Introduction
Cancer evolves through different abnormal molecular
signaling pathways, including chronic inflammation events.
Dysregulation of the inflammatory process might lead to chronic
inflammation that contributes to many diseases, including cancer,
cardiovascular, diabetes, lung diseases, Alzheimer's, and
autoimmune diseases (1,2). The relationship between cancer and
inflammation is well-established (3). Chronic inflammation is a hallmark of
tumorigenesis that significantly mediates various stages in cancer,
including cellular changes, initiation, promotion, proliferation,
survival, invasion, angiogenesis, and metastasis (1), leading to approximately 15 to 20% of
all cancer-related deaths worldwide (4).
Meanwhile, breast cancer (BC) is a heterogeneous
disease (5) that demonstrated its
link to inflammation (4). In the
United States, 15% of the diagnosed BC cases are classified as
basal-like BC subtype according to the molecular profile (6,7).
Triple-negative breast cancer (TNBC) is the most aggressive and
metastatic subgroup that comprises up to 75% of the basal-like BC
(8,9), and it is known to be more profound
among African American (AA) patients than Caucasian American (CA)
patients (10). In TNBC cells, the
absence of three specific receptors: Estrogen (ER), progesterone
(PR) and human epidermal growth factor (Her2/neu) is a difficult
challenge in treating the disease (11,12).
Even though, TNBC has an initial significant response to many
chemotherapy agents (13),
approximately 30% of the patients experience poor prognosis and
treatment failure after repeated exposure to these agents, leading
to a median survival of one year (14).
Chronic inflammation-associated cancer is
characterized by the presence of leukocyte infiltration,
prominently macrophages that produce chemokines and cytokines
(15). These cytokines are the
critical mediators of communication between neoplastic cells in the
inflammatory tumor microenvironment (16). Particularly in BC, two chemokines,
IL-8, and CCL2 regulate tumor angiogenesis (17,18),
survival, and metastasis of cancer cells (19). In addition to their chemotactic
role, these two chemokines enable the recruitment of other cells,
including monocytes, neutrophils, T lymphocytes, and NK cells
(20). Under normal conditions,
the pro-inflammatory cytokine, tumor necrosis factor-α (TNF-α),
mediates the inflammatory pathway and controls inflamed cells
(1). In an aggressive BC
environment, TNF-α is excessively produced by the tumor-associated
macrophages (TAMs) (21,22) and substantially upregulates many
genes involved in cancer cell proliferation, invasion, and
metastasis (23).
In normal tissue, homeostasis of cell number and
rational cell functions are precisely controlled and maintained
through the release of growth-promoting signals. Cell signal
dysregulation and cell evolvement to the neoplastic state may
impair growth suppressor genes that could lead to prolonging
proliferative signaling, cell death resistance, angiogenesis, and
even triggering invasion and metastasis (24). In the context of cancer, many
signaling pathways, including JAK-STAT, MAPK, PI3K-AKT, NF-қB,
Notch, and Wnt, have received substantial attention as targets in
cancer therapeutic intervention. The Janus kinase (JAK)-signal
transducer and activator of transcription (STAT) pathway is
involved in the cellular response to cytokines and play a pivotal
role in the growth factor signaling and apoptosis. Impaired
JAK-STAT signaling can lead to tumorigenesis, either directly or
indirectly (25). The
Mitogen-Activated Protein Kinase (MAPK) pathway (also known as ERK
pathway) is essential in stimulating survival, proliferation,
migration, and cell adhesion through signals transduction from
cytokines and growth factors (19). Parallel to the MAPK pathway, the
activated phosphoinositide 3-kinase-protein kinase B (PI3K-AKT)
pathway mediates cancer cell metabolism and regulates
proliferation. Additionally, this pathway activates other signaling
pathways, including Wnt and NF-қB (26), which in turn regulates different
genes involved in inflammation, cell viability, proliferation, and
apoptosis (27) in BC and many
others cancer types.
Plant-derived compounds have been thoroughly
screened for their potency in preventing and treating cancer
(28). Numerous studies have
demonstrated the medicinal importance of the polyphenol compound,
gossypol (GOSS) (2,20-binaphthalene)-8,80-dicarboxaldehyde,
1,10,6,60,7,70-hexahydroxy-5,50-diisopropyl-3,30-dimethyl, a minor
constituent of cotton (Gossypium hirsutum L.) seeds
(29–31). GOSS has various biological
activities, including antifertility, antiviral, antimicrobial, and
antioxidative activity (32).
Moreover, the anti-proliferative, anti-metastatic, and apoptotic
effects of GOSS have been documentd against several human cancers,
including colon, prostate, glioma, adrenal, leukemia (24,33–37),
in addition to breast cancer (28,38–40).
The drug combination is critical to accomplish a synergistic
therapeutic effect (41) and to
overcome the resistance mechanisms of many diseases, including
cancer (42). GOSS has been found
to induce apoptosis in various types of human cancer cells in
combination with low doses of dexamethasone (43), doxorubicin (44), taxanes (45), and valproic acid (46).
Many studies have demonstrated the anticancer effect
of GOSS in BC, including the TNBC subtype, MDA-MB-231 (MM-231)
cells. However, studying the racial perspective of the compound
effects on MDA-MB-468 (MM-468), and its gene-related mechanism of
action in comparison to MM-231 cells has never been addressed.
Moreover, the potential effect of GOSS on the proinflammatory
cytokines, IL-8 and CCL2 has not been reported prior to this work.
Therefore, the current study is designed to compare the anticancer
effect of GOSS on two TNF-α-stimulated human TNBC cell lines:
MM-231 and MM-468, representing Caucasian (CA) and African American
(AA) women, respectively (47). We
hypothesized that GOSS could modulate the expression of genes
involved in many cellular signaling pathways that mediate the
regulation of diverse cancer-related cytokines/chemokines.
Materials and methods
Materials
The compound GOSS (purity ≥90%) was purchased from
Santa Cruz Biotechnology, Inc. Trypsin-EDTA solution 0.25% and
Alamar Blue® (a sterile buffered solution of resazurin
fluorescence dye) were purchased from Sigma-Aldrich; Merck KGaA.
Dimethyl sulfoxide (DMSO), penicillin/streptomycin, and Dulbecco's
Phosphate Buffer Saline (DPBS) were obtained from the American Type
Culture Collection. Dulbecco's Modified Eagle Medium (DMEM),
heat-inactivated fetal bovine serum (FBS), and cell culture plates
were purchased from VWR International (Radnor). TNF-α, Human
Cytokine Antibody Array kit (cat. no. AAH-CYT-1000), Human ELISA
kits for C-C Motif Ligand 2 [CCL2, also known as monocyte
chemoattractant protein-1 (MCP-1), cat. no. ELH-MCP1] and
Interleukin-8 (IL-8, also known as CXCL-8, cat. no. ELH-IL-8) were
purchased from RayBiotech. TURBO DNA-free™ kit (cat. no. AM1907)
was purchased from Life Technologies, Inc. TRIzol®
reagent was purchased from Invitrogen; Thermo Fisher Scientific. An
iScript™ cDNA Synthesis kit (cat. no. 170-8891), SsoAdvanced™
Universal SYBR® Green Supermix (cat. no. 1725271), Human
PCR primers (CCL2, IKBKE, IL-8, STAT3, MAPK1, MAPK3, CCDC88A,
PIK3CD, and GAPDH) were purchased from Bio-Rad
Laboratories, Inc.
Cell culture
The two immortalized TNBC cell models: MM-231
(https://www.atcc.org/products/all/HTB-26.aspx) and
MM-468 (https://www.atcc.org/Products/All/HTB-132.aspx), were
purchased from ATCC. Both cell lines were grown in 75-cm TC-flasks
at 37°C in humidified 5% CO2 incubator and subculture as
required, using trypsin/EDTA (0.25%). The routinely used DMEM
growth medium contained 4 mM L-glutamine and was supplemented with
10% FBS (v/v), and 1% penicillin/streptomycin salt solution (100
U/ml and 0.1 mg/ml, respectively). The DMEM experimental medium was
the same, except it was phenol-free and was supplemented with 2.5%
FBS as previously reported by others (48).
Cell viability assay
In this experiment, cells were incubated overnight
at 37°C at a density of 5×104 cells/well in 96-well
microplates. GOSS powder was reconstituted in DMSO. Both types of
cells were treated for 24 h with 50 ng/ml of TNF-α, in addition to
the compound (concentration ranges of 0–100 µM in MM-231 or 0–50 µM
in MM-468 cells). Control wells were treated with DMSO at the
highest used concentration (<0.1%). Blank wells were treated in
the same manner but without cells. Alamar Blue® was used
to determine cell viability, as described in our previous study
(49). The fluorescent-fuchsia dye
of the reduced resazurin by viable cells was measured at an
excitation/emission of 530/590 nm using a Synergy HTX Multi-Reader
(BioTek).
Human cytokine/chemokine protein
microarray
Cytokine expression microarray analysis was designed
based on the data of the cytotoxicity assay. Four flasks for each
cell line were incubated overnight with a density of
10×106 cells/75-cm2 TC flask using the
experimental media with 2.5% FBS. Four flasks for each cell line
were incubated overnight with a density of 10×106
cells/75-cm2 TC flask using the experimental media with
2.5% FBS. On the next day, the media were discarded, and the cells
were treated with 50 ng/ml TNF-α and low concentrations of GOSS
that slightly impacted the cell viability. MM-231 cells were
treated as follows: TNF-α (50 ng/ml), GOSS (6.25 µM), and TNF-α +
GOSS (50 ng/ml + 6.25 µM, respectively). Similarly, MM-468 cells
were treated with TNF-α (50 ng/ml), GOSS (5 µM), and TNF-α + GOSS
(50 ng/ml + 5 µM, respectively). In both experimental sets, control
samples were exposed to only the solvent DMSO at a concentration of
<0.1%. After a 24 h exposure period, the cell-free supernatant
of each sample was collected, aliquoted, and stored at −80°C for
later use. At the same time, the cells from each flask were
pelleted and similarly stored at −80°C for RT-qPCR study. For each
cell line, a semi-quantitative method using antibody-coated array
membranes was established to measure chemokine/cytokine expression
in the cell-free supernatants. The assay was established following
the manufacturer protocols. Briefly, four membranes were carefully
placed in four chamber-incubation trays and blocked with the
provided buffer on a shaker for 30 min at RT. After that, the
blocking buffer was decanted, and replaced with 1 ml cell-free
supernatant from resting, GOSS-treated, TNF-α-stimulated or
cotreated cells, and the four membranes were then kept overnight on
a low-speed shaker at 4°C. On the following day, the supernatants
were removed from each chamber, and the membranes were washed with
the kit washing buffers. Next, 1 ml of freshly constituted
biotinylated antibody cocktail was pipetted to each membrane and
incubated at RT for 2 h, followed by washing with the same wash
buffers. The membranes were incubated again for another 2 h with 2
ml of diluted horseradish peroxidase-conjugated streptavidin
(HRP-Streptavidin) followed by the final washes. Cytokines
intensities on the blots were detected as spots using a
chemiluminescence cocktail. The blot images were captured using a
Flour-S Max Multiimager (Bio-Rad Laboratories, Inc.), and the spot
intensities were measured with the Quantity-One Software (Bio-Rad
Laboratories, Inc.). The Excel-based data analysis was established,
using the Human Cytokine Array software C1000 (CODE:
S02-AAH-CYT-1000) from RayBiotech.
