Introduction
Non-alcoholic fatty liver disease (NAFLD) refers to
a spectrum of liver pathologies with hepatic fat accumulation,
which are collectively the leading cause of chronic liver disease
(1–3). The pathogenesis of NAFLD is not fully
understood. The widely accepted ‘multiple hits’ hypothesis suggests
that excessive oxidative metabolites and lipid peroxidation occur
as a result of insulin resistance (IR). This causes oxidative
stress (OS) and continuously damages the mitochondria within
hepatocytes, while producing inflammatory mediators and cytokines
(1,3). Hepatocytic inflammation and necrosis
with steatosis is referred to as non-alcoholic steatohepatitis
(NASH). Without intervention, NASH can exacerbate IR and
inflammation, causing fatty liver accumulation and increasing the
risk of liver cirrhosis and cancer (1,3).
Currently, effective NAFLD treatment is still lacking (4).
The Notch family was initially discovered in
Drosophila and is expressed in a number of species. Notch
molecules serve an important role in the embryonic development of
organisms, as well as the occurrence of malignant tumors (5,6).
Previous studies predominantly focused on the role of Notch genes
in the regulation of cell differentiation, proliferation and
apoptosis (7,8). However, it has also been demonstrated
that Notch genes are involved in cell metabolism. Indeed, changes
in Notch gene expression leads to dysfunction in glucose and lipid
metabolism, which can lead to IR, lipid deposition and obesity
(9,10). Therefore, the relationship between
Notch genes and metabolic diseases has gradually become a research
focus.
NAFLD is the hepatic manifestation of metabolic
syndrome. With recent changes in living standards and lifestyle,
the incidence of NAFLD is rising rapidly, along with those of type
2 diabetes mellitus (T2DM) and overweight/obesity (11,12).
Nevertheless, the association of the Notch family with NAFLD is
rarely reported (13). Therefore,
the aim of the present study was to examine the dynamic association
between Notch and NAFLD, both in vitro and in vivo.
The time course of Notch gene expression was assessed in liver cell
lines treated with palmitic acid (PA), as well as a murine NAFLD
model induced by a methionine-choline-deficient (MCD) diet.
Materials and methods
Cell culture and treatments
The following hepatic cell lines were obtained from
The Beijing Union Medical College Resource Center and cultured in
DMEM: i) Hepatic stellate cell line LX2; ii) hepatocellular
carcinoma (HCC) cell line Huh7; and iii) human immortalized
hepatocytes MIHA. MIHA, LX2 and Huh7 cell lines were chosen so that
different stages of NAFLD (hepatocytes steatosis, fibrosis and
carcinoma, respectively), would be represented. PA (MedChemExpress)
was dissolved in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
containing 1% BSA (Sigma-Aldrich; Merck KGaA) and filtered through
a 0.22-µm filter, then added to the cells and incubated at 37°C to
80% confluency in DMEM with 10% FBS. Filter-sterilized complete
DMEM with 1% BSA without PA was used as a control. The
concentrations of PA used were 0.1, 0.25 or 0.5 mM, and the
incubation times were 12–72 h.
Cell proliferation and migration
assay
Cell proliferation under different PA concentrations
was assessed using a 5-bromo-2′-deoxyuridine assay kit
(Sigma-Aldrich; Merck KGaA), following the manufacturer's
instructions. Cell migration was evaluated using a wound healing
assay. A total of three cell lines were treated with or without PA.
A thick black line was drawn under each plate as a base line, and
shown in every field under a Leica DC 300F optical microscope
(magnification, ×400). The plate was scratched with the head of a
pipette to disrupt the cellular growth, creating a break in the
cells that simulates an injury. The scratch is vertical to the
black line under the plate. The wound width (WW) was measured in
three microscope fields. For every condition, the WW to initial WW
ratio was recorded and averaged. The lower the ratio, the faster
the cell migration. ImageJ software version 1.44p (National
Institutes of Health) was used for imaging.
Oil Red O (ORO) staining
All three cell lines were counted and seeded into
24-well tissue culture plates in DMEM with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C to 80% confluency, then exposed to
0.25 mM PA. Cells were observed 12–72 h after PA treatment. Cells
were washed with PBS, fixed with 4% paraformaldehyde for 10 min,
washed by 60% isopropanol for 1 min, and stained with fresh ORO
dissolved in 60% isopropanol (Sigma-Aldrich; Merck KGaA) for 30
min, as previously described (14). Then, cells were washed with
ddH2O, counterstained with hematoxylin for 1 min, and
mounted by Permount solution (Thermo Fisher Scientific, Inc.).
Finally, the cells were examined under a light microscope
(magnification, ×400) (Leica Microsystems GmbH) to observe lipid
droplets and identify steatotic cells.
