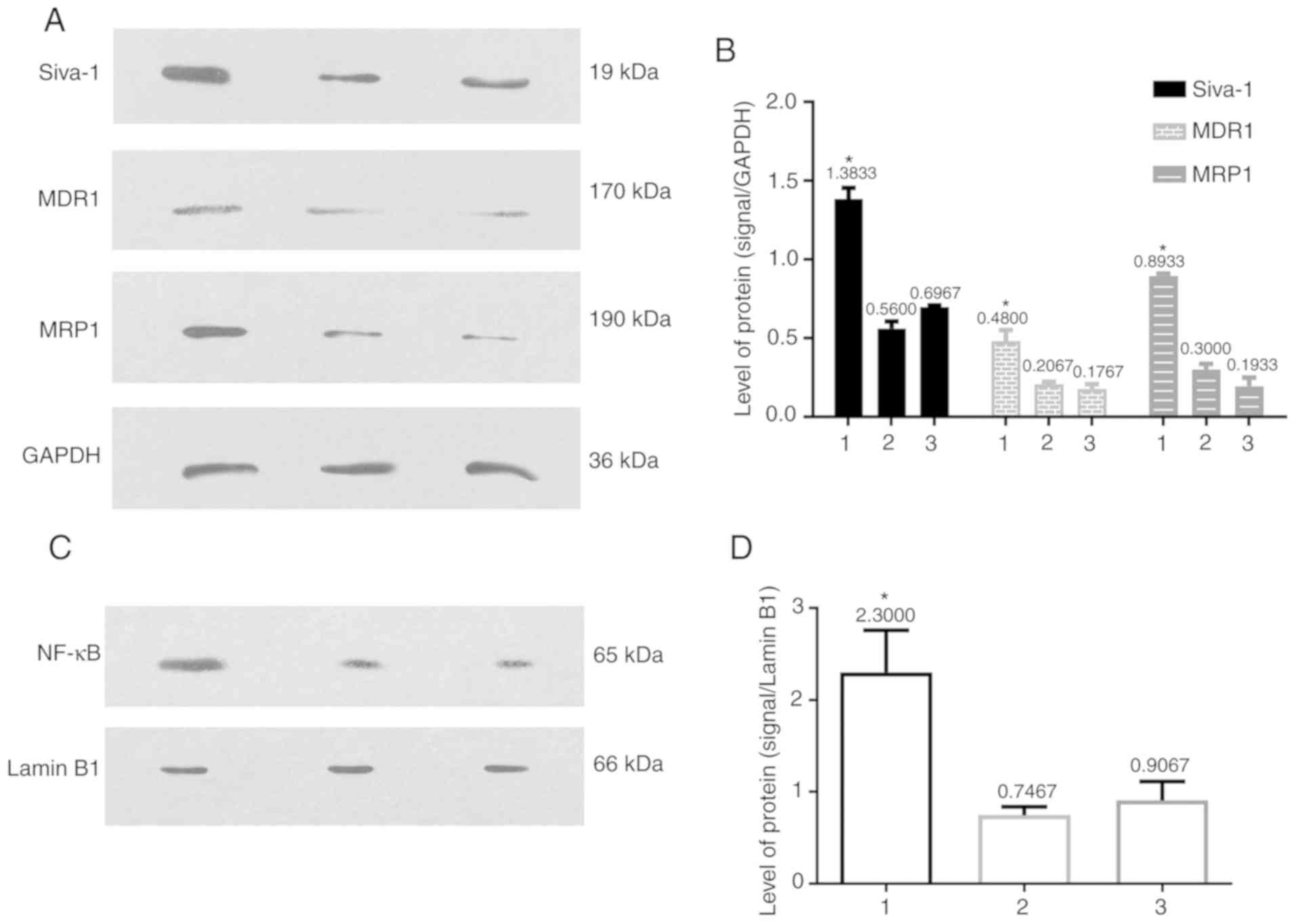 | Figure 2.Siva-1, NF-κB, MDR1 and MRP1 protein
expression is determined via western blotting. MDR1 and MRP1
protein levels were increased following Siva-1 overexpression and
NF-κB was active following its rapid translocation into the nucleus
after the same treatment. (A) Western blot analysis and (B)
subsequent semi-quantification of Siva-1, MDR1 and MRP1 protein
levels in the three groups. (C) Western blot analysis and (D)
subsequent semi-quantification of NF-κB protein levels in the three
groups. Expression was normalized to that of GAPDH or Lamin B1, and
presented as the mean ± standard deviation. *P<0.05 vs. group 2
and 3. 1, KATO III/VCR + LV-Siva-1; 2, KATO III/VCR + LV-NC; 3,
KATO III/VCR. MDR1, multidrug resistance 1; MRP1, multidrug
resistance protein 1; VCR, vincristine; LV, lentivirus; NC,
negative control. |
 | Table I.Relative expression of various
proteins determined by western blotting. |
Table I.