Human CCL2 and IL-8 chemokines ELISA
study
Enzyme-Linked Immunosorbent Assay (ELISA) kits were
used to measure the protein levels (pg/ml) for both CCL2 and IL-8
chemokines. Briefly, the standard curves, samples, and reagents
were prepared at RT for both chemokines. Standards and samples of
100 µl each were incubated with the antibody pre-coated 96-well
ELISA microplates for 2.5 h. The supernatant was replaced by 100 µl
of the freshly constituted biotinylated antibody for another hour,
then removed. Streptavidin solution (100 µl) was added for 45 min,
followed by the addition of 100 µl of the substrate reagent for
30-min incubation. Washes were always performed after each step
according to the manufacturer's protocol. The reaction was
terminated by the addition of 50 µl of a stop-solution, and the
intensity for the chemokines and the standard were measured at 450
nm using a Synergy HTX Multi-Reader (BioTek).
RNA isolation and cDNA synthesis
In this assay, the previously-80°C-frozen cell
pellets were used, as mentioned above in the Human
cytokine/chemokine protein microarray study. The total RNA was
extracted from each sample using 1 ml of Trizol® reagent
and sonicated for 30 sec at RT using VirTishear mechanical
homogenizer (LabWrench). Subsequently, 200 µl of chloroform was
added to each sample, vortexed, incubated at RT for 2–3 min and
centrifuged for 15 min at 10,000 × g and 2–8°C. The aqueous phase
was separated and mixed with 500 µl of isopropyl alcohol to
precipitate the RNA for different samples. The RNA pellets were
then reconstituted in approximately 30–50 µl of nuclease-free water
to measure the RNA concentration and purity in each sample using a
Nanodrop spectrophotometer (Thermo Fisher Scientific, Inc.).
Thereafter, the purified RNA was reverse transcribed into cDNA
using an iScript™ cDNA Synthesis Kit and PCR run as follows: 46°C
for 20 min and 95°C for 1 min.
Reverse transcription-quantitative PCR
(RT-qPCR)
In this study, we measured the expression of various
genes involved in CCL2 and IL-8 regulation, using qPCR of the
Bio-Rad CFX96 Real-Time System (Bio-Rad Laboratories, Inc.)
(50). Briefly, the freshly
synthesized cDNA (1 µl; 200 ng) was used for each sample in a final
volume of 20 µl (10 µl of the 2× real-time Master Mix, 8 µl
nuclease-free water, and 1 µl of primer mix). According to the
manufacturer's protocol, the PCR run was performed as follows:
Reactants were first incubated at 95°C for 2 min, and then 39
cycles of amplification (51) were
carried out with each cycle consisting of denaturing at 95°C for 10
sec, annealing at 60°C for 30 sec, and melting curve at 65–95°C for
5 sec. All qPCR reactions were performed in triplicates for each
primer. In this experiment, the used PCR primers were compatible
with the different genes under investigation and are summarized in
Table I. GAPDH was applied as a
reference gene to normalize the mRNA levels for genes of
interest.
 | Table I.List of primers used in reverse
transcription-quantitative PCR experiments. |
Table I.
List of primers used in reverse
transcription-quantitative PCR experiments.
| Name | GenBank accession
no. | Amplicon context
sequence |
|---|
|
MCP1/CCL2 | NM_002982.4 |
ACTGAAGCTCGCACTCTCGCCTCCAGCATGAAAGTCTCTGCCGCCCTTCTGT |
|
|
|
GCCTGCTGCTCATAGCAGCCACCTTCATTCCCCAAGGGCTCGCTCAGCCAGA |
|
|
|
TGCAATCAATGCCCCAGTCACCTGCTGTTATAACTTCACCAATAGGAAGATCT |
|
|
|
CAGTGCAGAGGCTCGCGAGCTAT |
| IL-8 | NM_000584.4 |
GAGCACTCCATAAGGCACAAACTTTCAGAGACAGCAGAGCACACAAGCTTC |
|
|
|
TAGGACAAGAGCCAGGAAGAAACCACCGGAAGGAACCATCTCACTGTGTG |
| IKBKE | NM_014002.4 |
GGCTTGGCTACAACGAGGAGCAGATTCACAAGCTGGATAAGGTGAATTTCAG |
|
|
|
TCATTTAGCCAAAAGACTCCTGCAGGTGTTCCAGGAGGAGTGCGTGCAGAA |
|
|
|
GTATCAAGCGTCCTTAGTCACACACGGCAAGAGGATGAGGGTGGTGCACGAG |
| MAPK1 | NM_002745.4 |
TTCAGCTGGTCAAGATAATGCTTCCCTGGAAAGATGGGCCTGTTAGAAAGCAT |
|
|
|
TTCTGCCAGAATGCAGCCTACAGACCAAATATCAATGGACTTGGTGTAGCCCT |
|
|
|
TGGAATTCAACATAATTTCTGGAGC |
| MAPK3 | NM_002746.3 |
TCTCCATCAGGTCCTGCACAATGTAGACATCTCTCATGGCTTCCAGGGTGGAC |
|
|
|
GCCCGCAGAATGTCTCGGATGCCGATGACATTCTCATGGCGGAAGCGCAGCA |
|
|
|
GGATCTGGATCTCCCGGAGCGTGCGCTGGCAGTAGGTCTGATGTTCGAAGGG |
|
|
|
GCTGATCTTCTTGATGGCCACGC |
| CCDC88A | NM_001365480.1 |
GGAGTTGGGCATTCTGGTTCATGAGTGAGGTACTTTGGGAATTAAGGGTGGA |
|
|
|
ATTTTCAACCTGAAGCTTGGCATTCTGTGTTTGAAGAGTGGTATTCTGTTCTTG |
|
|
|
TAATGACACTGTCTGCCTCTGAAGTGCAAGAATCTGAGCCTGCAAATTATTGT |
|
|
| TCTGTGTCT |
| STAT3 | NM_139276.2 |
GGTGTCACACAGATAAACTTGGTCTTCAGGTATGGGGCAGCGCTACCTGGGT |
|
|
|
CAGCTTCAGGATGCTCCTGGCTCTCTGGCCGACAATACTTTCCGAATGCCTCCT |
|
|
|
CCTTGGGAATGTCAGGATAGAGATAGACCAGTGGAGACACCAGGATATTGGT |
| PIK3CD | NM_005026.4 |
GCGGCTGGAGTTCGACATCAACATCTGCGACCTGCCCCGCATGGCCCGTCTCT |
|
|
|
GCTTTGCGCTGTACGCCGTGATCGAGAAAGCCAAGAAGGCTCGCTCCACCAA |
|
|
|
GAAGAAGTCCAAGAAGGCGGACTGCCCCATTGCCTGGG |
| GAPDH | NM_002046.7 |
GTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCAC |
|
|
|
ATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACA |
|
|
|
AGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACAC |
Statistical analysis
The quantitative data for this study were analyzed
using GraphPad Prism 6.2 software. All data points were obtained
from the average of at least two independent studies and are
expressed as the mean ± SEM. For the viability assay,
IC50s values were determined by nonlinear regression
model of log (inhibitor) vs. normalized response-variable slope on
the software with the R2 best fit and the lowest 95% confidence
interval. The significance of the difference between each control
and its related treatment groups was determined using one-way
ANOVA, followed by Bonferroni's multiple comparison tests. A
statistical difference was considered significant at P<0.05 and
less. For array blots and ELISA studies, independent (unpaired)
Student's t-test was used to verify the significance of the
difference between TNF-α vs. control groups, or between TNF-α vs.
TNF-α + GOSS groups. Quantitative mRNA expression was analyzed
using CFX 3.1 Manager software for Bio-Rad Laboratories, Inc. All
genes were normalized against the expression of the housekeeping
gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and
verified with unpaired Student's t-test.
Results
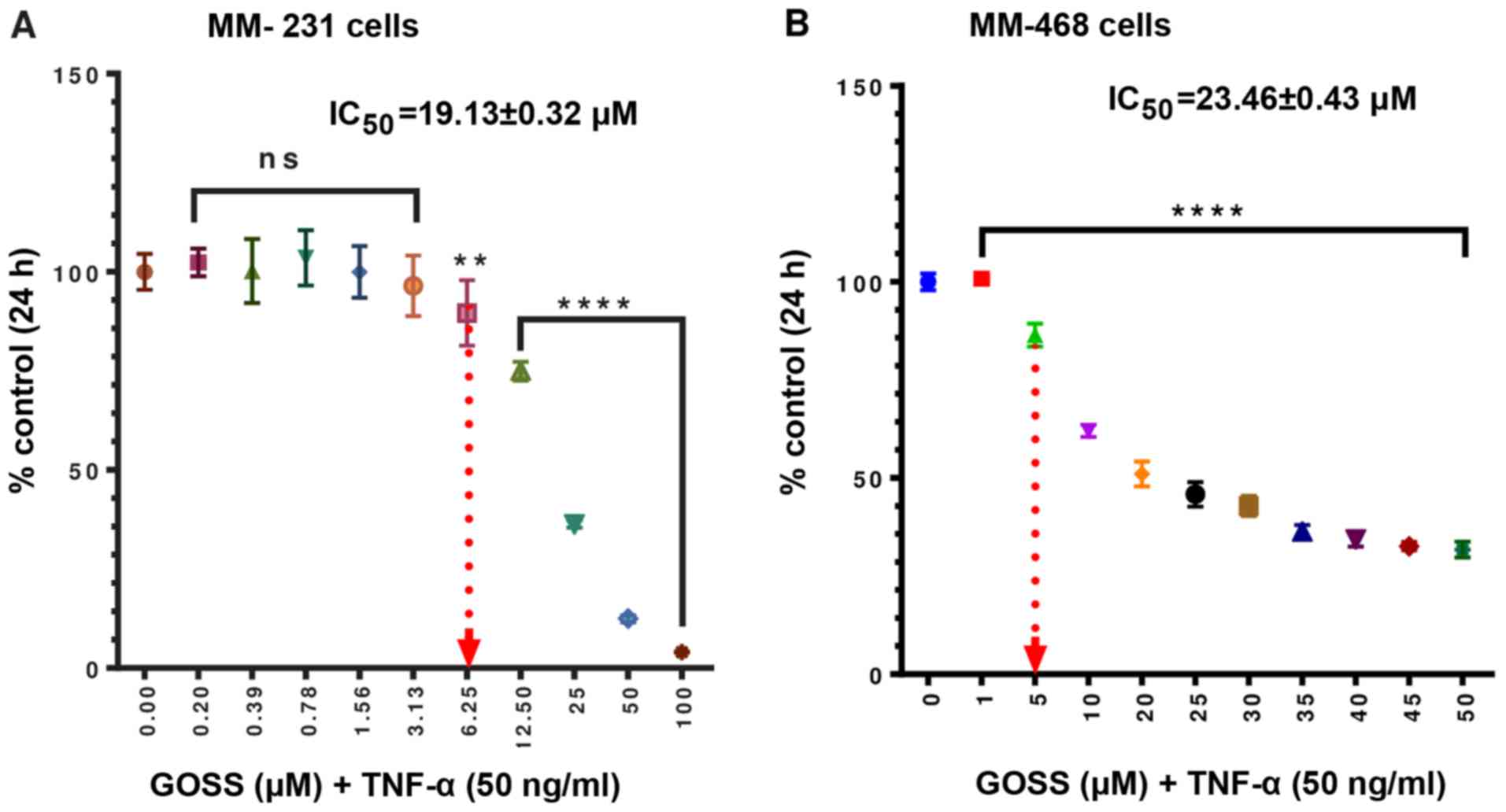
GOSS decreases cell viability in
MM-231 and MM-468 TNBC cells
The anticancer effects of GOSS were examined in the
presence of 50 ng/ml of TNF-α in both MM-231 and MM-468 cell lines.