Expression of Notch signaling pathway
genes
Total RNA was extracted from cell lines and primary
mouse liver tissues using an RNeasy Mini kit (Qiagen GmbH). Primers
and probes for qRT-PCR were purchased from Applied Biosystems
(Thermo Fisher Scientific, Inc.). Sequences of primers are listed
in Table SI. RNAase inhibitor,
dNTP and Oligo (dT) were from Toyobo Life Science. RNA was then
used for cDNA synthesis (42°C for 60 min, 70°C for 15 min, and 4°C)
by reverse transcription according to the manufacturer's
instructions. Reverse transcription-quantitative PCR (RT-qPCR) was
performed using a SYBR Green kit (Takara Biotechnology Co., Ltd.)
in an ABI 7500 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The standard conditions for qPCR were:
50°C for 10 min and 95°C for 15 and 45 sec annealing/elongation at
58°C or 60°C. GAPDH was used as an internal control. Relative genes
expressions were calculated using the 2−ΔΔCq method.
qRT-PCR experiments were repeated three times (15).
Animals and in vivo assay
A total of 80, 4–6-week-old, male C57BL/6 nude mice
(weight, 13–20 g) were obtained from The Institute of Laboratory
Animal Science (Chinese Academy of Medical Sciences). All animals
were housed in a controlled room with 22.0±2.0°C temperature and
65±5% humidity under a 12-h dark-light cycle. Mice were randomized
into 2 groups (n=40 in each group) receiving different diets. The
MCD group was used as the NAFLD model, and the
methionine-choline-sufficient (MCS) group served as a negative
control. Both groups were examined daily, then divided into four
subgroups (n=10 in each subgroup) euthanized at different time
points (days 5, 10, 21 and 70) (16,17).
All mice were subjected to overnight fasting, anesthetized by
intraperitoneal injection of 400 mg/kg chloral hydrate, then
euthanized by exsanguination performed by cardiac puncture. The
liver tissues were harvested only after the mice were confirmed to
lose the vital signs. No animal died accidently during the
experiment. All experiments were approved by the Ethics Committee
of Xin Hua Hospital (Shanghai Jiao Tong University School of
Medicine).
Harvested livers were frozen at −70°C for subsequent
examination of the expression levels of Notch1-4, hairy and
enhancer of split-1 (Hes1) and Hes-related family bHLH
transcription factor with YRPW motif 1 (Hey1) using RT-qPCR.
Statistical analysis
A total of three independent experiments were
performed. Data are presented as the mean ± SD. Multi-group
comparisons were carried out using one-way ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference. All data were analyzed with
SPSS 13.0 statistical package (SPSS, Inc.).
Results
Effect of PA on proliferation and
migration of hepatic cells
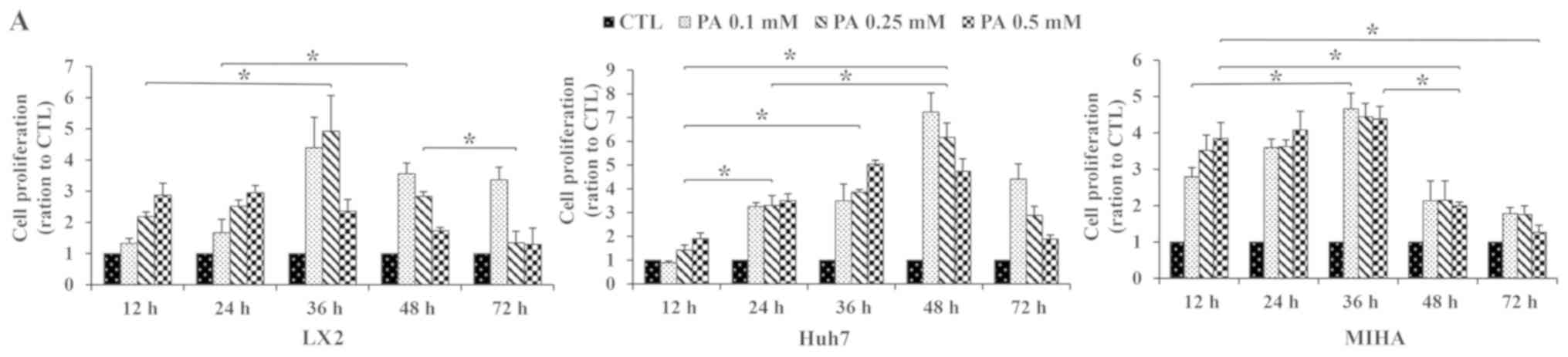
To determine the effects of PA exposure on
proliferation and migration of hepatic cells, Huh7, MIHA and LX2
cells were treated with 0.10, 0.25 or 0.50 mM PA for different
incubation times. After 12 h of exposure to PA, cell proliferation
increased in Huh7, MIHA and LX2 cells, compared with their
respective untreated controls. Proliferation reached a maximal
value at 36 h for LX2 and MIHA, and at 48 h Huh7 cells. Peak
proliferation was observed with 0.1 mM PA in the Huh7 and MIHA cell
line, as well as 0.25 mM for LX2. MIHA proliferation ratio seemed
close at certain timepoint whatever PA concentrations (Fig. 1A).
