Introduction
Acute myocardial infarction is the leading cause of
morbidity and mortality worldwide and >7 million new patients
have been diagnosed with acute myocardial infarction worldwide
annually (1,2). The primary therapeutic strategy for
acute myocardial ischemic injury is prompt and effective
reperfusion, including fibrinolytic therapy and primary
percutaneous coronary intervention; however, reperfusion itself
further induces myocardial injury (3). Moreover, the underlying mechanisms
responsible for myocardial ischemia/reperfusion (I/R) injury are
complicated and are yet to be completely elucidated; therefore,
myocardial I/R injury remains an ongoing unmet clinical issue.
Previous studies have reported that non-coding RNAs
serve an important role in regulating pathophysiological processes
of myocardial I/R injury, including cell differentiation,
proliferation, apoptosis, necrosis and autophagy (4–7).
Non-coding RNAs are functional RNAs that are transcribed from
non-coding DNAs and have important regulatory functions, although
they lack the potential of protein coding (8). Among non-coding RNAs, both microRNAs
(miRNAs/miRs) and long non-coding RNAs (lncRNAs) have gained
increased attention for their roles in myocardial I/R injury
(9). miRNAs are 20–25 nucleotides
in length and can inhibit the translation of mRNAs or induce
degradation of post-transcriptional RNAs (10). It has been revealed that a number
of miRNAs, such as hsa-miR-124-3 and hsa-miR-9-1, are involved in
myocardial I/R injury (11).
Furthermore, lncRNAs, ≥200 nucleotides in length, can regulate
miRNA functions by acting as endogenous sponges; and miRNAs have
also been shown to bind and regulate lncRNAs stability (12,13).
For example, the mitochondrial dynamic-related lncRNA has been
reported to act as an endogenous sponge by binding to miR-361,
downregulating its expression and inhibiting mitochondrial fission
and apoptosis in cardiomyocytes (14,15).
Despite these findings, the comprehensive expression profiles of
lncRNAs, miRNAs and mRNAs, as well as their individual potential
functions and the regulatory networks among them, are not well
characterized in myocardial I/R injury.
In the present study, the expression profiles of
lncRNAs, miRNAs and mRNAs in mice hearts after myocardial I/R were
investigated using microarray analysis. The potential functions of
these differentially expressed genes were analyzed via
bioinformatics, including Gene Ontology (GO) and pathway analyses.
Moreover, lncRNA-miRNA-mRNA regulatory networks were constructed
using competing endogenous RNA (ceRNA) analysis. The results
provided a series of novel areas for future research on lncRNAs,
miRNAs, mRNAs and their interactions in the process of myocardial
I/R injury, which is important to further understand the underlying
mechanisms and potential therapies for myocardial I/R injury.
Materials and methods
Animals
A total of 24 male C57BL/6J mice (age, 8 weeks,
20-25g) were purchased from the Department of Experimental Animals
of Shandong University (Jinan, China). All animal procedures were
in accordance with the US National Research Council Committee
Guidelines (16) and were approved
by the Animal Use and Care Committee of Shandong University. All
mice were housed in a temperature-controlled room under a 12-h
light-dark circadian cycle with temperature of 21.0±1.0°C, humidity
of 55.0±5.0% and had free access to standard chow and water.
The mice were randomly divided into sham-operation
(Sham) group (n=12) and I/R group (n=12). Among them, four mice in
Sham group and four mice in the I/R group were used for microarray
analysis. The other eight mice in each group were used to assess
the success of the I/R model and the results of microarray
analysis.
Mouse model of myocardial I/R
injury
The myocardial I/R model was performed in mice as
previously described (17). Mice
were anesthetized with 1.5% isoflurane mixed with 100% oxygen.
Following the skin incision, mice hearts were exposed via a left
thoracotomy in the third or fourth intercostal space. Ischemia was
performed by ligating the left anterior descending artery with an
8-0 silk ligature at 1.5–2.0 mm below the tip of the left auricle.