As indicated in Fig. 1A and B, a
highly significant cytotoxic effect (P<0.0001) was detected at
all the tested concentrations ranges 12.5–100 µM in MM-231 cells
and 1–50 µM in MM-468 cells. The obtained IC50s
(19.13±0.32 µM for MM-231 and 23.46±0.43 µM for MM-468) showed a
slightly higher response to the compound in MM-231 cells. Based on
the viability studies data, optimum doses of GOSS that causes less
than 20% cell death and 50 ng/ml of TNF-α were determined for
cytokine/chemokine expression study.
GOSS inhibits the release of CCL2 in
MM-231 cells and IL-8 in MM-468 cells
To understand the mechanism of the antitumor effects
of GOSS on TNF-α-stimulated MM-231 and MM-468 TNBC cells, we
measured the expression of different cytokines/chemokines released
in the cell-free supernatants in the presence and the absence of
GOSS and 50 ng/ml of TNF-α (52).
Low concentrations of GOSS that slightly impacted the cell
viability were used (6.25 µM in MM-231 cells and 5 µM in MM-468
cells).
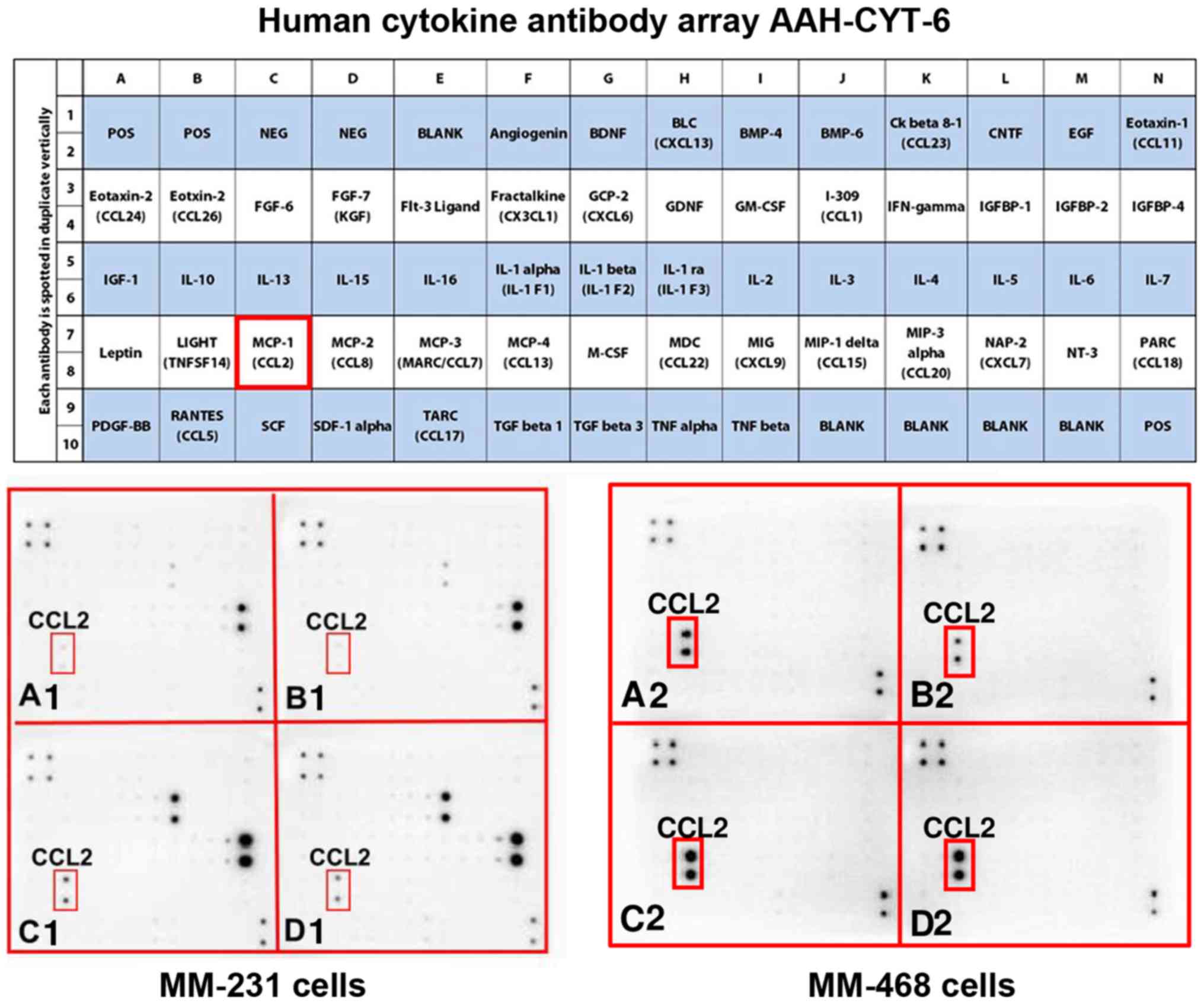
The obtained results show that GOSS repressed the
expression of two critical chemokines, CCL2 in MM-231 cells and
IL-8 in MM-468 cells, as indicated by the red frames on both the
maps and the blot arrays. In the cytokine blots, AAH-CYT-6, the
expression of the cytokine was scarcely noticeable on most of blots
presenting the supernatants of both control and GOSS-treated cells
(Fig. 2A and B). High expression
of CCL2 was observed in both TNF-α-treated cell lines (Fig. 2C1 and C2). The intensity of the spot was
attenuated in MM-231 cells by the effect of 6.25 µM GOSS (Fig. 2D1), the observation that was not
showed in MM-468 cells (Fig.
2D2).
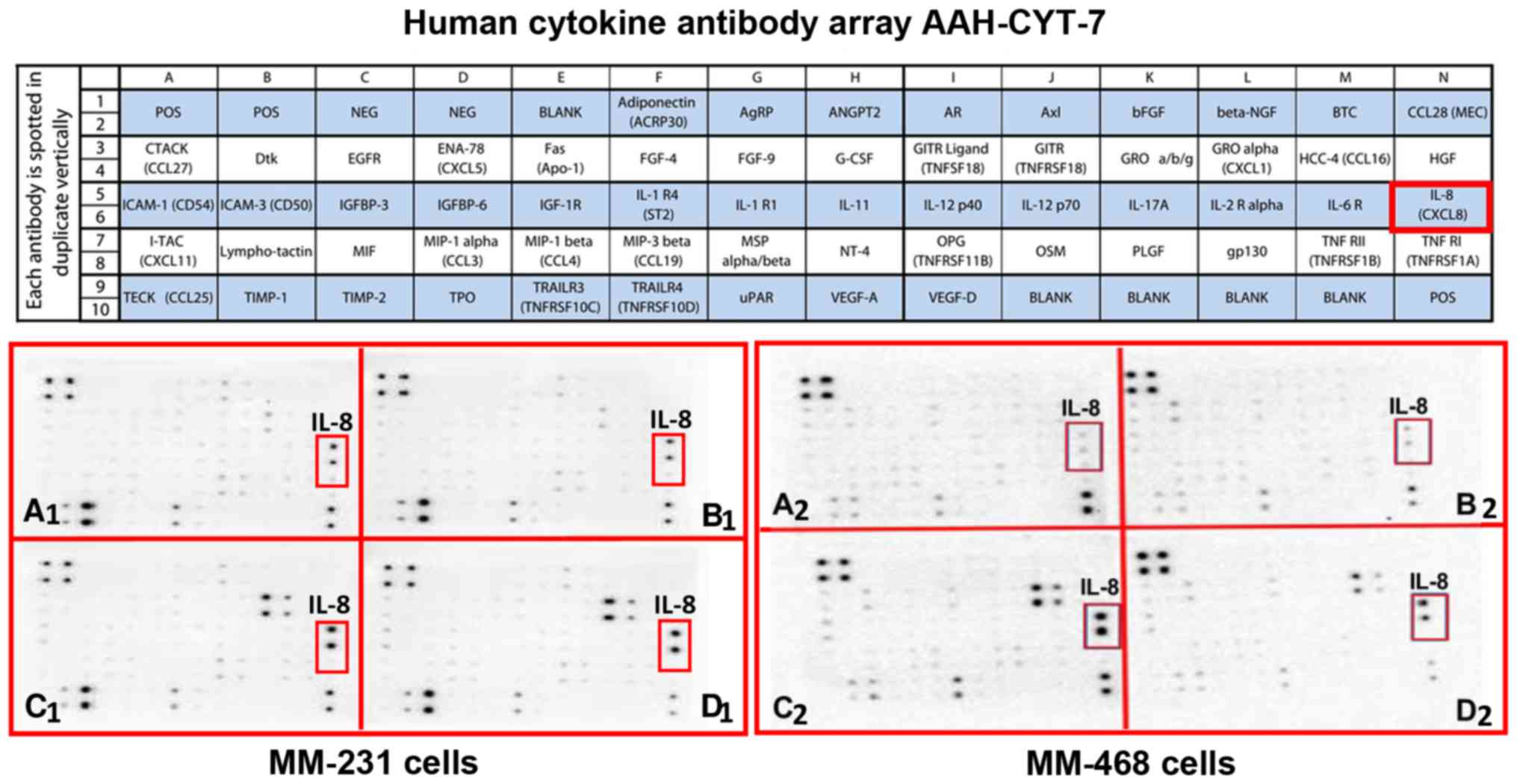
Meanwhile, the intensity of IL-8 was low on the
blots presenting the supernatants of both control and GOSS-treated
cells (Fig. 3A and B). Indeed,
TNF-α has increased IL-8 expression in both cell lines, as
indicated on the AAH-CYT-7 blots (Fig.
3C1 and C2). The compound GOSS
did not reduce the cytokine expression in MM-231 cells (Fig. 3D1). However, an observed decrease
was detected in MM-468 cells in the presence of 5 µM GOSS (Fig. 3D2).
Both chemokines, CCL2 and IL-8, were quantified as a
percentage relative to their control. TNF-α has induced an 8-fold
increase in CCL2 expression in MM-231 cells (P<0.001) (Fig. 4A). The data indicated that the
presence of both GOSS and TNF-α significantly attenuated CCL2 by
30% (P<0.01). Similarly, in MM-468 cells (Fig. 4B), a more than 8-fold increase in
the expression of IL-8 (P<0. 01) was quantified in
TNF-α-stimulated cells and then inhibited by 60% in the presence of
GOSS (P<0.05). The results show that the GOSS compound did not
attenuate IL-8 expression in MM-231 cells (Fig. 4C), nor CCL2 in MM-468 (Fig. 4D). Also, no changes were detected
when we compare the control to the GOSS-treated cells in both
chemokines of interest.