In addition, a wound healing assay indicated more
suspended (dead) LX2 cells in the medium at PA 0.25 mM after 48 h
(WW, 0.70±0.03), and 0.5 mM from 24 h (WW, 0.78±0.03) onward
(Fig. 1B; Table SII). Notable growth was observed
between 12 and 72 h following treatment with 0.25 mM PA
(P<0.05). The Huh7 cell line displayed increased growth rates
between 12 and 72 h at all concentrations of PA (Fig. 1B and C). The WW at 0.1 and 0.25 mM
PA were lower than that at 0.5 mM (Table SII). However, PA did not affect
the MIHA growth rate at any concentration (P>0.05). Therefore, a
dosage of 0.25 mM PA was used for subsequent experiments. It should
be noted that the shape of the suspended LX2 cells was different
from that of the adherent cells, and were excluded when WW was
calculated (Table SII).
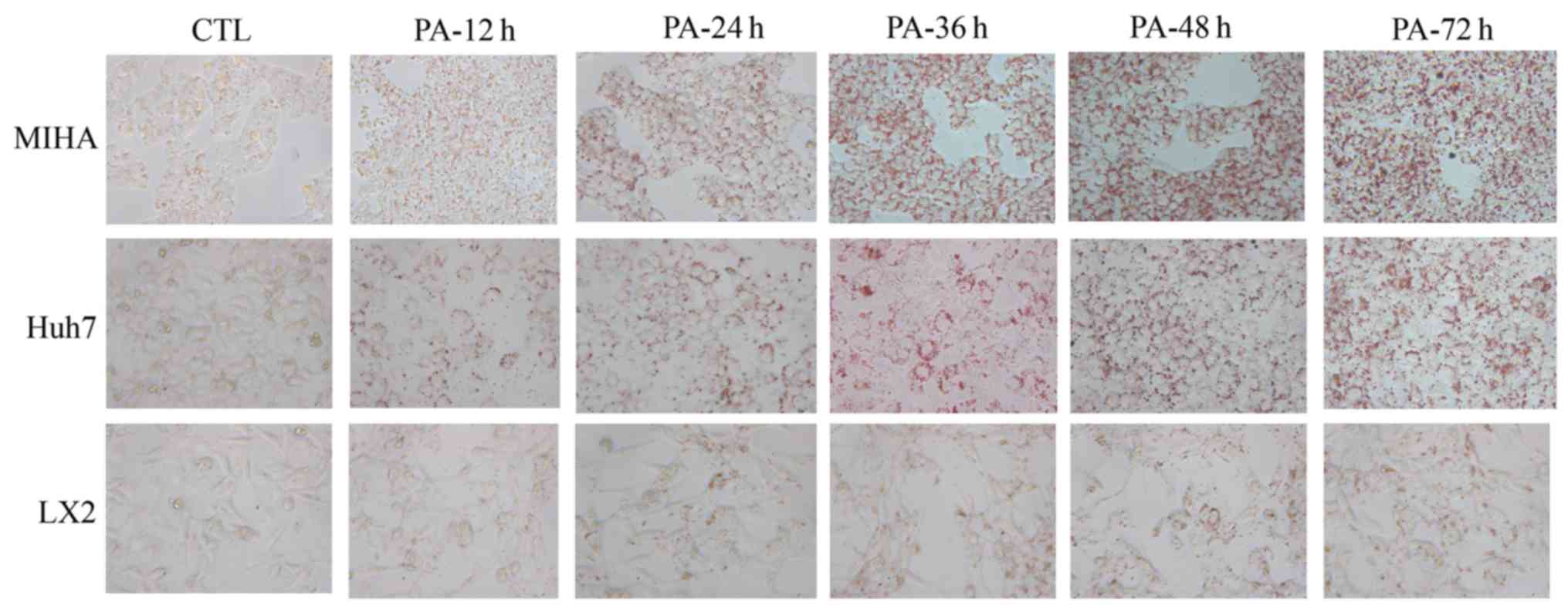
PA induces steatosis in hepatic
cells
ORO stains intracellular neutral lipids (18). Hepatic cells were stained with ORO
in the presence or absence of PA (Fig.
2). Following exposure to PA, notably more lipid droplets were
observed in three microscope fields, compared with the control. In
addition, a time-dependent accumulation of lipid appeared in LX2,
Huh7 and MIHA cells treated with 0.25 mM PA. These results
suggested that PA could enhance lipid synthesis and steatosis in
hepatic cell lines.