Relative expression of various
proteins determined by western blotting.
| Group | Siva-1 | MDR1 | MRP1 | NF-κB |
|---|
| KATO III/VCR +
LV-Siva-1 | 1.38±0.04 | 0.48±0.02 | 0.89±0.03 | 2.30±0.07 |
| KATO III/VCR +
LV-NC | 0.56±0.04 | 0.21±0.03 | 0.30±0.03 | 0.75±0.03 |
| KATO III/VCR | 0.70±0.06 | 0.18±0.02 | 0.19±0.07 | 0.91±0.03 |
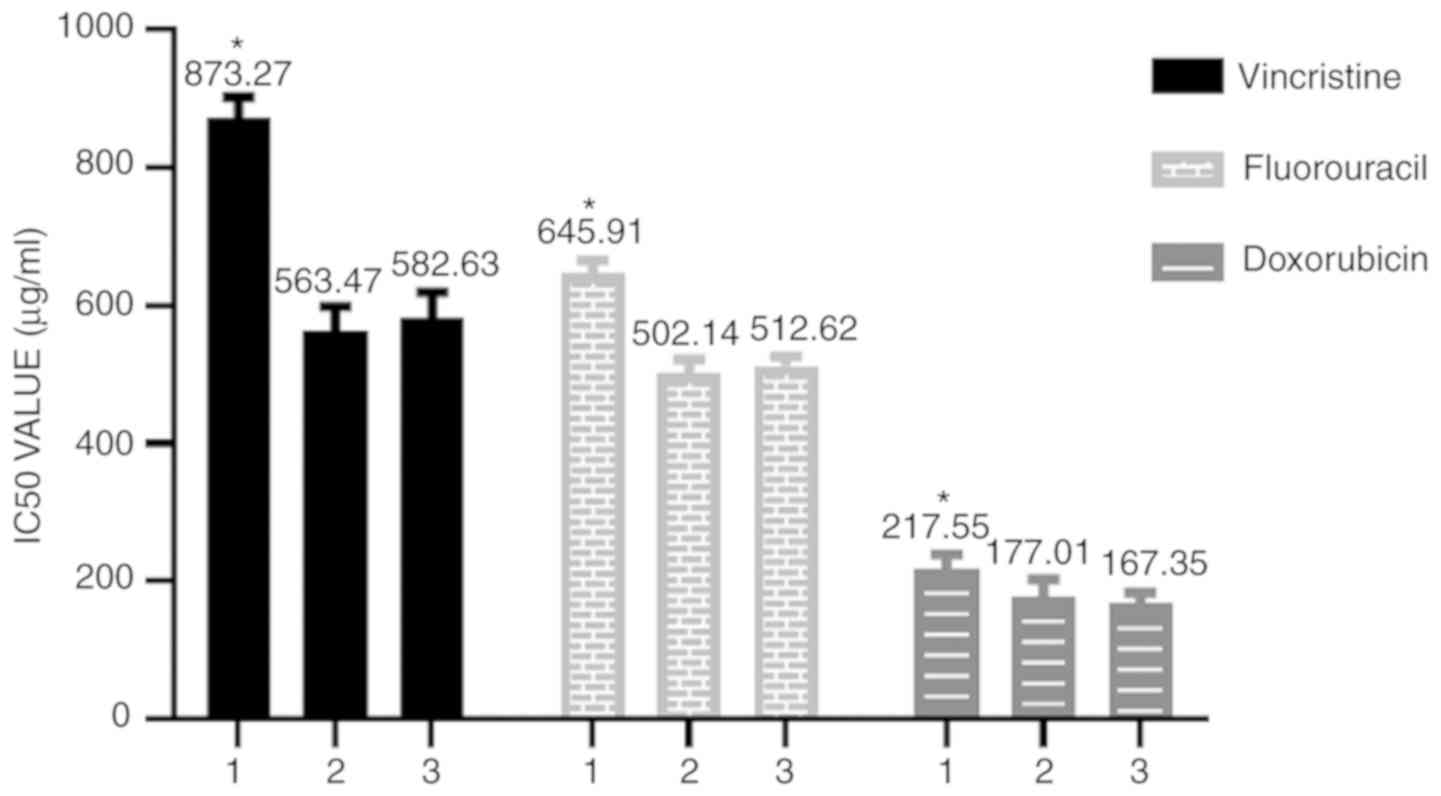
Siva-1 overexpression promotes
multidrug resistance
To elucidate the role of Siva-1 overexpression in
anticancer drug resistance, the IC50 values of KATO
III/VCR + LV-Siva-1 cells exposed to three clinical
chemotherapeutic drugs (VCR, 5-fluorouracil and doxorubicin) at
37°C were determined. The diluted VCR (1.8 µg/ml), 5-FU (20 µM) and
DOX (0.4 µg/ml) were added to each well for 48 h. The results
revealed that compared with the KATO III/VCR + LV-NC group and the
KATO III/VCR group, the KATO III/VCR + LV-Siva-1 group exhibited
significantly increased IC50 values for VCR,
5-fluorouracil and doxorubicin (P<0.05; Fig. 3 and Table II).
 | Table II.IC50 values were
determined for anticancer drugs applied to KATO III/VCR cells by a
MTT assay. |
Table II.
IC50 values were
determined for anticancer drugs applied to KATO III/VCR cells by a
MTT assay.