Mice in the Sham group underwent the same procedure without
occlusion of the left anterior descending artery. After 45 min of
occlusion, the suture was untied for reperfusion, and the chest
cavity and skin incision were closed. After 90 min of reperfusion,
mice were sacrificed by cervical dislocation and the heart was
quickly removed and cut into eight 1–2 mm thick slices from the
apex to the base. The peripheral region of infarct region was
selected as the area at risk (18). In addition, the two adjacent slices
below the site of occlusion were collected. One slice was used for
2, 3, 5-triphenyltetrazolium chloride (TTC) staining as a reference
for infarct area. Then, myocardial samples of 3×3×3 mm were
collected from the area at risk from the other slice based on the
reference of TTC staining. Blood samples (~1 ml) were obtained
using the retrobulbar technique (19). Blood samples were centrifuged at
1000 × g for 20 min at 4°C to collect plasma.
Detection of myocardial I/R
injury
In order to evaluate the success of the myocardial
I/R procedure, the infarct area was determined using staining with
1% TTC (cat. no. 17779; SigmaAldrich; Merck KGaA). Fresh myocardial
slices of 1–2 mm were incubated with 1% TTC at 37°C protected from
light for 30 min and then observed using an Olympus SZ61
stereoscopic microscope (Olympus Corporation) at ×10 magnification.
In addition, plasma lactate dehydrogenase (LDH) and creatine
kinase-MB (CK-MB) levels were detected using ELISA kits (cat. nos.
SEB864Ra and SEA479Ra, respectively; Cloud-Clone Corp.), according
to the manufacturer's instructions.
RNA extraction
RNA was extracted from myocardial tissue using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The quantification and quality of RNA were measured using
NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.), and the
integrity of RNA was further tested by 1% denaturing agarose gel
electrophoresis. For spectrophotometry, the optical density (OD)
A260/A280 ratio was ~2.0 for pure RNA (ratios between 1.8 and 2.1
are acceptable) (20) and the OD
A260/A230 ratio was >1.8.
Acquisition and analysis of microarray
data
The Arraystar Mouse lncRNA array v3.0 (8×60K;
Arraystar Inc.) was designed to profile lncRNAs and mRNAs in the
mouse genome. A total of 35,923 lncRNAs and 24,881 mRNA were
collected from authoritative data sources, including RefSeq
(https://www.ncbi.nlm.nih.gov/refseq/)
(21), University of California
Santa Cruz Known Genes (https://genome.ucsc.edu/) (22), Ensembl (http://ensemblgenomes.org/) (22), Fantom5 Cat, Gencode and
BIGTranscriptome (23,24). The 7th generation miRCURY LNA™
miRNA array (Exiqon; Qiagen) was designed to profile miRNA in the
mice genome, it contains 3,100 capture probes, covering all human,
mouse and rat miRNAs annotated in the miRBase 19.0 (http://microrna.sanger.ac.uk/) (25), as well as all viral miRNAs related
to these species.
The Agilent Feature Extraction software (version
11.0.1.1; Agilent Technologies, Inc.) was used to analyze acquired
microarray images. Quantile normalization and subsequent data
processing were performed using the GeneSpring GX v12.1 software
package (Agilent Technologies Inc.). After quantile normalization
of the raw data, lncRNAs, miRNAs and mRNAs, of which ≥4/8 samples
had flags in Present or Marginal (‘All Targets Value’) which
indicated good quality of data, were selected for further data
analysis. Differentially expressed lncRNAs, miRNAs and mRNAs with
statistical significance between the two groups were identified via
P-value filtering (P<0.05) and fold change filtering (fold
change >2). Hierarchical clustering and combined analysis were
performed using the Multiple Experiment Viewer 4.9.0 (http://mev.tm4.org/) (26).
Reverse transcription-quantitative
(RT-q)PCR
RT-qPCR was performed to assess the results of the
microarray analysis. Total RNAs, extracted from four samples in the
Sham group and four samples in the I/R group using miRcute miRNA
isolation kit (cat. no. DP501; Tiangen Biotech. Co.) for miRNA and
TRIzol (cat. no. DP405; Tiangen Biotech. Co.) for mRNA and lncRNA,
were reverse transcribed with a High-Capacity RNA-to-cDNATM kit
(cat. no. 4387406; Invitrogen; Thermo Fisher Scientific, Inc.) at
42°C for 60 min followed by 95°C for 3 min. RT-qPCR was performed
with the IQ-SYBR® Green Supermix in a CFX96 apparatus
(Bio-Rad Laboratories, Inc.). The following thermocycling
conditions were used: initial denaturation at 95°C for 10 min,
followed by 40 cycles at 95°C for 10 sec and 60°C for 60 sec, and
final extension at 72°C for 5 min. Both β-actin and U6 small
nuclear (sn)RNA were used as housekeeping genes for normalization.