Notch family gene expression in
vitro
RT-qPCR assays were carried out to examine the
expression of Notch pathway genes in LX2, Huh7 and MIHA cells
following 0.25 mM PA exposure. Notch pathway members Notch1, −2,
−3, −4, Jagged1, Jagged2, Δ-like canonical Notch ligand (DLL) 1 and
DLL3, Hey1 and Hes1 were affected by PA exposure. The expression of
these signaling molecules followed varying trends over the course
of the experiment (Fig. 3). After
12 h PA treatment, the expression of Notch4 was significantly
upregulated in Huh7 lines, compared with control. The expression of
Notch3 was similar in LX2 and Huh7, but significantly downregulated
in MIHA cells at 12 h. At the 24-h time point, Notch2, −3 and −4
genes were upregulated only in the hepatic stellate cell line LX2,
compared with the control. At this time, point, Notch1 and −4 were
significantly downregulated in MIHA cells, while, Notch3 was
upregulated in Huh7 cells. However, the expression of these four
Notch genes partly recovered slowly with continuous PA exposure at
the endpoint of the experiment [CTL vs. 36 h: Notch1 (Huh7), Notch2
(MIHA), Notch3 (LX2), Notch4 (LX2), P<0.05] [CTL vs. 48 h:
Notch3 (Huh7, MIHA), Notch4 (LX2, Huh7), P<0.05]. For Jagged1/2,
DLL1/3, Hey1 and Hes1, there were no regular trend changes found in
the three cell lines, although their expression was statistically