| Group | Vincristine
(µg/ml) | 5-fluorouracil
(µg/ml) | Doxorubicin
(µg/ml) |
|---|
| KATO III/VCR +
LV-Siva-1 | 873.27±29.31 | 645.91±20.37 | 217.55±21.12 |
| KATO III/VCR +
LV-NC | 563.47±35.25 | 502.14±19.57 | 177.01±25.91 |
| KATO III/VCR | 582.63±37.25 | 512.62±13.72 | 167.35±16.52 |
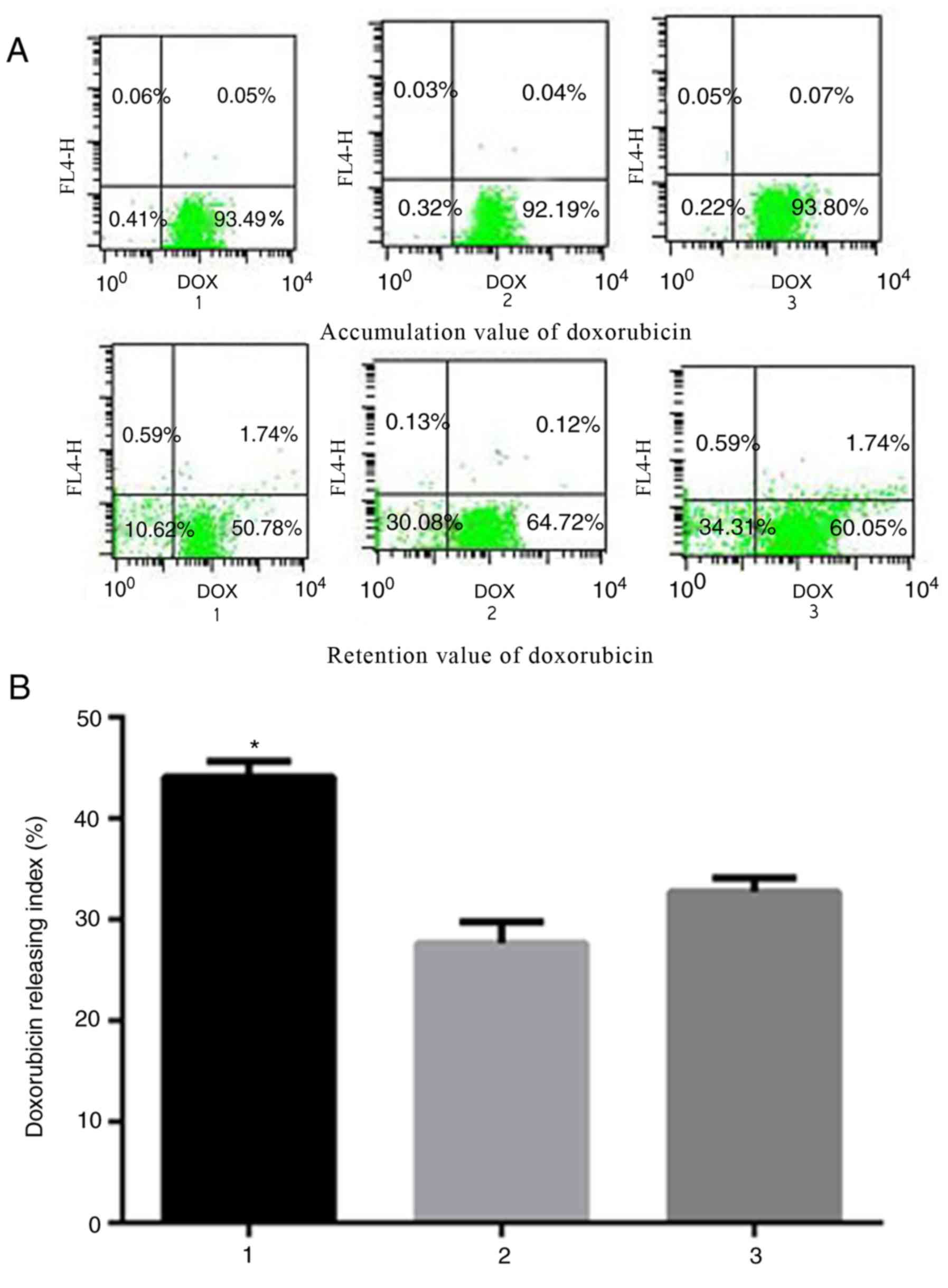
Siva-1 overexpression increases the
pump rate of doxorubicin
MRP1 is best known for its contributions to
chemoresistance, serving a role in anticancer drug efflux (23). Intracellular drug accumulation and
retention were evaluated using doxorubicin as a probe in gastric
cancer cells. As indicated in Fig.
4, the pump rates of doxorubicin in KATO III/VCR + LV-Siva-1
vs. KATO III/VCR + LV-NC cells and KATO III/VCR cells were
44.12±1.54% vs. 27.66±2.12% and 32.72±1.36%, respectively
(P<0.05). The results indicated that the KATO III/VCR +
LV-Siva-1 group exhibited significantly decreased doxorubicin
accumulation and retention, as well as higher release indices of
doxorubicin, which suggested that Siva-1 overexpression increased
drug efflux in gastric cancer cells and promoted drug
resistances.
Siva-1 overexpression prevents
cellular apoptosis and promotes KATO III/VCR cell
proliferation
To verify the hypothesis that Siva-1 overexpression
suppressed gastric cancer cell apoptosis and promoted
vincristine-resistant human gastric cancer cell proliferation, the
effect of LV-Siva-1-GFP on vincristine-induced gastric cancer cell
apoptosis was determined by calculating the apoptosis index. Cells
were stained with Annexin V PE and 7-AAD, and analyzed by flow
cytometry. The results revealed that the apoptotic rate of the KATO
III/VCR + LV-Siva-1 group was 8.03±0.2%, which was significantly
lower than that of the KATO III/VCR + LV-NC (18.99±0.34%) and KATO
III/VCR groups (17.93±0.29%; P<0.05; Fig. 5A and B). Furthermore, the results
of the colony formation assay indicated that Siva-1-overexpressing
KATO III cells increased colony formation (21.00±2.00) compared
with control cells (11.33±2.52 and 10.67±3.06, respectively;
P<0.05; Fig. 5C and D).