The expression levels of lncRNA, miRNA and mRNA were measured in
terms of CT and then normalized to β-actin and U6 snRNA using the
2−ΔΔCq method (27).
The primers used for RT-qPCR were as follows: lncRNA-AK087886
forward, 5′-ACTTACGTCTGCGACCACG-3′ and reverse,
3′-GGCGGAACAAACTTCAACCT-5′; miRNA-30e-3p forward,
5′-GCCTTTCAGTCGGATGTTTACAGC-3′ and reverse,
3′-GCATGTTGTCACAGCTTGTGT-5′; mRNA-Olfml1 forward,
5′-GCCGAGCACCCATCTATCAA-3′ and reverse, 3′-GCCACCGGAACTGTAGACAA-5′;
U6 snRNA forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′; β-actin forward
5′-CTGTCTGGCGGCACCACCAT-3′ and reverse,
3′-GCAACTAAGTCATAGTCCGC-5′.
GO and pathway analysis
The GO database (http://www.geneontology.org) was used to identify
significantly overrepresented ‘biological processes’. The pathway
analysis was based on the Kyoto Encyclopedia of Genes and Genomes
database (KEGG; http://www.genome.ad.jp/kegg/), which was performed to
evaluate the potential module function.
ceRNA network analysis
lncRNAs were selected from the top ten up- and
downregulated lncRNAs for ceRNA analysis with rigorous standards,
including fold change >2, P<0.05, raw intensity >2,000,
exon sense overlapping and natural antisense. Then, the
lncRNA-miRNA-mRNA ceRNA network was constructed based on the ceRNA
hypothesis that all types of RNA transcripts, such as mRNAs,
transcribed pseudogenes and lncRNAs, could communicate via a new
‘language’ mediated by miRNA binding sites (‘microRNA response
elements’) (28). To construct the
ceRNA network, the interaction between the miRNAs and lncRNAs was
predicted using Arraystar miRNA target prediction software
(29) based on TargetScan 7.2
(http://www.targetscan.org/vert_72/)
and miRanda (http://www.microrna.org/microrna/home.do) (30–32).
High-confidence miRNA-lncRNA pairs had a TargetScan cumulative
weighted context score <-0.3, total context score <-0.3,
miRanda energy score <-10 and structure score >140.
Interactions between miRNAs and mRNAs were predicted using
TargetScan and miRDB (http://www.mirdb.org/) (miRDB score >70).
Hypergeometric distribution was performed for each ceRNA pair
separately (P<0.05) and the interactions were then combined with
the microarray data to construct the ceRNA relationship. The ceRNA
network was visualized using Cytoscape v2.8.3 software (https://cytoscape.org/).
Statistical analysis
The data relating to infarct size, LDH, CK-MB and
RT-qPCR were presented as the mean ± SEM from at least three
independent experiments. An unpaired Student's t-test was used to
analyze the data between the two groups. P<0.05 was considered
to indicate a statistically significant difference. Statistical
analysis was performed using GraphPad Prism 6 software (GraphPad
Software, Inc.).