different at some time points.
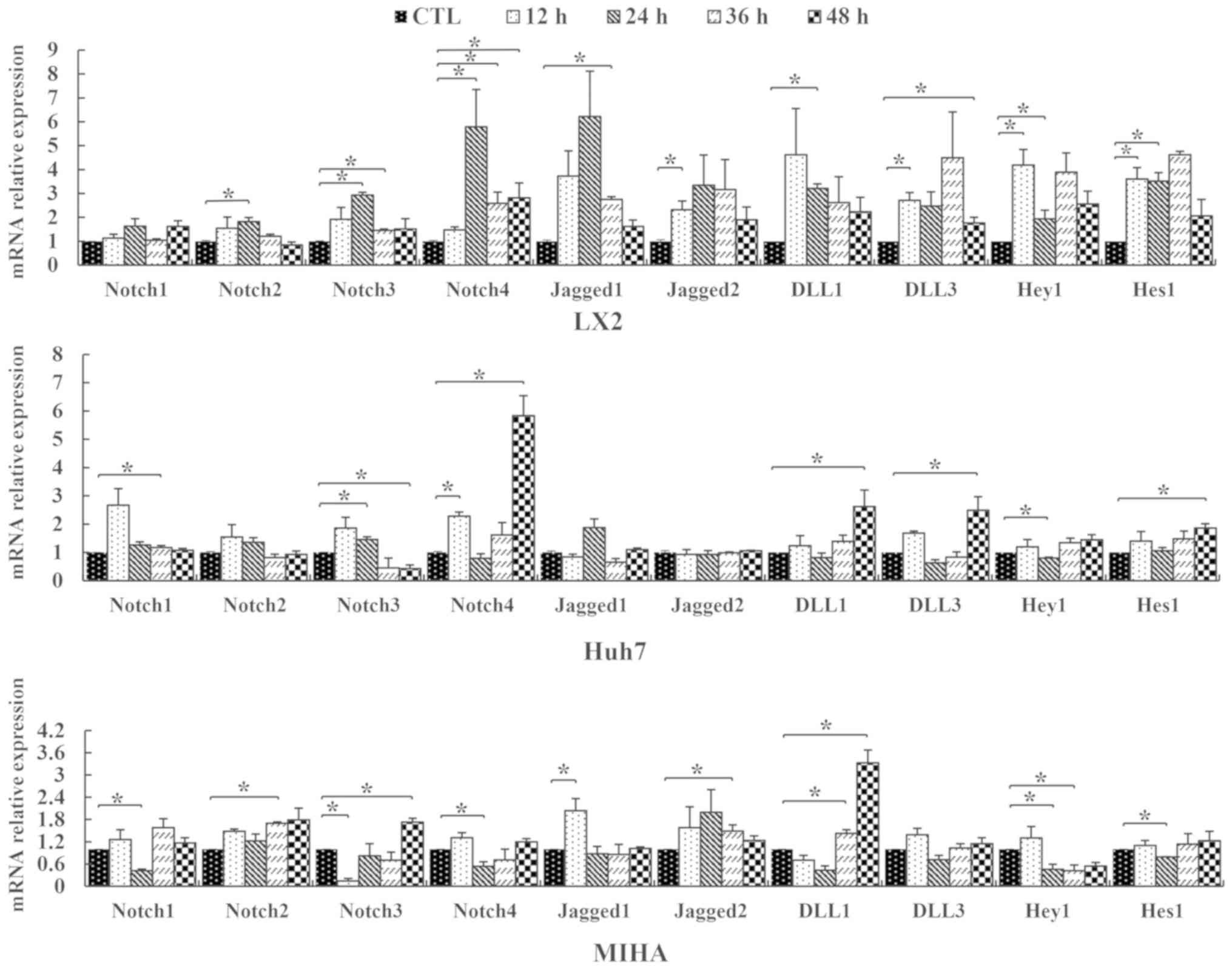 | Figure 3.Relative expression levels of Notch
genes in hepatic cell lines. LX2, Huh7 and MIHA cells were treated
without PA (CTL) or 0.25 mM palmitic acid, and RT-qPCR was
performed to assess expression levels of Notch1, −2, −3, −4,
Jagged1, Jagged2, DLL1, DLL3, Hey1 and Hes1 following various
durations of treatment (12, 24, 36 or 48 h). Data are presented as
the mean ± SD. *P<0.05. CTL, control; DLL, delta-like ligand;
Hey1, hes-related family bHLH transcription factor with YRPW motif
1; Hes1, hairy and enhancer of split-1. |
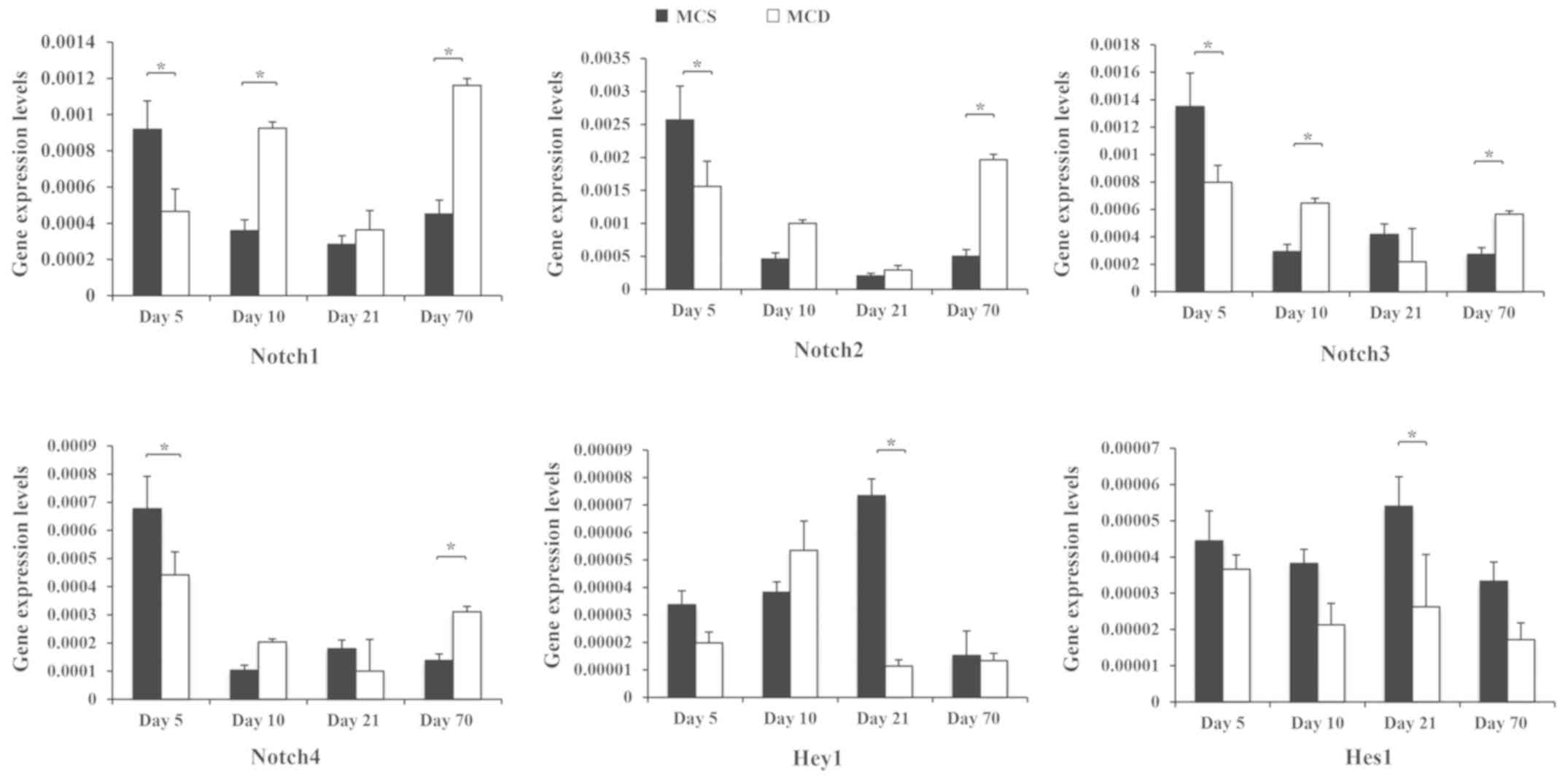
Abnormal expression of Notch signaling
pathway genes during NAFLD in vivo
C57BL/6 mice were fed an MCD diet (Fig. 4). The MCD model is mainly
characterized by steatosis and inflammatory responses of
hepatocytes at the third week, while liver fibrosis begins at week
8 (16,17). Notch1, −2, −3 and −4 mRNA decreased
significantly in MCD-fed mice at the early stage of NAFLD, compared
with MCS mice (day 5; P<0.05). This trend changed from day 10,
at which point, the expression levels of Notch1 and −3 were
significantly upregulated (P<0.05). At the final time point, the
expression of all Notch genes was significantly higher in MCD-fed
mice, compared with MCS controls (day 70; P<0.05).