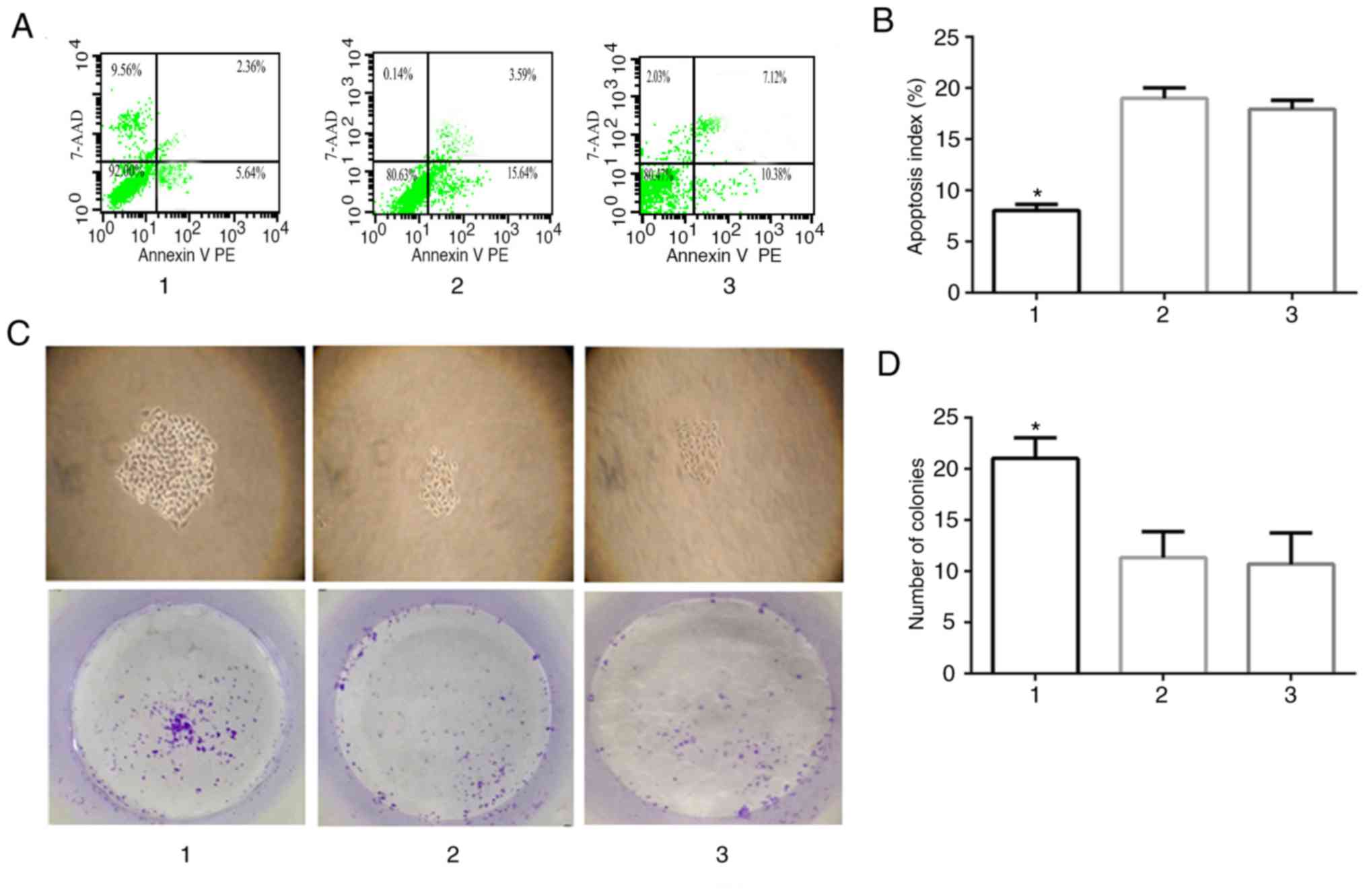 | Figure 5.Effect of Siva-1 overexpression on
KATO III/VCR cell growth. (A and B) Apoptotic rate in Siva-1
overexpressed-KATO III/VCR cells was analyzed by flow cytometry. (C
and D) KATO III/VCR + LV-Siva-1 cells, KATO III/VCR + LV-NC cells
and KATO III/VCR cells were plated in 6-well plates at a density of
200 cells/well, after which colony growth was observed under an
optical microscope following 14 days (magnification, ×40). The
surviving fraction of cells (visible colonies) was stained with
gentian violet and counted manually. Data are presented as the mean
± standard deviation from 3 independent experiments. *P<0.05 vs.
group 2 and 3. 1, KATO III/VCR + LV-Siva-1; 2, KATO III/VCR +
LV-NC; 3, KATO III/VCR; VCR, vincristine; LV, lentivirus; NC,
negative control; 7-AAD, 7-amino-actinomyosin D. |
Siva-1 promotes migration and invasion
in vitro
Stable Siva-1 overexpression was induced in KATO
III/VCR, KATO III/VCR + LV-Siva-1 and KATO III/VCR + LV-NC cells to
ascertain the role of Siva-1 in cell migration and invasion. The
results of the wound healing assay revealed that Siva-1
overexpression in KATO III/VCR cells significantly enhanced wound
healing by increasing wound closure and cell migration (P<0.05;
Fig. 6A and B). In addition, the
results of Transwell assays indicated that cells with higher Siva-1
expression demonstrated significantly increased invasion
(P<0.05; Fig. 6C and D). These
data suggested that Siva-1 overexpression increased the invasive
abilities of VCR-resistant human gastric cancer cells in
vitro.