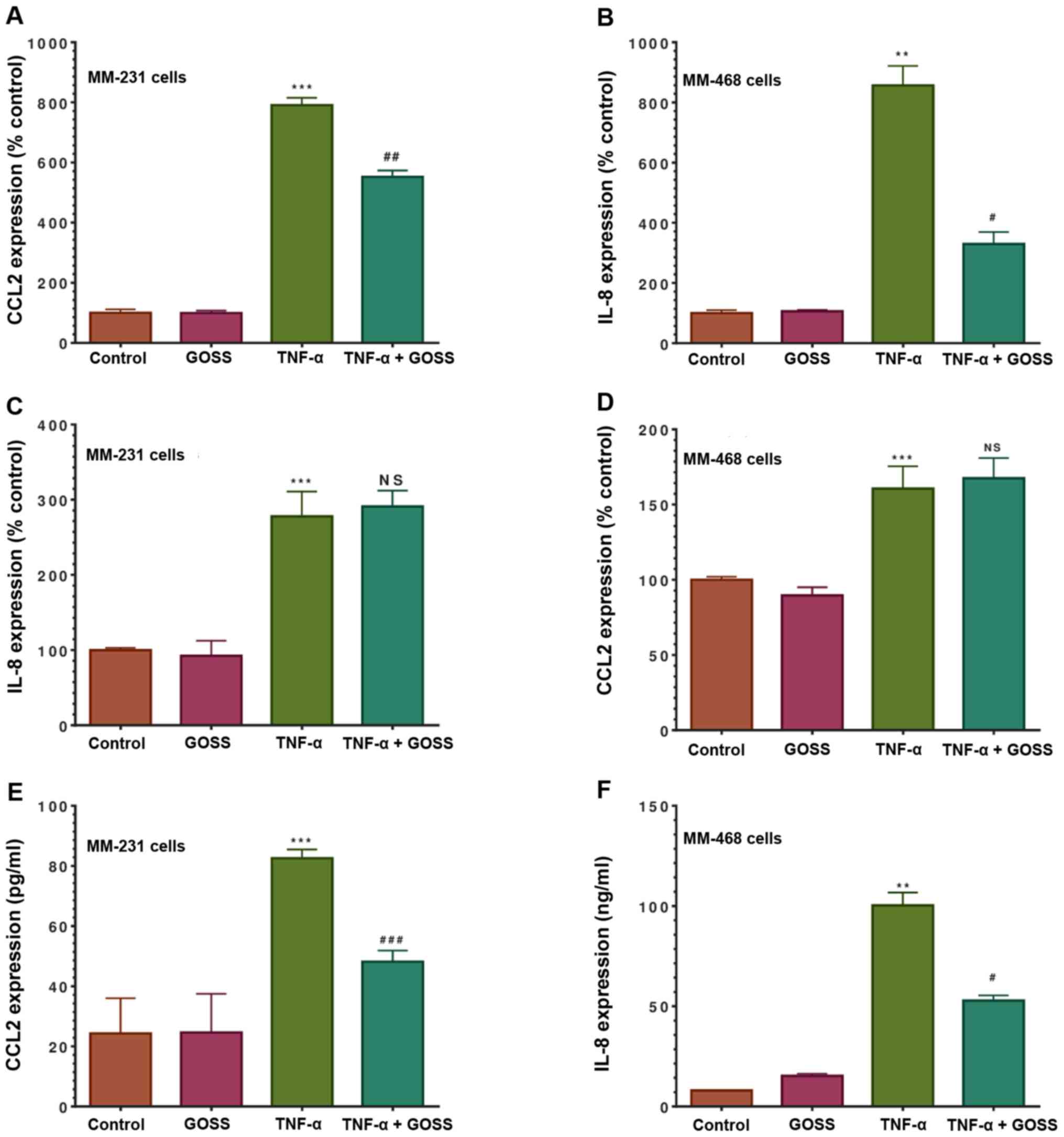 | Figure 4.Cytokine array quantification of
chemokines inhibited by GOSS in TNF-α-stimulated triple-negative
breast cancer cell lines. The effect of GOSS on the extracellular
cytokine expression was assessed as follows: CCL2 release from (A)
MM-231 cells and (D) MM-468 cells, IL-8 release from (B) MM-468 and
(C) MM-231 cells. ELISA quantifications (E) for CCL2 released from
MM-231 cells and (F) for IL-8 released from MM-468 cells. The
normalized data show the cytokine expression in four sets of
experimental cell supernatants, namely control, GOSS-treated (6.25
µM in MM-231 cells and 5 µM in MM-468 cells), 50 ng/ml
TNF-α-stimulated, and co-treated cells (TNF-α + GOSS). The data are
presented as the mean ± SEM of three independent experiments. The
dot intensities are expressed as percent relative to control. The
significant difference between TNF-α-stimulated cells vs. resting
cells (*) or between TNF-α-stimulated vs. TNF-α + GOSS-treated
cells (#) groups was determined by an unpaired t-test. P<0.05
was considered to indicate a statistically significant difference.
**P<0.01, ***P<0.001, ##P<0.01 and
###P<0.001. NS, not significant; GOSS, gossypol;
TNF-α, tumor necrosis factor α. |
We further validated the microarray data for only
the significantly attenuated chemokines. Protein expression
quantification for CCL2 (pg/ml) and IL-8 (ng/ml) was measured using
two independent ELISA assays. Overall, the data for cytokine arrays
and ELISA were consistent in both cell lines. The assay indicated a
significant increase in CCL2 (P<0.001) and IL-8 (P<0.01) in
TNF-α-treated vs. resting cells (Fig.
4E and F). The simultaneous presence of GOSS and TNF-α
attenuated the expression of CCL2 by ~40% in MM-231 cells
(P<0.001), and IL-8 expression was also reduced by 50% in MM-468
cells (P<0.05).
GOSS alters the expression of genes
regulating the release of CCL2 in MM-231 cells and IL-8 in MM-468
cells
RT-qPCR was performed to determine the relevance of
our data and to elucidate the mechanism by which GOSS impact CCL2
and IL-8 regulation in MM-231 and MM-468 cells, respectively
(Figs. 5 and 6). Profiling the normalized expression of
mRNAs in both cell lines provides precise insights into the
influence of the compound on signaling pathways-related genes.
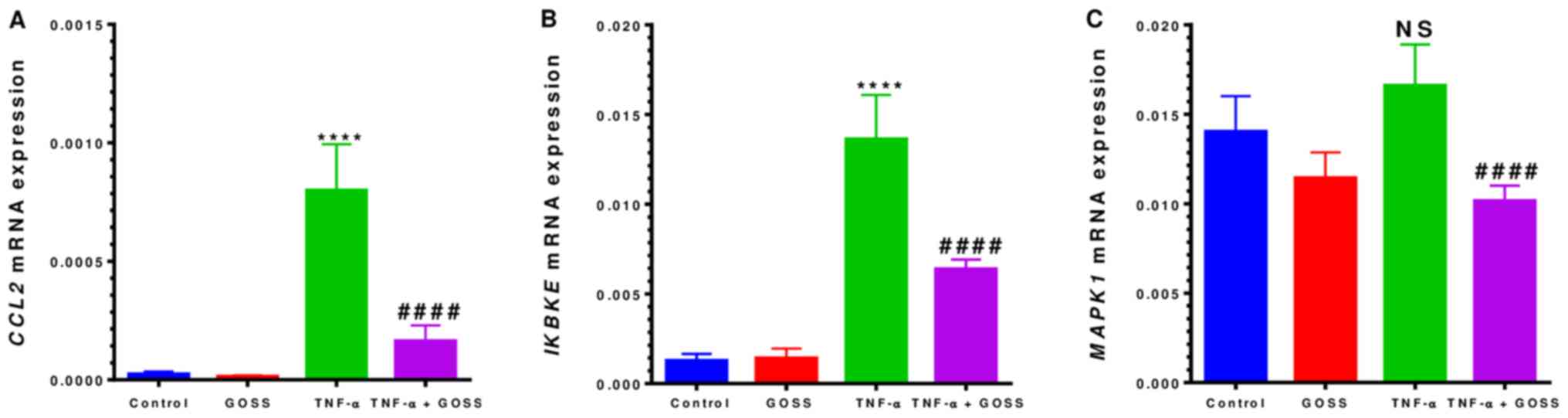
The RT-qPCR data presented in Figs. 5A and 6A indicated that mRNAs expressions of
CCL2 and IL-8 were consistent with those of cytokine
microarray and ELISA protein studies in both MM-231 and MM-468 cell
lines, respectively. The mRNA's data showed that both cell lines
responded to TNF-α and TNF-α + GOSS. In TNF-α-stimulated MM-231
cells, CCL2 mRNA expression exhibited a highly significant
32-fold up-regulation (P<0.0001) (Fig. 5A and Table II). This gene was dramatically
repressed by 80% in the presence of 6.25 µM GOSS (P<0.0001).
Furthermore, GOSS repressed two more genes, IKBKE and
MAPK1, which are involved in the signaling pathways of
TNF-α-induced CCL2 release. IKBKE showed a significant
increase in expression of ~11-fold by TNF-α that was repressed to
~53% by GOSS (Fig. 5B and Table II). The detected up-regulation in
MAPK1 was non-significant in the TNF-α-stimulated cells.
However, GOSS was able to suppress significantly (P<0.0001) its
expression by ~39% (Fig. 5C and
Table II).
 | Table II.Gene expression changes in MM-231
triple-negative breast cancer. |
Table II.
Gene expression changes in MM-231
triple-negative breast cancer.
| Control vs.
TNF-α | TNF-α vs. TNF-α +
GOSS |
|---|
|
|
|---|
| Target gene | Fold (+) | P-value | Target gene | Inhibition (%) | P-value |
|---|
| CCL2 | 32.72 | <0.0001 | CCL2 | 79.46 | <0.0001 |
| IKBKE | 10.60 | <0.0001 | IKBKE | 53.06 | <0.0001 |
| MAPK1 | 1.18 | 0.0648 | MAPK1 | 38.81 | <0.0001 |
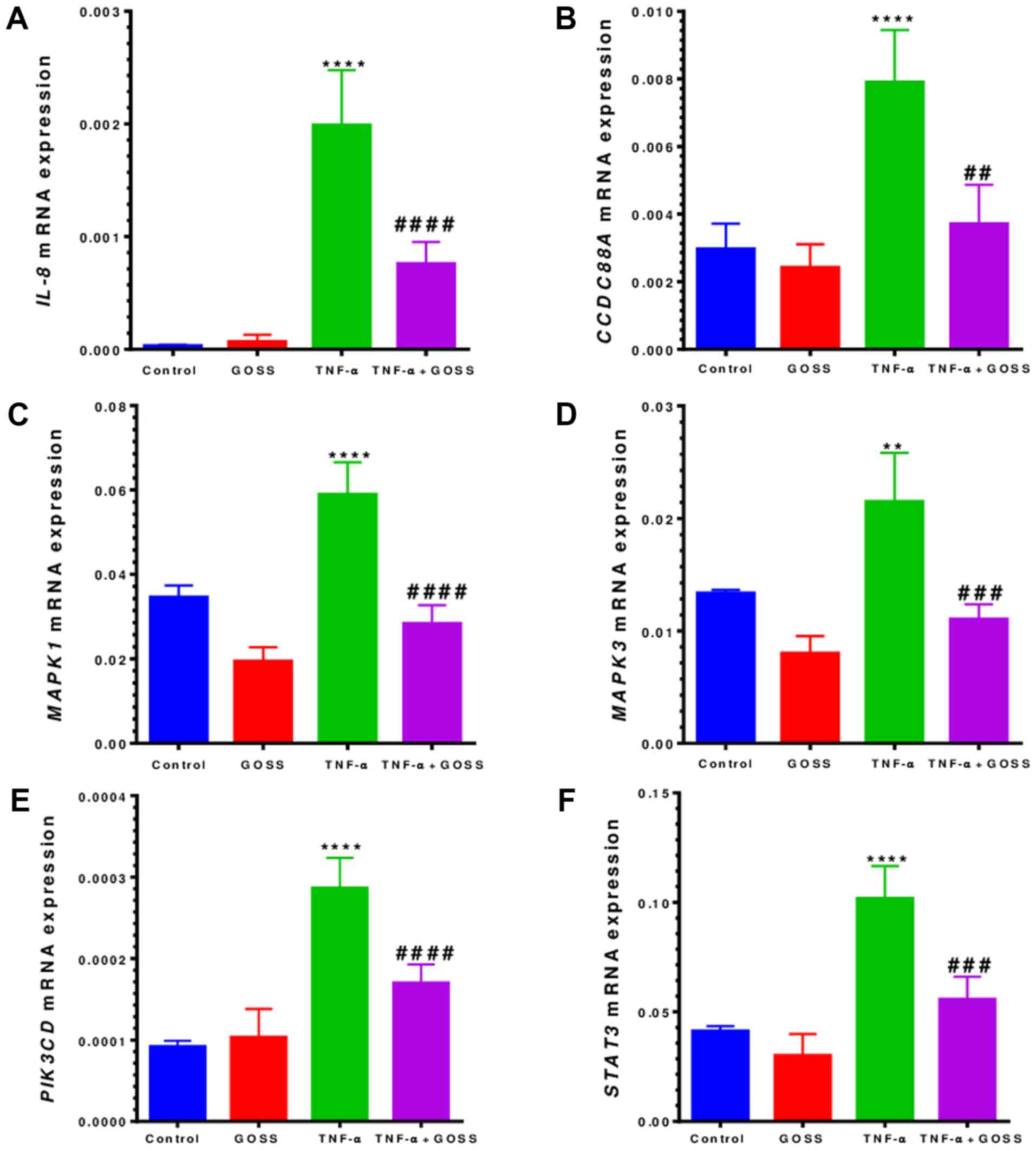
Similarly, in TNF-α-stimulated MM-468 cells, IL-8
mRNA was upregulated by ~65-fold (P<0.0001) followed by 60%
downregulation (P<0.001) when combined with 5.0 µM GOSS
(Fig. 6A and Table III). Moreover, a significant
fold-increase (P<0.01-P<0.0001) was also found in five more
genes (Fig. 6B-F and Table III), including MAPK1, MAPK3,
CCDC88A, STAT3, and PIK3CD that showed the highest
increase (3-fold). GOSS repressed the expression of these five
genes by almost 40–50%, as shown in Fig. 6B-F and Table III.
 | Table III.mRNA gene expression changes in
MM-468 triple-negative breast cancer. |
Table III.
mRNA gene expression changes in
MM-468 triple-negative breast cancer.
| Control vs.