Discussion
NAFLD is the main cause of chronic liver disease and
cirrhosis worldwide, irrespective of age (19). The spectrum of NAFLD includes
steatohepatitis, NASH and associated cirrhosis, and HCC (1,2).
Although steatosis presents minimal clinical manifestations, NASH
with lobular inflammation is regarded as a driving force in the
progression of NAFLD. The ‘multi-hit’ theory is the most widely
accepted theory accounting for the complex pathophysiology of NAFLD
(12,13,16).
The onset of NAFLD is characterized by the production of reactive
oxygen species, reduced levels of β-oxidation and increased
lipogenesis, followed by lipid accumulation in hepatocytes along
with cellular imflammation (20).
In addition, adaptive immunity, dysfunction of Notch signaling,
vitamin D deficiency and sleep deprivation have been reported as
additional factors promoting liver inflammation or injury during
NAFLD process (11,13,21–23).
In the present study, an NAFLD cell line was established and
verified using ORO staining. In order to examine the PA dosage and
treatment duration required in this model, cell proliferation and
migration were assessed in liver cells treated with PA over time.
Peak proliferation was observed at 36 h (LX2, MIHA) or 48 h (Huh7)
with 0.1 and 0.25 mM PA, respectively. In addition, increased cell
death was observed in LX2 cells when PA was 0.25 mM (≥48 h) and 0.5
mM (from 24 h onward). Thus, a concentration of 0.25 mM PA was used
in subsequent experiments, with a 36 to 48-h treatment time.
The Notch signaling pathway regulates downstream
genes and plays important roles in the physiological and
pathological processes of cell differentiation, proliferation,
apoptosis, embryonic development and tumor formation (24,25).
Previous studies suggested that the effects of Notch signaling vary
between different types of tumor, or within the same tumor during
different periods. In the liver, the Notch family is associated
with the onset and development of a number of liver diseases
(26,27). Aimaiti et al (28) demonstrated that LY450139, an
inhibitor of the Notch pathway, could decrease the expression of
Notch1 and myofibroblast markers in hepatic stellate cells via
transforming growth factor-β1, providing novel insight into
stellate cell activation by Notch signalling (28).
To investigate the role played by Notch signaling in
the liver, hepatic cell lines LX2, Huh7 and MIHA were used in the
present study to assess the expression of Notch genes. Changes in
the expression of Notch signaling genes occurred at different time
points in these cell lines. The levels of all Notch mRNA was
altered in all cell lines starting at 24 h of PA treatment. Notch4
was then upregulated at the latest time point in Huh7 cells.
Expression of Notch3 increased gradually in MIHA cells with PA
exposure time. In LX2 cells, Notch3 was upregulated at 12–24 h and
downregulated from 36 h; in Huh7 cells, it was upregulated at 12 h
but downregulated after 24 h. In the present animal experiment, the
expression levels of these genes increased at days 10 and 70.
According to the progress of NAFLD, the early stage primarily
comprises inflammation of hepatocytes and steatosis, whereas the
later stage involves liver fibrosis and/or cirrhosis (1,4,15).
Therefore, it is generally considered that 12–24 h in vitro
corresponds to days 5 and 10 in vivo, and 48 h in
vitro corresponds to day 70 in vivo. It may be
hypothesized that Notch1, Notch2 and Notch4 primarily participated
in inflammatory responses (early stage) and fibrosis (late stage)
during NALFD, while Notch3 might be primarily involved in steatosis
and inflammatory lesions.
Several studies have investigated the associations
between Notch signaling, liver fibrosis, cirrhosis and carcinoma,
both in humans and animal models. For instance, Zhang et al
(29) demonstrated that ring
finger protein 187 (RNF187) and Notch1 promoted invasion and
metastasis in HCC, and prognosis was poorer for patients who
exhibited Notch1-RNF187 activation. In patients with NASH, Notch
activity in hepatocytes was associated with severity and
responsiveness to treatment (30).