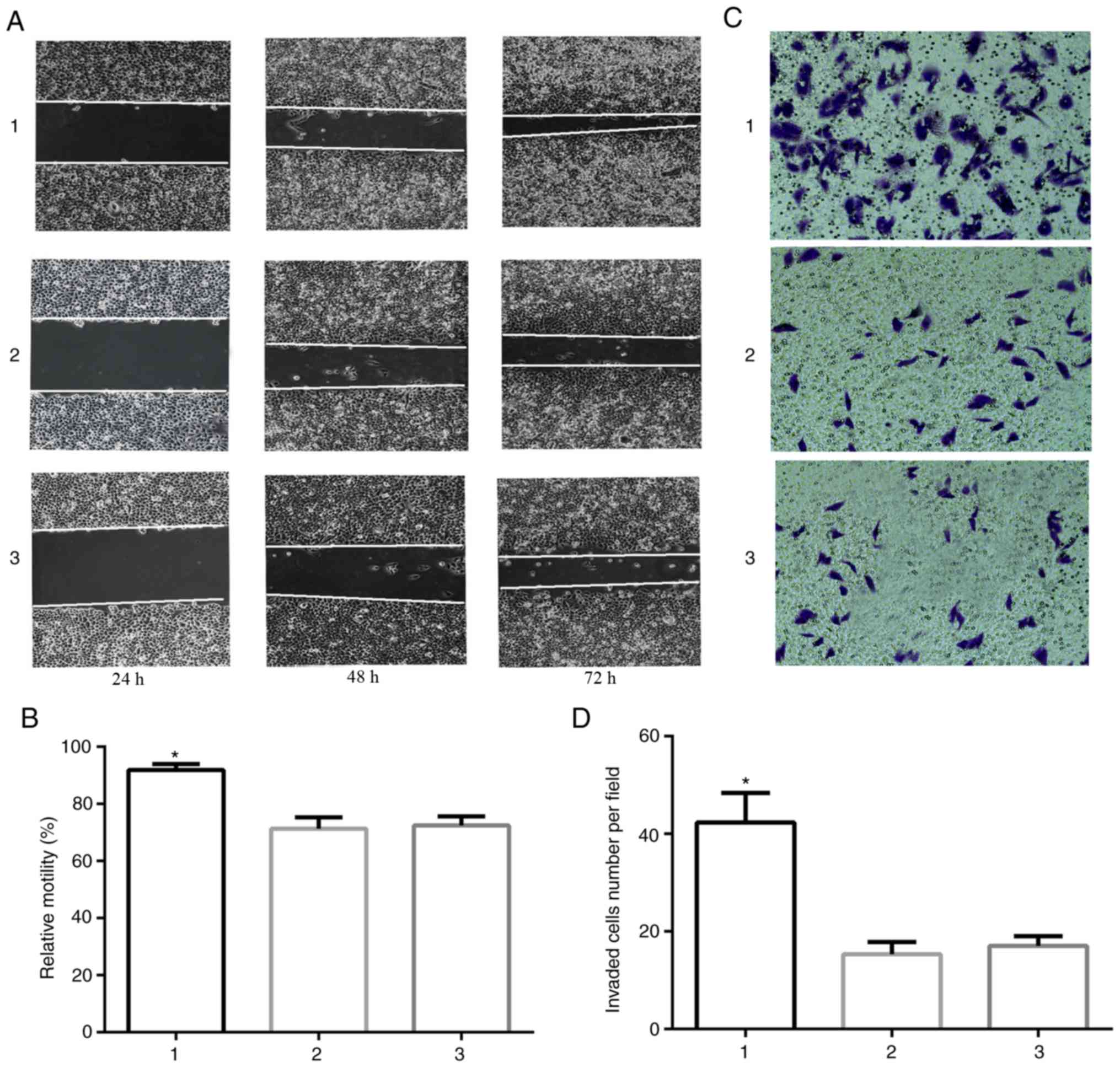 | Figure 6.Siva-1 overexpression increases KATO
III/VCR cell migration and invasion. (A and B) KATO III/VCR +
LV-Siva-1 cells, KATO III/VCR + LV-NC cells and KATO III/VCR cells
were cultured to confluence on 6-well plates, after which a central
linear wound was created with a 200-µl sterile pipette tip. The
wound was imaged over a 4-day interval (magnification, ×40). (C and
D) KATO III/VCR + LV-Siva-1 cells, KATO III/VCR + LV-NC cells and
KATO III/VCR cells were loaded into the upper chambers of a
Matrigel-coated Transwell plate. Filtrated cells on the
undersurface of the polycarbonate membranes were stained and
counted under an optical microscope after 48 h (magnification,
×200). *P<0.05 vs. group 2 and 3. 1, KATO III/VCR + LV-Siva-1;
2, KATO III/VCR + LV-NC; 3, KATO III/VCR. VCR, vincristine; LV,
lentivirus; NC, negative control. |
Siva-1 overexpression increases NF-κB,
MDR1 and MRP1 expression
To investigate the mechanism by which LV-Siva-1-GFP
induces MDR in KATO III/VCR cells, levels of several well-known
multidrug resistance-associated proteins, including MDR1 and MRP1,
were determined by western blotting. A cell fractionation kit was
used to obtain nuclear lysates, after which western blotting was
performed using antibodies against NF-κB in the nucleus. The
results revealed that MDR1, MRP1 and nuclear NF-κB levels were
higher in the KATO III/VCR + LV-Siva-1 group compared with the KATO
III/VCR + LV-NC and KATO III/VCR groups (P<0.05). However, no
significant differences were identified between the latter two
groups (Fig. 2).
Discussion
Gastric cancer is one of the most common types of
digestive tract malignancy worldwide with the highest incidence and
mortality rates (24) Although the
surgical removal of lesions is currently the main treatment for
patients with gastric cancer, chemotherapy still serves a key role
post-surgery in eradicating malignant cells as the majority of
patients are diagnosed at an advanced stage. Multidrug resistance
is usually associated with the poor prognosis of patients with
gastric cancer (25,26). Therefore, preventing multidrug
resistance to improve the efficacy of chemotherapy is imperative.
Chemoresistance represents an event whereby cancer cells exhibit
tolerance to a specific chemotherapeutic agent or class of
pharmaceutical drug. The development of chemoresistance is a major
obstacle for successful anticancer therapy. Understanding the
molecular mechanisms underlying chemoresistance is therefore
necessary to improve the therapeutic efficacy of cytotoxic
drugs.