Results
Infarct size, and LDH and CK-MB levels
in myocardial I/R injury
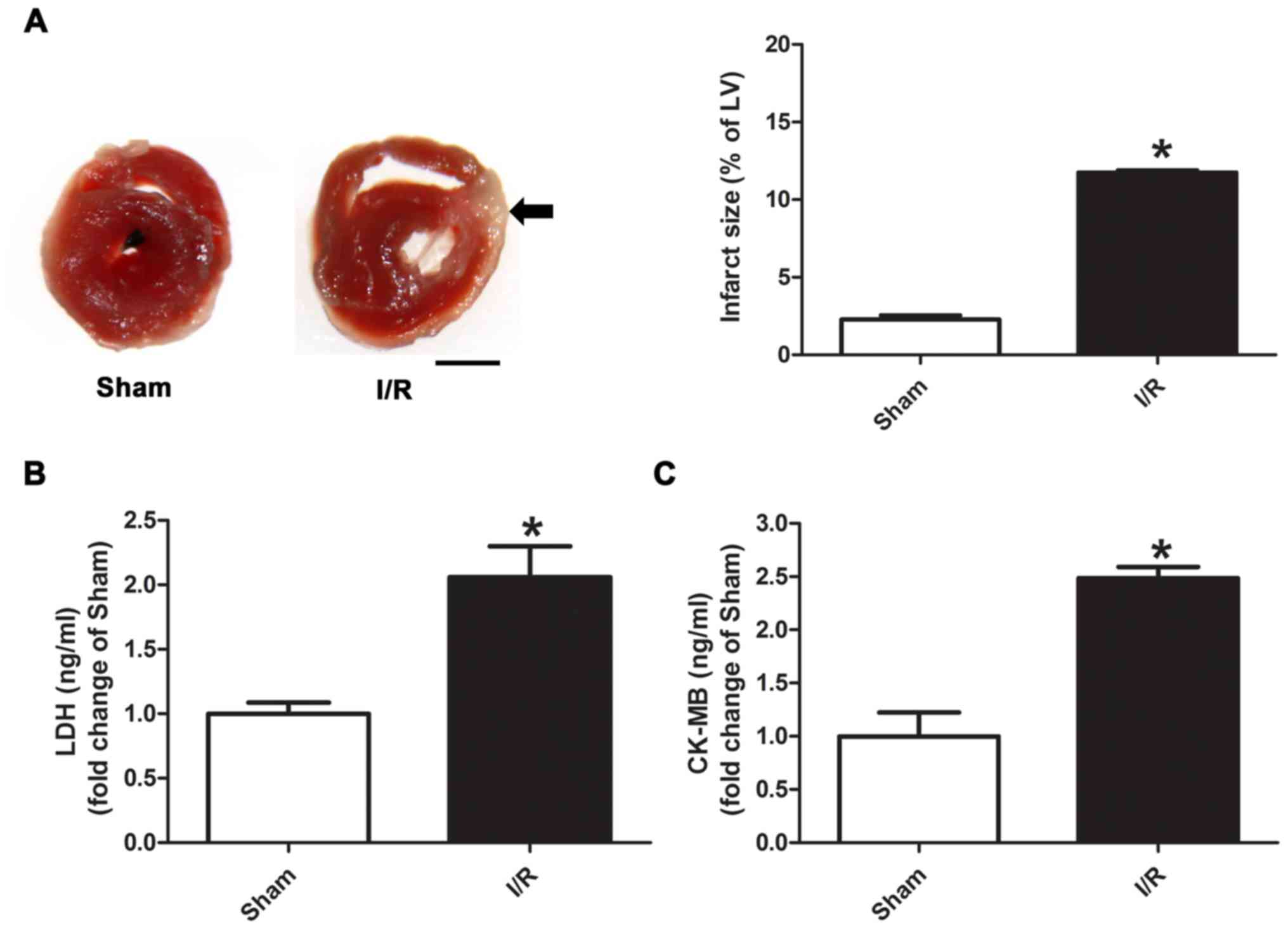
TTC staining was used to evaluate infarct size (% of
left ventricle), which was significantly higher in the myocardial
I/R group compared with the Sham group (Fig. 1A; 11.7 vs. 2.3%). It was also
identified that the distal area from the site of occlusion was more
severe than the proximal area after I/R injury. Moreover, the
levels of plasma LDH and CK-MB were significantly increased in the
myocardial I/R group compared with the Sham group (Fig. 1B and C).