In Notch loss-of-function mouse models, hepatocyte-specific liver
inflammation and fibrosis are reduced, suggesting maladaptive
hepatocytic Notch response in NASH-associated liver fibrosis
(30). In the present study, the
dynamic effect of Notch genes during NAFLD development in
vivo was evaluated. Similar results were obtained in both MCD
mice and cell experiments. At the onset of NAFLD (day 10),
expression of Notch1, Notch2, Notch3 and Notch4 mRNA increased. In
the NASH period (week 3), Notch1 and Notch2 levels were higher in
the MCD group than in the MCS group, although these were not
significant. In a previous study, aggravation of fatty degeneration
and OS, liver fibrosis appeared in week 10 (17), and expression of these four genes'
mRNA rose again significantly. Thus, changes in Notch gene
expression may be related to the development of NAFLD. Notch genes,
and the associated upstream and downstream pathways, could
represent therapeutic targets for patients with NAFLD.
However, the present study has some limitations. The
MCD dietary model differed from the high-fat dietary (HFD) model,
and also from the human NASH microenvironment. MCD was not used to
simulate metabolic syndrome or IR, which often occur in the HFD
animal model and human patients with NAFLD. Future experiments
should focus on clinical samples or HFD animal models to verify the
conclusion that Notch expression levels change dynamically in NAFLD
and are differentially distributed across disease stages. Moreover,
while the same dynamic between Notch genes and NAFLD was observed
both in vitro and in vivo, the underlying mechanism
and potential associations between different Notch molecules
require extensive in-depth studies. Future studies should focus on
these aspects.
In conclusion, the present study demonstrated that
PA or MCD regulated the levels of Notch signaling genes during
NAFLD. The Notch family participated in the development of NAFLD.
These findings might provide a direction for molecular-targeted
therapy and early detection of NAFLD. However, additional studies
are needed to assess these possibilities.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
National Natural Science Foundation of China (grant nos. 81400610,
81400799 and 81470840), the Cross-Institute Research Fund of
Shanghai Jiao Tong University (grant no. YG2016MS72), The Project
Sponsored by the Scientific Research Foundation for the Returned
Overseas Chinese Scholars, State Education Ministry (grant no.
20144802) and The Shanghai Municipal Commission of Health and
Family Planning for Youth (grant no. 20134Y043).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WJD performed the experiments. JGF and LQ
conceptualized and designed the study. WJD, YWC and HBC performed
data analysis and interpretation. WJD and WJW drafted the
manuscript. WJW generated and revised the figures. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Ethics
Committee of Xin Hua Hospital affiliated to Shanghai Jiao Tong
University School of Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kleiner DE, Brunt EM, Wilson LA, Behling
C, Guy C, Contos M, Cummings O, Yeh M, Gill R, Chalasani N, et al:
Association of histologic disease activity with progression of
nonalcoholic fatty liver disease. JAMA Netw Open. 2:e19125652019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rhee EJ: Nonalcoholic fatty liver disease
and diabetes: An epidemiological perspective. Endocrinol Metab
(Seoul). 34:226–233. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gomaraschi M, Fracanzani AL, Dongiovanni
P, Pavanello C, Giorgio E, Da Dalt L, Norata GD, Calabresi L,
Consonni D, Lombardi R, et al: Lipid accumulation impairs lysosomal
acid lipase activity in hepatocytes: Evidence in NAFLD patients and
cell cultures. Biochim Biophys Acta Mol Cell Biol Lipids.
1864:1585232019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Wong GL, He FP, Sun J, Chan AW,
Yang J, Shu SS, Liang X, Tse YK, Fan XT, et al: Quantifying and
monitoring fibrosis in non-alcoholic fatty liver disease using
dual-photon microscopy. Gut. 69:1116–1126. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang MH, Chang KJ, Li B and Chen WS:
Arsenic trioxide suppresses tumor growth through antiangiogenesis
via notch signaling blockade in small-cell lung cancer. Biomed Res
Int. 2019:46472522019.PubMed/NCBI
|
|
6
|
Yamamoto S, Schulze KL and Bellen HJ:
Introduction to Notch signaling. Methods Mol Biol. 1187:1–14. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao W, Chen X and He M: Inhibition of the
Jagged/Notch pathway inhibits retinoblastoma cell proliferation via
suppressing the PI3K/Akt, Src, p38MAPK and Wnt/β-catenin signaling
pathways. Mol Med Rep. 10:453–458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aithal MGS and Rajeswari N: Bacoside a
induced Sub-G0 arrest and early apoptosis in human glioblastoma
cell line U-87 MG through Notch signaling pathway. Brain Tumor Res
Treat. 7:25–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jensen CH, Kosmina R, Rydén M, Baun C,
Hvidsten S, Andersen MS, Christensen LL, Gastaldelli A, Marraccini
P, Arner P, et al: The imprinted gene Delta like non-canonical
notch ligand 1 (Dlk1) associates with obesity and triggers insulin
resistance through inhibition of skeletal muscle glucose uptake.