The Siva-1 protein serves a crucial role in certain
extrinsic and intrinsic apoptosis signaling pathways (16). However, Siva-1 has been
demonstrated to serve contradictory roles in previous studies. For
example, Siva-1 is downregulated in colorectal cancer and breast
cancer (27–29), but as an important regulator of
apoptosis and metastasis, Siva-1 is also highly expressed and
facilitates tumorigenesis in a number of malignant tumors,
including ovarian cancer (30),
osteosarcoma (18), non-small cell
lung cancer (17) and gastric
cancer (29). Although Siva-1 was
initially identified as a promoter of apoptosis (7), the underlying molecular mechanism
requires further investigation. The results of the present study
indicated that Siva-1 overexpression inhibited apoptosis and
enhanced multidrug resistance. It was also revealed in this study
that Siva-1 increased the colony formation and invasion of cells,
potentially by acting to decrease the expression of NF-κB. NF-κB is
a transcription factor that regulates the expression of a wide
variety of genes involved in various cellular events, including
inflammation, immune response, proliferation, apoptosis and
multidrug resistance (31–33). Additionally, NF-κB serves a key
role in cancer development and metastasis (34). NF-κB is inactive in the cytoplasm
when bound to IκB. When IκB is ubiquitinated and subsequently
degraded, NF-κB is exposed to a nuclear localization sequence on
the NF-κB subunit RelA (p65), transferring the molecule to the
nucleus (35,36). The results of the current study
indicated that Siva-1 functioned as a regulator of MDR1 and MRP1
gene expression in gastric cancer cells via promotion of NF-κB
expression. Overexpression of Siva-1 in VCR-resistant cell lines
decreased the sensitivity of KATO III/VCR cells towards VCR by
enhancing the activity of NF-κB and thereby increasing the
expression of MDR1 and MRP1 to enhance chemoresistance. This result
is consistent with a previous report in which NF-κB activated the
overexpression of antiapoptotic genes (37).
The findings of the present study indicated that
Siva-1 may serve as a regulator for drug-related signal proteins,
MDR1 and MRP1, in vitro. These results are consistent with
additional in vivo analyses performed in a separate study:
KATO III/VCR cells were implanted subcutaneously into the flanks of
BALB/c nude mice. When the resulting tumor measured 5 mm in
diameter, animals were administered an intratumoral injection of
LV-Siva-1-GFP or LV-GFP. VCR was administered via intraperitoneal
injection. The tumor volumes were monitored and analyzed (38,39).
The above results (unpublished data) of the procedure in
vivo was consistent with the results of the current study. This
may also indicate the role served by Siva-1 in overcoming drug
resistance in gastric cancer.
MDR1 and MRP1 have been widely investigated as
multidrug resistance proteins and are associated with cancer
therapeutic resistance. MDR1 and MRP1 have also been identified as
the major drug efflux pumps responsible for multidrug resistance
(40). The MDR1 gene sequence was
examined by Bentires-Alj et al (41) and a putative NF-κB binding site
(CCTTTCGGGG) was identified in the first intron of the MDR1 gene
promoter. The expression of MDR1 (also termed P-glycoprotein) RNA
can be reduced by promoting the expression of NF-κB so that
sensitivity to chemotherapy can be enhanced in digestive malignant
cells (42). Drug efflux
transporters, including MRP1, can significantly influence the
transfer of drugs. The major roles of MRP1 include the efflux of
endogenous metabolites, the transport of inflammatory mediators and
the development of drug resistance in a variety of diseases
(43). MRP1, an ATP-dependent
transmembrane glycoprotein, is ubiquitously expressed and
participates in the multidrug resistance of various types of tumor
cell (44,45).
The current study hypothesized that suppressed NF-κB
levels mediated by Siva-1 could be used to treat patients with
gastric cancer and multidrug resistance. The study indicated that
Siva-1 acts as a cancer-promoting factor and a antiapoptotic
protein. MDR1 and MRP1 gene regulation were analyzed by
overexpressing Siva-1 and subsequently upregulating NF-κB in the
KATO III gastric cancer cell lines. Given the crucial role of
Siva-1 in the regulation of apoptosis and tumor metastasis, it may
represent a potential target to address the major challenges of
therapeutic intervention in patients with cancer, including cancer
relapse and chemotherapy resistance.
Acknowledgements
The authors would like to thank Mrs. Chang Wang
(Guangxi Medical University, Taiyuan Langdong Hospital) for her
assistance and contribution to the team.
Funding
The current study was supported by grants from the
Scientific Research Project of Guangxi Health Commission (grant no.
Z20180739), the Youth Foundation of People's Hospital of Guangxi
Zhuang Autonomous Region (grant no. QN2018-22) and the Natural
Science Foundation of China (grant no. 81660416).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FBK performed the western blot experiments and made
substantial contributions to writing the manuscript. QMD
established the gastric cancer cell line with stable overexpression
of Siva-1. HQD measured the IC50 of gastric cells to
vincristine, 5-fluorouracil and doxorubicin. CCD performed the flow
cytometry experiments. LL performed the colony formation assay and
wound healing assay. CGH performed the Transwell assay. XTW
acquired funding for the project and performed the western blot
experiments. SX drafted the manuscript and contributed to the
conception and design. WM analyzed the data generated during the
study. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Emmings E, Mullany S, Chang Z, Landen CN
Jr, Linder S and Bazzaro M: Targeting mitochondria for treatment of
chemoresistant ovarian cancer. Int J Mol Sci. 20:2292019.