TNF-α | TNF-α vs. TNF-α +
GOSS |
|---|
|
|
|---|
| Target gene | Fold (+) | P-value | Target gene | Inhibition (%) | P-value |
|---|
| IL-8 | 64.6 | <0.0001 | IL-8 | 60.78 | <0.0001 |
| CCDC88A | 2.66 | <0.0001 | CCDC88A | 53.04 | 0.0013 |
| MAPK1 | 1.70 | <0.0001 | MAPK1 | 51.90 | <0.0001 |
| MAPK3 | 1.61 | 0.0062 | MAPK3 | 48.61 | 0.0002 |
| PIK3CD | 3.12 | <0.0001 | PIK3CD | 40.70 | <0.0001 |
| STAT3 | 2.47 | <0.0001 | STAT3 | 45.28 | <0.0001 |
Indeed, the obtained data in this study, including
those of the cytokine microarray blots, ELISA, and PCR evaluation,
have validated the up-regulation of CCL2 and IL-8 pro-inflammatory
proteins and their mRNAs and assured GOSS ability to inhibit both
of them.
Discussion
One of the significant goals in anticancer drug
development is targeting signaling pathways regulating cancer
cells' survival and proliferation through the inhibition of
distinctive gene expressions. The present study examined the
anticancer mechanism of the natural polyphenol GOSS in two racially
different TNBC cell models. The cytotoxic effects of GOSS with a
slight but significantly higher response against MM-231 cells than
MM-468 TNBC cells indicate possible different mechanistic effects
between the two cell lines. However, the effect of gossypol on
normal breast cells was not established; a previous study on
another cell line indicated the safety of the gossypol at a similar
dose (53). This compound
repressed the TNF-α-mediated upregulation of the two BC-dominant
proinflammatory chemokines (CCL2) in MM-231 and (IL-8) in MM-468.
Mechanisms of inhibitions involved the attenuation of mRNA
expression of divers, active genes crucial in regulating pivotal
signaling pathways. GOSS impacted MAPK, JAK-STAT, and NF-қB
pathways in both cell lines, in addition to the PI3K-AKT pathway
only in MM-468 (Fig. 7).
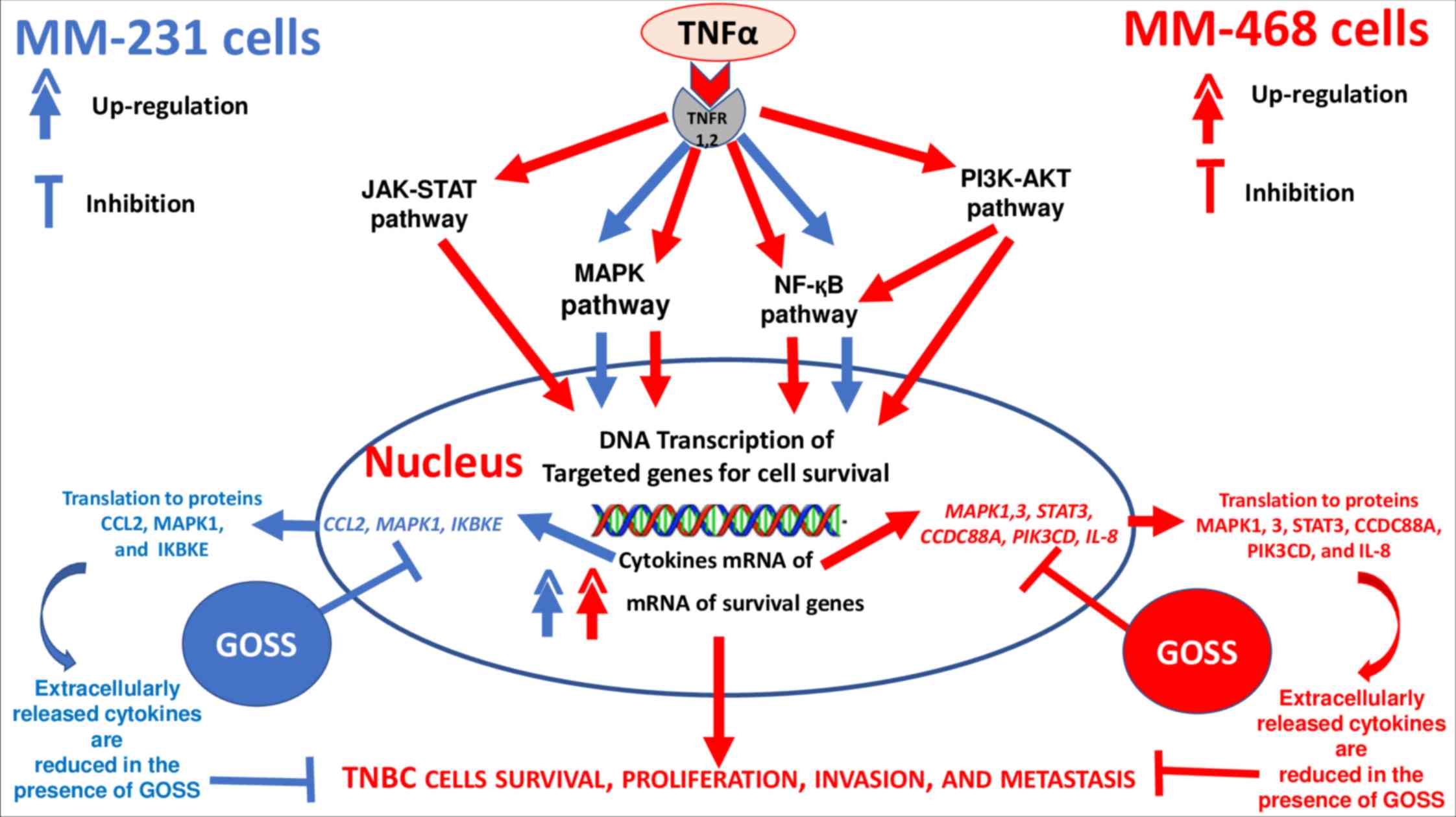 | Figure 7.Schematic diagram illustrating
pro-inflammatory genes involved in different signaling pathways
that mediate the release of CCL2 and IL-8 in TNF-α-stimulated
MM-231 (blue color) and MM-468 (red color) triple-negative breast
cancer cells, respectively. In MM-231 cells, the most
down-regulated genes by GOSS are CCL2, IKBKE, and MAPK1. In
MM-468 cells, the most down-regulated genes are IL-8, CCDC88A,
MAPK1 and 3, PIK3CD, and STAT3. GOSS, gossypol; TNF-α,
tumor necrosis factor α. |
The high expression of TNF-α has been reported in BC
patients (21). Whether it is
endogenous or is administered in a high dose, TNF-α can function as
tumor necrosis or tumor-promoting factor, respectively (54). In cancer, the highly expressed
TNF-α stimulates the release of many chemokines, including CCL2 and
IL-8 (5,55–57),
that are highly expressed in cancer.
The most attenuated chemokines, CCL2, and IL-8 have
been known to share some common characters. Both chemokines are
belonging to a superfamily of small, soluble, and secreted proteins
(58). They both are released into
the cancer microenvironment by the endothelial cells and
fibroblasts (59,60) to activate lymphocytes and
macrophages (61,62).
Treating MM-231 cells with TNF-α upregulated the
expression of CCL2, a finding that is consistent with previous
studies (17,63). In the presence of GOSS (low dose of
6.25 µM), CCL2 expression was dramatically downregulated in the
TNF-α-activated cells. Cytokine CCL2 is the most cancer-related
member of the CC chemokine family (64). In BC cells, upregulated CCL2
(65) promotes tumorigenesis and
metastasis, and its suppression significantly reduced tumor
aggressiveness (66). In MM-231
and other malignant tumors, the upregulated CCL2 (51,59)
was linked to the decreased survival of TNBC patients (6). Hence, the reduction of CCL2
expression following antibody administration or genetic mutation
(67) led to reduced metastasis
and enhanced survival (51), which
hold promise in treating TNBC (6).
Previous investigations have highlighted the impact
of CCL2 on different signaling pathways regulation. CCL2 and its
main receptor CCR2 control BC cell survival through MAPK- and
Smad3-dependent mechanisms (19).
Moreover, in various tissues, CCL2 activates Notch signaling
pathway and regulates cancer stem cells (68), which are vital for BC progression
(69,70). CCL2 gene silencing in MM-231
led to various consequences, including inhibition of CCL2
expression, decreased cell proliferation, increased necrosis and
autophagy, inhibition of self-renewal, and inhibition of the
primary and secondary invasion of the TNBC xenograft tumor, and
decreases M2 macrophage recruitment (6).
Our current RT-qPCR gene expression analyses
indicate that TNF-α significantly increased the mRNA expression of
two genes: CCL2 and its gene regulator IKBKE. At the
same time, the data obtained show the ability of GOSS to attenuate
the expression of these two genes as well as MAPK1. These
three genes are involved in many signaling pathways, including the
NF-қB, JAK-STAT, and MAPK, which regulate the expression of CCL2.
Nevertheless, NF-қB and MAPK signaling pathways are used by both
normal and cancer cells dominating these signaling pathways to
enhance their survival and invasion (71).
The gene IKBKE (also known as IKKε) is a
member of the IKK family of kinases (72). Extensive studies have reported the
correlation between IKBKE down-regulation and the decrease
of CCL2 release (49,73). Our current data supported these
previous findings and interpreted the mechanism of CCL2 inhibition
by GOSS. In TNBC cells, the IKBKE gene is expressed in 60%
of BC tissue (74) and acts as an
oncogene to support cell viability. Furthermore, the up-regulation
of IKBKE enhances resistance to anticancer drugs and
protects the BC cells from Tamoxifen-induced apoptosis (75). Indeed, IKBKE is an upstream
regulator of the transcription factor NF-kB and JAK-STAT pathways,
which have been actively involved in the pathogenesis of TNBC
(76–79). Expectedly, the silencing of
IKBKE in BC cells has been reported to reduce the NF-қB
activity and inhibit proliferation, clonogenicity, migration, and
invasion (80–84).