EBioMedicine. 46:368–380. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang KC, Chuang PY, Yang TY, Huang TW and
Chang SF: Hyperglycemia inhibits osteoblastogenesis of rat bone
marrow stromal cells via activation of the Notch2 signaling
pathway. Int J Med Sci. 16:696–703. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Trovato FM, Martines GF, Brischetto D,
Catalano D, Musumeci G and Trovato GM: Fatty liver disease and
lifestyle in youngsters: Diet, food intake frequency, exercise,
sleep shortage and fashion. Liver Int. 36:427–433. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen C, Zhu Z, Mao Y, Xu Y, Du J, Tang X
and Cao H: HbA1c may contribute to the development of non-alcoholic
fatty liver disease even at normal-range levels. Biosci Rep.
40:BSR201939962020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Romeo S: Notch and Nonalcoholic fatty
liver and fibrosis. N Engl J Med. 380:681–683. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Niture S, Gyamfi MA, Kedir H, Arthur E,
Ressom H, Deep G and Kumar D: Serotonin induced hepatic steatosis
is associated with modulation of autophagy and notch signaling
pathway. Cell Commun Signal. 16:782018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamada T, Obata A, Kashiwagi Y, Rokugawa
T, Matsushima S, Hamada T, Watabe H and Abe K:
Gd-EOB-DTPA-enhanced-MR imaging in the inflammation stage of
nonalcoholic steatohepatitis (NASH) in mice. Magn Reson Imaging.
34:724–729. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Larter CZ, Yeh MM, Williams J,
Bell-Anderson KS and Farrell GC: MCD-induced steatohepatitis is
associated with hepatic adiponectin resistance and adipogenic
transformation of hepatocytes. J Hepatol. 49:407–416. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gu LY, Qiu LW, Chen XF, Lü L and Mei ZC:
Oleic acid induced hepatic steatosis is coupled with downregulation
of aquaporin 3 and upregulation of aquaporin 9 via activation of
p38 signaling. Horm Metab Res. 13:125–129. 2014.
|
|
19
|
Shakir AK, Suneja U, Short KR and Palle S:
Overview of pediatric nonalcoholic fatty liver disease: A guide for
general practitioners. J Okla State Med Assoc. 111:806–811.
2018.PubMed/NCBI
|
|
20
|
Lee J, Park JS and Roh YS: Molecular
insights into the role of mitochondria in non-alcoholic fatty liver
disease. Arch Pharm Res. 42:935–946. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sutti S and Albano E: Adaptive immunity:
An emerging player in the progression of NAFLD. Nat Rev
Gastroenterol Hepatol. 17:81–92. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Z, Thorne JL and Moore JB: Vitamin D
and nonalcoholic fatty liver disease. Curr Opin Clin Nutr Metab
Care. 22:449–458. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trovato FM, Castrogiovanni P, Szychlinska
MA, Purrello F and Musumeci G: Early effects of high-fat diet,
extra-virgin olive oil and vitamin D in a sedentary rat model of
non-alcoholic fatty liver disease. Histol Histopathol.
33:1201–1213. 2018.PubMed/NCBI
|
|
24
|
Aleĭnik AN and Kondakova IV: The Notch
signaling systemand oncogenesis. Vopr Onkol. 58:593–597. 2012.(In
Russian). PubMed/NCBI
|
|
25
|
Dang TP: Notch, apoptosis and cancer. Adv
Exp Med Biol. 727:199–209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Geisler F and Strazzabosco M: Emerging
roles of Notch signaling in liver disease. Hepatology. 61:382–392.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun L, Sun G, Yu Y and Coy DH: Is Notch
signaling a specific target in hepatocellular carcinoma? Anticancer
Agents Med Chem. 15:809–815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aimaiti Y, Yusufukadier M, Li W,
Tuerhongjiang T, Shadike A, Meiheriayi A, Gulisitan, Abudusalamu A,
Wang H, Tuerganaili A, et al: TGF-β1 signaling activates hepatic
stellate cells through Notch pathway. Cytotechnology. 71:881–891.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Chen J, Yong J, Qiao L, Xu L and
Liu C: An essential role of RNF187 in Notch1 mediated metastasis of
hepatocellular carcinoma. J Exp Clin Cancer Res. 38:3842019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu C, Kim K, Wang X, Bartolome A, Salomao
M, Dongiovanni P, Meroni M, Graham MJ, Yates KP, Diehl AM, et al:
Hepatocyte Notch activation induces liver fibrosis in nonalcoholic
steatohepatitis. Sci Transl Med. 10:eaat03442018. View Article : Google Scholar : PubMed/NCBI
|