View Article : Google Scholar
|
|
2
|
Kongsema M, Wongkhieo S, Khongkow M, Lam
EW, Boonnoy P, Vongsangnak W and Wong Ekkabut J: Molecular
mechanism of Forkhead box M1 inhibition by thiostrepton in breast
cancer cells. Oncol Rep. 42:953–962. 2019.PubMed/NCBI
|
|
3
|
Zeng L, Liao Q, Zou Z, Wen Y, Wang J, Liu
C, He Q, Weng N, Zeng J, Tang H, et al: Long non-coding RNA
XLOC_006753 promotes the development of multidrug resistance in
gastric cancer cells through the PI3K/AKT/mTOR signaling pathway.
Cell Physiol Biochem. 51:1221–1236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen S, Wu J, Jiao K, Wu Q, Ma J, Chen D,
Kang J, Zhao G, Shi Y, Fan D, et al: MicroRNA-495-3p inhibits
multidrug resistance by modulating autophagy through GRP78/mTOR
axis in gastric cancer. Cell Death Dis. 9:10702018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kozovska Z, Gabrisova V and Kucerova L:
Colon cancer: cancer stem cells markers, drug resistance and
treatment. Biomed Pharmacother. 68:911–916. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alamolhodaei NS, Tsatsakis AM, Ramezani M,
Hayes AW and Karimi G: Resveratrol as MDR reversion molecule in
breast cancer: An overview. Food Chem Toxicol. 103:223–232. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu T, Ma Y, Wang Z, Zhang W and Yang X:
Siva 1 inhibits cervical cancer progression and its clinical
prognosis significance. Cancer Manag Res. 12:303–311. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Prasad KV, Ao Z, Yoon Y, Wu MX, Rizk M,
Jacquot S and Schlossman SF: CD27, a member of the tumor necrosis
factor receptor family, induces apoptosis and binds to Siva, a
proapoptotic protein. Proc Natl Acad Sci USA. 94:6346–6351. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen JY, Yang LX and Huang ZF: The N
terminal 33 amino acid domain of Siva 1 is sufficient for nuclear
localization. Braz J Med Biol Res. 46:1021–1027. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cottle DL, McGrath MJ, Wilding BR, Cowling
BS, Kane JM, D'Arcy CE, Holdsworth M, Hatzinisiriou I, Prescott M,
Brown S, et al: SLIMMER (FHL1B/KyoT3) interacts with the
proapoptotic protein Siva-1 (CD27BP) and delays skeletal myoblast
apoptosis. J Biol Chem. 284:26964–26977. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chu F, Borthakur A, Sun X, Barkinge J,
Gudi R, Hawkins S and Prasad KV: The Siva-1 putative amphipathic
helical region (SAH) is sufficient to bind to BCL-XL and sensitize
cells to UV radiation induced apoptosis. Apoptosis. 9:83–95. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Balci-Peynircioglu B, Waite AL, Hu C,
Richards N, Staubach-Grosse A, Yilmaz E and Gumucio DL: Pyrin,
product of the MEFV locus, interacts with the proapoptotic protein,
Siva. J Cell Physiol. 216:595–602. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin FT, Lai YJ, Makarova N, Tigyi G and
Lin WC: The lysophosphatidic acid 2 receptor mediates
down-regulation of Siva-1 to promote cell survival. J Biol Chem.
282:37759–37769. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rapisarda V, Ledda C, Matera S, Fago L,
Arrabito G, Falzone L, Marconi A, Libra M and Loreto C: Absence of
t(14;18) chromosome translocation in agricultural workers after
short term exposure to pesticides. Mol Med Rep. 15:3379–3382. 2107.
View Article : Google Scholar
|
|
15
|
Li T, Lv M, Chen X, Yu Y, Zang G and Tang
Z: Plumbagin inhibits proliferation and induces apoptosis of
hepatocellular carcinoma by downregulating the expression of SIVA.
Drug Des Devel Ther. 13:1289–1300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vachtenheim J Jr, Lischke R and
Vachtenheim J: Siva 1 emerges as a tissue specific oncogene beyond
its classic role of a proapoptotic gene. Onco Targets Ther.
11:6361–6367. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Van Nostrand JL, Brisac A, Mello SS,
Jacobs SB, Luong R and Attardi LD: The p53 target gene SIVA enables
non-small cell lung cancer development. Cancer Discov. 5:622–635.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du W, Jiang P, Li N, Mei Y, Wang X, Wen L,
Yang X and Wu M: Suppression of p53 activity by Siva1. Cell Death
Differ. 6:1493–1504. 2009. View Article : Google Scholar
|
|
19
|
Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv
L, Liu J, Xu Y, Shen Y and Yang M: Noncoding RNAs in gastric
cancer: Implications for drug resistance. Mol Cancer. 19:622020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang XT, Xie YB and Xiao Q: Lentivirus
mediated RNA interference targeting E2F 1 inhibits human gastric
cancer MGC 803 cell growth in vivo. Exp Mol Med. 43:638–645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan LH, Wang XT, Yang J, Lian C, Kong FB,
Wei WY, Luo W, Xiao Q and Xie YB: Reversal of multidrug resistance
in gastric cancer cells by CDX2 downregulation. World J
Gastroenterol. 19:4155–4165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan LH, Wei WY, Cao WL, Zhang XS, Xie YB
and Xiao Q: Overexpression of CDX2 in gastric cancer cells promotes
the development of multidrug resistance. Am J Cancer Res.
5:321–332. 2014.PubMed/NCBI
|
|
23
|
Lorendeau D, Dury L, Genoux Bastide E,
Lecerf Schmidt F, Simões Pires C, Carrupt PA, Terreux R, Magnard S,
Di Pietro A, Boumendjel A, et al: Collateral sensitivity of
resistant MRP1 overexpressing cells to flavonoids and derivatives
through GSH efflux. Biochem Pharmacol. 90:235–245. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang ZZ, Yu WX, Zheng M, Liao XH, Wang
JC, Yang DY, Lu WX, Wang L, Zhang S, Liu HK, et al: PIN1 Inhibition
Sensitizes Chemotherapy in Gastric Cancer Cells by Targeting Stem
Cell-like Traits and Multiple Biomarkers. Mol Cancer Ther.