GOSS inhibited the expression of MAPK1 (also
known as ERK) that was highly expressed in unstimulated
cells. The obtained data are consistent with those from previous
studies (85,86) that related the lower survival rate
in TNBC to highly expressed MAPKs. These serine/threonine
protein kinases are involved in various intracellular metabolism,
including differentiation, proliferation, apoptosis, and cellular
stress responses (87,88). Deregulated ERK signaling pathway
leads to uncontrolled cell proliferation and a reduction in
apoptosis (89). In MM-231 BC
cells, upregulated p38 pathway is linked to the increased
proliferation and migration (71)
and contribute to cancer cell invasion (90). Therefore, highlighting GOSS effects
on MAPKs implies its particularity for TNBC.
Our data indicated that TNF-α-stimulated the release
of IL-8 In MM-468 cells, the finding that agrees with a previous
study (56). Indeed, a significant
positive relationship has been reported between IL-8 release and
TNF-α that can synergistically affect cancer initiation and
progression (55). The
double-functional IL-8 can boost the immunoregulatory ability or
alter the cell microenvironment to enhance tumorigenesis (91). The chemokine IL-8 is a member of
the CXC chemokine family of multi-functional proinflammatory
cytokines (92). IL-8 is highly
expressed in many types of tumors and has the ability to promote
cancer cell proliferation (93).
In BC patients, elevated levels of serum IL-8 is a predictive
marker (55,94). Moreover, IL-8 is positively
correlated with angiogenesis and metastasis (95) and contributes to multidrug
resistance in human BC cells (96). Particularly in TNBC, IL-8 is highly
expressed compared with other BC subtypes (97), and it is associated with a poor
prognosis (55,92,98).
Meaningfully, the poorer prognosis and treatment resistance are
linked to IL-8 and its highly expressed receptors, CXCR1
(IL-8RA) and CXCR2 (IL-8RB) in BC patients
(95,99,100). Therefore, targeting CXCR
signaling is considered a promising approach in in-vivo
models of BC (101) as it
attenuates the distress effect of IL-8 (102). Here, the TNF-α-stimulated IL-8
expression was followed by a substantial attenuation of the
chemokine in the presence of only 5 µM of GOSS.
In MM-468 cells, GOSS repressed many highly
expressed genes, including IL-8, MAPK1, MAPK3, CCDC88A,
STAT3, and PIK3CD. The number and type of the affected
genes in MM-468 were divergent from its counterpart mRNA data for
MM-231. The repressed genes modulate major signaling pathways,
including MAPK, PI3K-AKT, JAK-STAT, and NF-қB. Those oncogenic
signaling pathways regulate IL-8 expression and potentiate tumor
cell migration or invasion (103,104). The data indicated high
expressions of MAPK1 and MAPK3 in unstimulated MM-468 cells;
an observation which is consistent with its counterpart MM-231 and
agrees with those previously reported findings (85,86).
The reported link between IL-8 mRNA stabilization, its
protein production, and the MAPKs (105–107) is compatible with GOSS effects in
our MM-468 study.
Furthermore, GOSS inhibited the expression of the
CCDC88A gene [also known as AKT phosphorylation enhancer
(APE), or AKT-binding protein Girdin], that is highly expressed in
BC and other types of cancer (108). The gene augmenting the PI3K-AKT
signaling pathway and regulate cell migration (109), proliferation, and apoptosis
(110). Also, upregulated AKT
expression enhances the NF-қB signaling pathway, promoting tumor
progression and metastasis (111). GOSS repressed the overexpression
of PIK3CD, the gene encoding phosphoinositide 3-kinase
(PI3K). The activated PI3K signaling is involved in
metabolism and is essential in promoting tumorigenesis and cell
proliferation (112–114). In normal cells, PI3K-AKt/mTOR
signaling pathways are critical for cell survival and
proliferation. Meanwhile, PI3K, AKT, and mTOR proteins are highly
upregulated in BC and other cancers. Inhibitors of these kinases
are found to induce apoptosis and to synergize the effect of
anticancer drugs to inhibit the tumor progression (112–114). Thus, the obtained data indicate
GOSS targeting ability of both PIK3CD and CCDC88A by
indirectly attenuating NFқB and, consequently, the overexpression
of IL-8 in MM-468 BC cells. The mRNA expression inhibition of the
signal transducer and activator of transcription3 (STAT3)
signifies the GOSS role against BC. STAT3 is a member of the
STAT family proteins, and it is highly triggered in more than 50%
of BC patients (115).
Furthermore, Targeting STAT3 impacts other cancer-associated
pro-inflammatory cytokines (116)
and many genes linked to apoptosis, proliferation, and angiogenesis
(117). Moreover, phosphorylation
of STAT3, AKT, and ERK1/2 stimulate BC cells to undergo
epithelial-mesenchymal transition, up-regulate the expression of
IL-8 and stimulate BC metastasis (104), inhibiting the mRNA expressions of
these proteins in MM-468 cells may further elucidate previously
reported anticancer mechanisms of GOSS through inhibiting their
phosphorylation (37). Thus,
inhibiting STAT3 activation by GOSS is a substantial target in
treating different types of cancer.
In summary, the comparative effect of gossypol on
different cytokines release from TNBC cells has never been reported
before. Also, gossypol effects on the aggressive TNBC of MM-468
cells, derived from AA women (118), are not reported. The current
study elucidated a novel mechanism targeting TNBC and highlight the
possible gene-related signaling pathways. However, lacking protein
production assessments, such as using Western blotting, is a
limitation in this research and needs further broader range
investigation. The polyphenol compound, GOSS, impacts MM-231 and
MM-468 cells differently. In MM-231 cells, the compound attenuated
the expression of the cytokine CCL2 through influencing its
encoding gene, CCL2, as well as two more regulatory genes
(IKBKE and MAPK1), which are involved in NF-қB,
JAK-STAT, and MAPK signaling pathways. Similarly, GOSS repressed
the cytokine IL-8 in MM-468 cells by targeting IL-8 mRNA
expression. However, more genes were inhibited by GOSS, including
MAPK1, MAPK3, PIK3CD, STAT3, and CCDC88A. These genes
are regulating different interactive signaling pathways mediating
IL-8 releases such as PI3K-AKT, JAK-STAT, and MAPK. In conclusion,
the data obtained in this study indicate that the polyphenol
compound GOSS may provide a valuable tool in TNBC therapies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Institute of Minority Health and Health Disparity (grant nos. U54
MD007582 and P20 MD006738).
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
The conceptualization of this study was
accomplished by SSM and KFAS. The research methodology was designed
by SSM, NOZ, PM, CC and KFAS. Data analysis was conducted by SSM,
NOZ, PM, CC and KFAS. Funding acquisition was fulfilled by KFAS.
Project administration was performed by KFAS, and study resources
were collected by SSM, NOZ, PM, CC and KFAS. Software analysis of
data and figures was conducted by SSM, NOZ, PM and CC, and
supervision of the research was conducted by KFAS. Writing of the
original draft was undertaken by SSM, and writing, review, and
editing of the manuscript were carried out by SSM, NOZ, PM and
KFAS. All authors read and approved the final manuscript and accept
to be responsible for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work is properly
considered.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aggarwal BB: Nuclear factor-kappaB: The
enemy within. Cancer Cell. 6:203–208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balkwill F and Mantovani A: Inflammation
and cancer: Back to virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kuper H, Adami HO and Trichopoulos D:
Infections as a major preventable cause of human cancer. J Intern
Med. 248:171–183. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Balkwill F: TNF-alpha in promotion and
progression of cancer. Cancer Metastasis Rev. 25:409–416. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fang WB, Yao M, Brummer G, Acevedo D,
Alhakamy N, Berkland C and Cheng N: Targeted gene silencing of CCL2
inhibits triple negative breast cancer progression by blocking
cancer stem cell renewal and M2 macrophage recruitment. Oncotarget.
7:49349–49367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niemeier LA, Dabbs DJ, Beriwal S, Striebel
JM and Bhargava R: Androgen receptor in breast cancer: Expression
in estrogen receptor-positive tumors and in estrogen
receptor-negative tumors with apocrine differentiation. Mod Pathol.
23:205–212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anders CK and Carey LA: Biology,
metastatic patterns, and treatment of patients with triple-negative
breast cancer. Clin Breast Cancer. 2 (Suppl 2):S73–S81. 2009.
View Article : Google Scholar
|
|
10
|
Albain KS, Unger JM, Crowley JJ, Coltman
CA Jr and Hershman DL: Racial disparities in cancer survival among
randomized clinical trials patients of the Southwest oncology
group. J Natl Cancer Inst. 101:984–992. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Beaumont T and Leadbeater M: Treatment and
care of patients with metastatic breast cancer. Nurs Stand.
25:49–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fernandez Y, Cueva J, Palomo AG, Ramos M,
de Juan A, Calvo L, García-Mata J, García-Teijido P, Peláez I and
García-Estévez L: Novel therapeutic approaches to the treatment of
metastatic breast cancer. Cancer Treat Rev. 36:33–42. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu P, Kumar IS, Brown S, Kannappan V,
Tawari PE, Tang JZ, Jiang W, Armesilla AL, Darling JL and Wang W:
Disulfiram targets cancer stem-like cells and reverses resistance
and cross-resistance in acquired paclitaxel-resistant
triple-negative breast cancer cells. Br J Cancer. 109:1876–1885.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Craig DW, O'Shaughnessy JA, Kiefer JA,
Aldrich J, Sinari S, Moses TM, Wong S, Dinh J, Christoforides A,
Blum JL, et al: Genome and transcriptome sequencing in prospective
metastatic triple-negative breast cancer uncovers therapeutic
vulnerabilities. Mol Cancer Ther. 12:104–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Allavena P, Garlanda C, Borrello MG, Sica
A and Mantovani A: Pathways connecting inflammation and cancer.
Curr Opin Genet Dev. 18:3–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Candido J and Hagemann T: Cancer-related
inflammation. J Clin Immunol. 33 (Suppl 1):S79–S84. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saji H, Koike M, Yamori T, Saji S, Seiki
M, Matsushima K and Toi M: Significant correlation of monocyte
chemoattractant protein-1 expression with neovascularization and
progression of breast carcinoma. Cancer. 92:1085–1091. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bieche I, Chavey C, Andrieu C, Busson M,
Vacher S, Corre LL, Guinebretière JM, Burlinchon S, Lidereau R and
Lazennec G: CXC chemokines located in the 4q21 region are
up-regulated in breast cancer. Endocr Relat Cancer. 14:1039–1052.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang WB, Jokar I, Zou A, Lambert D,
Dendukuri P and Cheng N: CCL2/CCR2 chemokine signaling coordinates
survival and motility of breast cancer cells through Smad3 protein-
and p42/44 mitogen-activated protein kinase (MAPK)-dependent
mechanisms. J Biol Chem. 287:36593–36608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Perrot-Applanat M, Vacher S, Toullec A,
Pelaez I, Velasco G, Cormier F, El Sheikh Saad H, Lidereau R, Baud
V and Bièche I: Similar NF-kB gene signatures in TNF-α treated
human endothelial cells and breast tumor biopsies. PLoS One.
6:e215892011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leek RD, Landers R, Fox SB, Ng F, Harris
AL and Lewis CE: Association of tumour necrosis factor alpha and
its receptors with thymidine phosphorylase expression in invasive
breast carcinoma. Br J Cancer. 77:2246–2251. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang N, Liang H and Zen K: Molecular
mechanisms that influence the macrophage m1-m2 polarization
balance. Front Immunol. 5:6142014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yin Y, Chen X and Shu Y: Gene expression
of the invasive phenotype of TNF-alpha-treated MCF-7 cells. Biomed
Pharmacother. 63:421–428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Voss V, Senft C, Lang V, Ronellenfitsch
MW, Steinbach JP, Seifert V and Kögel D: The pan-Bcl-2 inhibitor
(−)-gossypol triggers autophagic cell death in malignant glioma.