19:906–919. 2020.PubMed/NCBI
|
|
26
|
Peng L, Li Y, Wei S, Li X, Dang Y, Zhang W
and Zhang G: LAMA4 activated by Androgen receptor induces the
cisplatin resistance in gastric cancer. Biomed Pharmacother.
124:1096672020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barkinge JL, Gudi R, Sarah H, Chu F,
Borthakur A, Prabhakar BS and Prasad KV: The p53-induced Siva-1
plays a significant role in cisplatin-mediated apoptosis. J
Carcinog. 8:22009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okuno K, Yasutomi M, Nishimura N, Arakawa
T, Shiomi M, Hida J, Ueda K and Minami K: Gene expression analysis
in colorectal cancer using practical DNA array filter. Dis Colon
Rectum. 44:295–299. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li N, Jiang P, Du W, Wu Z, Li C, Qiao M,
Yang X and Wu M: Siva1 suppresses epithelial-mesenchymal transition
and metastasis of tumor cells by inhibiting stathmin and
stabilizing microtubules. Proc Natl Acad Sci USA. 108:12851–12856.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei SH, Lin F, Wang X, Gao P and Zhang HZ:
Prognostic significance of stathmin expression in correlation with
metastasis and clinicopathological characteristics in human ovarian
carcinoma. Acta Histochem. 110:59–65. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Huang S, Jiang Y, Liu Y, Song T,
Li D and Yang L: Reactive astrocytes increase the expression of P
gp and Mrp1 via TNF α and NF κB signaling. Mol Med Rep.
17:1198–1204. 2017.PubMed/NCBI
|
|
32
|
Cui X, Shen D, Kong C, Zhang Z, Zeng Y,
Lin X and Liu X: NF-κB suppresses apoptosis and promotes bladder
cancer cell proliferation by upregulating survivin expression in
vitro and in vivo. Sci Rep. 7:407232017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen MY, Wang Y, Cui SY, Wu XL, Guo Y and
Xu RR: MicroRNA-125a regulates proliferation and apoptosis of acute
myeloid leukemia through targeting NF-κB pathway. Eur Rev Med
Pharmacol Sci. 23:3594–3601. 2019.PubMed/NCBI
|
|
34
|
Huber MA, Azoitei N, Baumann B, Grünert S,
Sommer A, Pehamberger H, Kraut N, Beug H and Wirth T: NF κB is
essential for epithelial mesenchymal transition and metastasis in a
model of breast cancer progression. J Clin Investig. 114:569–581.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nakanishi C and Toi M: Nuclear
factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat
Rev Cancer. 5:297–309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu T, Wei R, Zhang Y, Chen W and Liu H:
Association between NF-κB expression and drug resistance of liver
cancer. Oncol Lett. 17:1030–1034. 2018.PubMed/NCBI
|
|
37
|
Tanaka T, Hozumi Y, Iino M and Goto K:
NAP1L1 regulates NF-κB signaling pathway acting on anti-apoptotic
Mcl-1 gene expression. Biochim Biophys Acta Mol Cell Res.
1864:1759–1768. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei W, Cao W, Zhan Z, Yan L, Xie Y and
Xiao Q: miR-1284 suppresses gastric cancer progression by targeting
EIF4A1. OncoTargets Ther. 12:3965–3976. 2019. View Article : Google Scholar
|
|
39
|
Cao W, Wei W, Zhan Z, Xie D, Xie Y and
Xiao Q: Regulation of drug resistance and metastasis of gastric
cancer cells via the microRNA647 ANK2 axis. Int J Mol Med.
41:1958–1966. 2018.PubMed/NCBI
|
|
40
|
Strouse JJ, Ivnitski Steele I, Njus HM,
Foutz TD, Yao T, Weiner WS, Schroeder CE, Simpson DS, Maki BE, Li
K, et al: Selective Efflux Inhibition of ATP binding Cassette Sub
family G Member 2. Probe Reports from the NIH Molecular Libraries
Program (Internet) (Bethesda (MD)). National Center for
Biotechnology Information (US). Nov 30–2010.9:(updated Feb 25,
2013).
|
|
41
|
Bentires-Alj M, Barbu V, Fillet M, Chariot
A, Relic B, Jacobs N, Gielen J, Merville MP and Bours V: NF-kappaB
transcription factor induces drug resistance through MDR1
expression in cancer cells. Oncogene. 22:90–97. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yuan Z, Liang X, Zhan Y, Wang Z, Xu J, Qiu
Y, Wang J, Cao Y, Le VM, Ly HT, et al: Targeting CD133 reverses
drug resistance via the AKT/NF κB/MDR1 pathway in colorectal
cancer. Br J Cancer. 122:1342–1353. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang AK, Zhou ZW, Wei MQ, Liu JP and Zhou
SF: Modulators of multidrug resistance associated proteins in the
management of anticancer and antimicrobial drug resistance and the
treatment of inflammatory diseases. Curr Top Med Chem.
10:1732–1756. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sisodiya SM, Lin WR, Harding BN, Squier MV
and Thom M: Drug resistance in epilepsy: Expression of drug
resistance proteins in common causes of refractory epilepsy. Brain.
125:22–31. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Han F, Liu G, Sun C and Wei J: Ailanthone
reverses multidrug resistance by inhibiting the P glycoprotein
mediated efflux in resistant K562/A02 cells. Cell Mol Biol (Noisy
le grand). 64:55–61. 2018. View Article : Google Scholar : PubMed/NCBI
|