Mol Cancer Res. 8:1002–1016. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bromberg J: Stat proteins and oncogenesis.
J Clin Invest. 109:1139–1142. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dreesen O and Brivanlou AH: Signaling
pathways in cancer and embryonic stem cells. Stem Cell Rev. 3:7–17.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zubair A and Frieri M: Role of nuclear
factor-kB in breast and colorectal cancer. Curr Allergy Asthma Rep.
13:44–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhong S, Leong J, Ye W, Xu P, Lin SH, Liu
JY and Lin YC: (−)-Gossypol-enriched cottonseed oil inhibits
proliferation and adipogenesis of human breast pre-adipocytes.
Anticancer Res. 33:949–955. 2013.PubMed/NCBI
|
|
29
|
Cao H, Sethumadhavan K and Bland JM:
Isolation of cottonseed extracts that affect human cancer cell
growth. Sci Rep. 8:104582018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He Z, Zhang H and Olk DC: Chemical
composition of defatted cottonseed and soy meal products. PLoS One.
10:e01299332015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sharifi-Rad M, Fokou PVT, Sharopov F,
Martorell M, Ademiluyi AO, Rajkovic J, Salehi B, Martins N, Iriti M
and Sharifi-Rad J: Antiulcer agents: From plant extracts to
phytochemicals in healing promotion. Molecules. 23:17512018.
View Article : Google Scholar
|
|
32
|
Wang X, Howell CP, Chen F, Yin J and Jiang
Y: Gossypol-a polyphenolic compound from cotton plant. Adv Food
Nutr Res. 58:215–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang M, Liu H, Guo R, Ling Y, Wu X, Li B,
Roller PP, Wang S and Yang D: Molecular mechanism of
gossypol-induced cell growth inhibition and cell death of HT-29
human colon carcinoma cells. Biochem Pharmacol. 66:93–103. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pang X, Wu Y, Wu Y, Lu B, Chen J, Wang J,
Yi Z, Qu W and Liu M: (−)-Gossypol suppresses the growth of human
prostate cancer xenografts via modulating VEGF signaling-mediated
angiogenesis. Mol Cancer Ther. 10:795–805. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Flack MR, Pyle RG, Mullen NM, Lorenzo B,
Wu YW, Knazek RA, Nisula BC and Reidenberg MM: Oral gossypol in the
treatment of metastatic adrenal cancer. J Clin Endocrinol Metab.
76:1019–1024. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moon DO, Kim MO, Lee JD and Kim GY:
Gossypol suppresses NF-kappaB activity and NF-kappaB-related gene
expression in human leukemia U937 cells. Cancer Lett. 264:192–200.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Benvenuto M, Mattera R, Masuelli L,
Taffera G, Andracchio O, Tresoldi I, Lido P, Giganti MG, Godos J,
Modesti A and Bei R: (±)-Gossypol induces apoptosis and autophagy
in head and neck carcinoma cell lines and inhibits the growth of
transplanted salivary gland cancer cells in BALB/c mice. Int J Food
Sci Nutr. 68:298–312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moon DO, Choi YH, Moon SK, Kim WJ and Kim
GY: Gossypol decreases tumor necrosis factor-α-induced
intercellular adhesion molecule-1 expression via suppression of
NF-κB activity. Food Chem Toxicol. 49:999–1005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gilbert NE, O'Reilly JE, Chang CJ, Lin YC
and Brueggemeier RW: Antiproliferative activity of gossypol and
gossypolone on human breast cancer cells. Life Sci. 57:61–67. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu YF, Chang CJ, Brueggemeier RW and Lin
YC: Gossypol inhibits basal and estrogen-stimulated DNA synthesis
in human breast carcinoma cells. Life Sci. 53:PL433–PL438. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chou TC: Drug combination studies and
their synergy quantification using the Chou-Talalay method. Cancer
Res. 70:440–446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoshida R, Niki M, Jyotaki M, Sanematsu K,
Shigemura N and Ninomiya Y: Modulation of sweet responses of taste
receptor cells. Semin Cell Dev Biol. 24:226–231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheng W, Zhao YQ, Li YM and Yang DJ:
Effects of gossypol acetate on apoptosis in primary cultured cells
from patients with lymphoid leukemia and its synergy with
dexamethasone. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 20:229–234.
2012.(In Chinese). PubMed/NCBI
|
|
44
|
Baoleri X, Dong C, Zhou Y, Zhang Z, Lu X,
Xie P and Li Y: Combination of L-gossypol and low-concentration
doxorubicin induces apoptosis in human synovial sarcoma cells. Mol
Med Rep. 12:5924–5932. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Karaca B, Atmaca H, Uzunoglu S, Karabulut
B, Sanli UA and Uslu R: Enhancement of taxane-induced cytotoxicity
and apoptosis by gossypol in human breast cancer cell line MCF-7. J
BUON. 14:479–485. 2009.PubMed/NCBI
|
|
46
|
Zhao GX, Xu LH, Pan H, Lin QR, Huang MY,
Cai JY, Ouyang DY and He XH: The BH3-mimetic gossypol and
noncytotoxic doses of valproic acid induce apoptosis by suppressing
cyclin-A2/Akt/FOXO3a signaling. Oncotarget. 6:38952–38966. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tate CR, Rhodes LV, Segar HC, Driver JL,
Pounder FN, Burow ME and Collins-Burow BM: Targeting
triple-negative breast cancer cells with the histone deacetylase
inhibitor panobinostat. Breast Cancer Res. 14:R792012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sato N, Beitz JG, Kato J, Yamamoto M,
Clark JW, Calabresi P, Raymond A and Frackelton Jr AR:
Platelet-derived growth factor indirectly stimulates angiogenesis
in vitro. Am J Pathol. 142:1119–1130. 1993.PubMed/NCBI
|
|
49
|
Messeha SS, Zarmouh NO, Mendonca P,
Alwagdani H, Kolta MG and Soliman KFA: The inhibitory effects of
plumbagin on the NF-kB pathway and CCL2 release in racially
different triple-negative breast cancer cells. PLoS One.
13:e02011162018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yamamoto H, Omelchenko I, Shi X and
Nuttall AL: The influence of NF-kappaB signal-transduction pathways
on the murine inner ear by acoustic overstimulation. J Neurosci
Res. 87:1832–1840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gallelli L, Falcone D, Cannataro R, Perri
M, Serra R, Pelaia G, Maselli R, Savino R, Spaziano G and
D'Agostino B: Theophylline action on primary human bronchial
epithelial cells under proinflammatory stimuli and steroidal drugs:
A therapeutic rationale approach. Drug Des Dev Ther. 11:265–272.
2017. View Article : Google Scholar
|
|
53
|
Barba-Barajas M, Hernandez-Flores G,
Lerma-Diaz JM, Ortiz-Lazareno PC, Domínguez-Rodríguez JR,
Barba-Barajas L, de Celis R, Jave-Suarez LF, Aguilar-Lemarroy AC,
Guevara-Barraza MG and Bravo-Cuellar A: Gossypol induced apoptosis
of polymorphonuclear leukocytes and monocytes: Involvement of
mitochondrial pathway and reactive oxygen species. Immunopharmacol
Immunotoxicol. 31:320–330. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Anderson GM, Nakada MT and DeWitte M:
Tumor necrosis factor-alpha in the pathogenesis and treatment of
cancer. Curr Opin Pharmacol. 4:314–320. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ma Y, Ren Y, Dai ZJ, Wu CJ, Ji YH and Xu
J: IL-6, IL-8 and TNF-α levels correlate with disease stage in
breast cancer patients. Adv Clin Exp Med. 26:421–426. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
John M, Au BT, Jose PJ, Lim S, Saunders M,
Barnes PJ, Mitchell JA, Belvisi MG and Chung KF: Expression and
release of interleukin-8 by human airway smooth muscle cells:
Inhibition by Th-2 cytokines and corticosteroids. Am J Respir Cell
Mol Biol. 18:84–90. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Grund EM, Kagan D, Tran CA, Zeitvogel A,
Starzinski-Powitz A, Nataraja S and Palmer SS: Tumor necrosis
factor-alpha regulates inflammatory and mesenchymal responses via
mitogen-activated protein kinase kinase, p38, and nuclear factor
kappaB in human endometriotic epithelial cells. Mol Pharmacol.
73:1394–1404. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
An J, Xue Y, Long M, Zhang G, Zhang J and
Su H: Targeting CCR2 with its antagonist suppresses viability,
motility and invasion by downregulating MMP-9 expression in
non-small cell lung cancer cells. Oncotarget. 8:39230–39240. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Strieter RM, Wiggins R, Phan SH, Wharram
BL, Showell HJ, Remick DC, Chensue SW and Kunkel SL: Monocyte
chemotactic protein gene expression by cytokine-treated human
fibroblasts and endothelial cells. Biochem Biophys Res Commun.
162:694–700. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sticherling M, Hetzel F, Schroder JM and
Christophers E: Time- and stimulus-dependent secretion of
NAP-1/IL-8 by human fibroblasts and endothelial cells. J Invest
Dermatol. 101:573–576. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zachariae CO, Anderson AO, Thompson HL,
Appella E, Mantovani A, Oppenheim JJ and Matsushima K: Properties
of monocyte chemotactic and activating factor (MCAF) purified from
a human fibrosarcoma cell line. J Exp Med. 171:2177–2182. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Krupa A, Fol M, Dziadek BR, Kepka E,
Wojciechowska D, Brzostek A, Torzewska A, Dziadek J, Baughman RP,
Griffith D and Kurdowska AK: Binding of CXCL8/IL-8 to mycobacterium
tuberculosis modulates the innate immune response. Mediators
Inflamm. 2015:1247622015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ueno T, Toi M, Saji H, Muta M, Bando H,
Kuroi K, Koike M, Inadera H and Matsushima K: Significance of
macrophage chemoattractant protein-1 in macrophage recruitment,
angiogenesis, and survival in human breast cancer. Clin Cancer Res.
6:3282–3289. 2000.PubMed/NCBI
|
|
64
|
Matsushima K, Larsen CG, DuBois GC and
Oppenheim JJ: Purification and characterization of a novel monocyte
chemotactic and activating factor produced by a human
myelomonocytic cell line. J Exp Med. 169:1485–1490. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Espinoza-Sanchez NA, Chimal-Ramirez GK,
Mantilla A and Fuentes-Panana EM: IL-1β, IL-8, and matrix
metalloproteinases-1,-2, and −10 are enriched upon monocyte-breast
cancer cell cocultivation in a matrigel-based three-dimensional
system. Front Immunol. 8:2052017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Nam JS, Kang MJ, Suchar AM, Shimamura T,
Kohn EA, Michalowska AM, Jordan VC, Hirohashi S and Wakefield LM:
Chemokine (C-C motif) ligand 2 mediates the prometastatic effect of
dysadherin in human breast cancer cells. Cancer Res. 66:7176–7184.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Qian BZ, Li J, Zhang H, Kitamura T, Zhang
J, Campion LR, Kaiser EA, Snyder LA and Pollard JW: CCL2 recruits
inflammatory monocytes to facilitate breast-tumour metastasis.
Nature. 475:222–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tsuyada A, Chow A, Wu J, Somlo G, Chu P,
Loera S, Luu T, Li XA, Wu X, Ye W, et al: CCL2 mediates cross-talk
between cancer cells and stromal fibroblasts that regulates breast
cancer stem cells. Cancer Res. 72:2768–2779. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Koury J, Zhong L and Hao J: Targeting
signaling pathways in cancer stem cells for cancer treatment. Stem
Cells Int. 2017:29258692017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Pires BR, DE Amorim ÍS, Souza LD,
Rodrigues JA and Mencalha AL: Targeting cellular signaling pathways
in breast cancer stem cells and its implication for cancer
treatment. Anticancer Res. 36:5681–5691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Huth HW, Santos DM, Gravina HD, Resende
JM, Goes AM, de Lima ME and Ropert C: Upregulation of p38 pathway
accelerates proliferation and migration of MDA-MB-231 breast cancer
cells. Oncol Rep. 37:2497–2505. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Peters RT, Liao SM and Maniatis T:
IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex.
Mol Cell. 5:513–522. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bauer D, Redmon N, Mazzio E and Soliman
KF: Apigenin inhibits TNFα/IL-1α-induced CCL2 release through
IKBK-epsilon signaling in MDA-MB-231 human breast cancer cells.
PLoS One. 12:e01755582017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Williams V, Grosset AA, Zamorano Cuervo N,
St-Pierre Y, Sylvestre MP, Gaboury L and Grandvaux N: Detection of
IKKepsilon by immunohistochemistry in primary breast cancer:
Association with EGFR expression and absence of lymph node
metastasis. BMC Cancer. 17:3562017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Guo JP, Shu SK, Esposito NN, Coppola D,
Koomen JM and Cheng JQ: IKK phosphorylation of estrogen receptor α
Ser-167 and contribution to tamoxifen resistance in breast cancer.
J Biol Chem. 291:228572016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Berishaj M, Gao SP, Ahmed S, Leslie K,
Al-Ahmadie H, Gerald WL, Bornmann W and Bromberg JF: Stat3 is
tyrosine-phosphorylated through the interleukin-6/glycoprotein
130/Janus kinase pathway in breast cancer. Breast Cancer Res.
9:R322007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li L and Shaw PE: Autocrine-mediated
activation of STAT3 correlates with cell proliferation in breast
carcinoma lines. J Biol Chem. 277:17397–17405. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Walker SR, Nelson EA, Zou L, Chaudhury M,
Signoretti S, Richardson A and Frank DA: Reciprocal effects of
STAT5 and STAT3 in breast cancer. Mol Cancer Res. 7:966–976. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Marotta LL, Almendro V, Marusyk A,
Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ,
Choudhury SA, Maruyama R, et al: The JAK2/STAT3 signaling pathway
is required for growth of CD44(+)CD24(−) stem cell-like breast
cancer cells in human tumors. J Clin Invest. 121:2723–2735. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
House CD, Grajales V, Ozaki M, Jordan E,
Wubneh H, Kimble DC, James JM, Kim MK and Annunziata CM: IΚΚε
cooperates with either MEK or non-canonical NF-kB driving growth of
triple-negative breast cancer cells in different contexts. BMC
Cancer. 18:5952018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Boehm JS, Zhao JJ, Yao J, Kim SY,
Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S,
Richardson AL, et al: Integrative genomic approaches identify IKBKE
as a breast cancer oncogene. Cell. 129:1065–1079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hutti JE, Shen RR, Abbott DW, Zhou AY,
Sprott KM, Asara JM, Hahn WC and Cantley LC: Phosphorylation of the
tumor suppressor CYLD by the breast cancer oncogene IKKepsilon
promotes cell transformation. Mol Cell. 34:461–472. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Barbie TU, Alexe G, Aref AR, Li S, Zhu Z,
Zhang X, Imamura Y, Thai TC, Huang Y, Bowden M, et al: Targeting an
IKBKE cytokine network impairs triple-negative breast cancer
growth. J Clin Invest. 124:5411–5423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Qin B and Cheng K: Silencing of the IKKε
gene by siRNA inhibits invasiveness and growth of breast cancer
cells. Breast Cancer Res. 12:R742010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Bartholomeusz C, Gonzalez-Angulo AM, Liu
P, Hayashi N, Lluch A, Ferrer-Lozano J and Hortobágyi GN: High ERK
protein expression levels correlate with shorter survival in
triple-negative breast cancer patients. Oncologist. 17:766–774.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Seddighzadeh M, Zhou JN, Kronenwett U,
Shoshan MC, Auer G, Sten-Linder M, Wiman B and Linder S: ERK
signalling in metastatic human MDA-MB-231 breast carcinoma cells is
adapted to obtain high urokinase expression and rapid cell
proliferation. Clin Exp Metastasis. 17:649–654. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kyriakis JM and Avruch J: Mammalian MAPK
signal transduction pathways activated by stress and inflammation:
A 10-year update. Physiol Rev. 92:689–737. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Samatar AA and Poulikakos PI: Targeting
RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug
Discov. 13:928–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wen S, Hou Y, Fu L, Xi L, Yang D, Zhao M,
Qin Y, Sun K, Teng Y and Liu M: Cancer-associated fibroblast
(CAF)-derived IL32 promotes breast cancer cell invasion and
metastasis via integrin β3-p38 MAPK signalling. Cancer Lett.
442:320–332. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Benoy IH, Salgado R, Van Dam P, Geboers K,
Van Marck E, Scharpé S, Vermeulen PB and Dirix LY: Increased serum
interleukin-8 in patients with early and metastatic breast cancer
correlates with early dissemination and survival. Clin Cancer Res.
10:7157–7162. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Guo Y, Zang Y, Lv L, Cai F, Qian T, Zhang
G and Feng Q: IL8 promotes proliferation and inhibition of
apoptosis via STAT3/AKT/NFkB pathway in prostate cancer. Mol Med
Rep. 16:9035–9042. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Snoussi K, Mahfoudh W, Bouaouina N, Ahmed
SB, Helal AN and Chouchane L: Genetic variation in IL-8 associated
with increased risk and poor prognosis of breast carcinoma. Hum
Immunol. 67:13–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zuccari DA, Leonel C, Castro R, Gelaleti
GB, Jardim BV, Moscheta MG, Regiani VR, Ferreira LC, Lopes JR, Neto
Dde S and Esteves JL: An immunohistochemical study of interleukin-8
(IL-8) in breast cancer. Acta Histochem. 114:571–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kim S, Jeon M, Lee JE and Nam SJ: MEK
activity controls IL-8 expression in tamoxifen-resistant MCF-7
breast cancer cells. Oncol Rep. 35:2398–2404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kim S, You D, Jeong Y, Yu J, Kim SW, Nam
SJ and Lee JE: Berberine down-regulates IL-8 expression through
inhibition of the EGFR/MEK/ERK pathway in triple-negative breast
cancer cells. Phytomedicine. 50:43–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yao C, Lin Y, Chua MS, Ye CS, Bi J, Li W,
Zhu YF and Wang SM: Interleukin-8 modulates growth and invasiveness
of estrogen receptor-negative breast cancer cells. Int J Cancer.
121:1949–1957. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Park SH, Das BB, Casagrande F, Tian Y,
Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM
and Opella SJ: Structure of the chemokine receptor CXCR1 in
phospholipid bilayers. Nature. 491:779–783. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Singh JK, Simoes BM, Clarke RB and Bundred
NJ: Targeting IL-8 signalling to inhibit breast cancer stem cell
activity. Expert Opin Ther Targets. 17:1235–1241. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Singh JK, Simoes BM, Howell SJ, Farnie G
and Clarke RB: Recent advances reveal IL-8 signaling as a potential
key to targeting breast cancer stem cells. Breast Cancer Res.
15:2102013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Alfaro C, Teijeira A, Onate C, Pérez G,
Sanmamed MF, Andueza MP, Alignani D, Labiano S, Azpilikueta A,
Rodriguez-Paulete A, et al: Tumor-produced interleukin-8 attracts
human myeloid-derived suppressor cells and elicits extrusion of
neutrophil extracellular traps (NETs). Clin Cancer Res.
22:3924–3936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Elliott CL, Allport VC, Loudon JA, Wu GD
and Bennett PR: Nuclear factor-kappa B is essential for
up-regulation of interleukin-8 expression in human amnion and
cervical epithelial cells. Mol Hum Reprod. 7:787–790. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang L, Tang C, Cao H, Li K, Pang X, Zhong
L, Dang W, Tang H, Huang Y, Wei L, et al: Activation of IL-8 via
PI3K/Akt-dependent pathway is involved in leptin-mediated
epithelial-mesenchymal transition in human breast cancer cells.
Cancer Biol Ther. 16:1220–1230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lian S, Xia Y, Ung TT, Khoi PN, Yoon HJ,
Kim NH, Kim KK and Jung YD: Carbon monoxide releasing molecule-2
ameliorates IL-1β-induced IL-8 in human gastric cancer cells.
Toxicology 361–362. 24–38. 2016. View Article : Google Scholar
|
|
106
|
Singh RK, Gutman M, Radinsky R, Bucana CD
and Fidler IJ: Expression of interleukin 8 correlates with the
metastatic potential of human melanoma cells in nude mice. Cancer
Res. 54:3242–3247. 1994.PubMed/NCBI
|
|
107
|
Fernando RI, Hamilton DH, Dominguez C,
David JM, McCampbell KK and Palena C: IL-8 signaling is involved in
resistance of lung carcinoma cells to erlotinib. Oncotarget.
7:42031–42044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Jiang P, Enomoto A, Jijiwa M, Kato T,
Hasegawa T, Ishida M, Sato T, Asai N, Murakumo Y and Takahashi M:
An actin-binding protein girdin regulates the motility of breast
cancer cells. Cancer Res. 68:1310–1318. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lin C, Ear J, Pavlova Y, Mittal Y,
Kufareva I, Ghassemian M, Abagyan R, Garcia-Marcos M and Ghosh P:
Tyrosine phosphorylation of the Gα-interacting protein GIV promotes
activation of phosphoinositide 3-kinase during cell migration. Sci
Signal. 4:ra642011. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ni W, Fang Y, Tong L, Tong Z, Yi F, Qiu J,
Wang R and Tong X: Girdin regulates the migration and invasion of
glioma cells via the PI3K-Akt signaling pathway. Mol Med Rep.
12:5086–5092. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Romashkova JA and Makarov SS: NF-kappaB is
a target of AKT in anti-apoptotic PDGF signalling. Nature.
401:86–90. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
112
|
Jiang BH and Liu LZ: PI3K/PTEN signaling
in tumorigenesis and angiogenesis. Biochim Biophys Acta.
1784:150–158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Abraham RT: Chemokine to the rescue:
Interleukin-8 mediates resistance to PI3K-pathway-targeted therapy
in breast cancer. Cancer Cell. 22:703–705. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Darnell JE: Validating Stat3 in cancer
therapy. Nat Med. 11:595–596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Subramaniam A, Shanmugam MK, Perumal E, Li
F, Nachiyappan A, Dai X, Swamy SN, Ahn KS, Kumar AP, Tan BKH, et
al: Potential role of signal transducer and activator of
transcription (STAT)3 signaling pathway in inflammation, survival,
proliferation and invasion of hepatocellular carcinoma. Biochim
Biophys Acta. 1835:46–60. 2013.PubMed/NCBI
|
|
117
|
Rajendran P, Li F, Shanmugam MK, Kannaiyan
R, Goh JN, Wong KF, Wang W, Khin E, Tergaonkar V, Kumar AP, et al:
Celastrol suppresses growth and induces apoptosis of human
hepatocellular carcinoma through the modulation of STAT3/JAK2
signaling cascade in vitro and in vivo. Cancer Prev Res (Phila).
5:631–643. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Samson ME, Porter NG, Hurley DM, Adams SA
and Eberth JM: Disparities in breast cancer incidence, mortality,
and quality of care among African American and European American
women in South Carolina. South Med J. 109:24–30. 2016. View Article : Google Scholar : PubMed/NCBI
|