Differentially expressed RNAs in
myocardial I/R injury
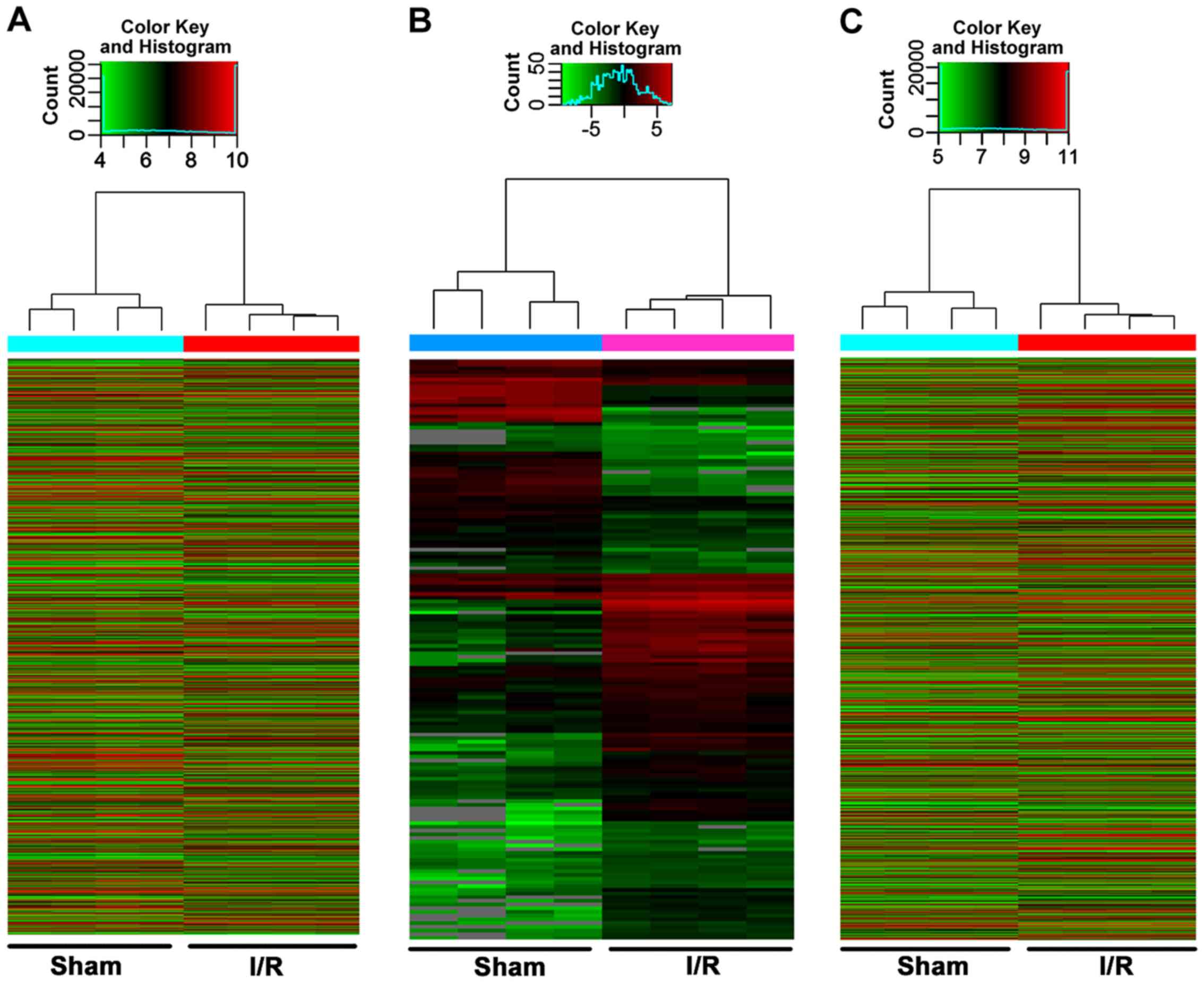
The integrity of RNAs for microarray analysis was
assessed using NanoDrop ND-1000 (Table SI) and denaturing agarose gel
electrophoresis (Fig. S1). Using
microarray analysis, 14,366 differentially expressed lncRNAs were
identified in the myocardial I/R group compared with the Sham group
(fold change >2 and P<0.05) (data not shown). Among them,
9,259 lncRNAs (64.45%) were upregulated and 5,107 lncRNAs (35.55%)
were downregulated. A total of 151 miRNAs were also differentially
expressed; 98 (64.90%) were upregulated and 53 (35.10%) were
downregulated. In addition, 9,377 mRNAs were differentially
expressed; 6,472 (69.02%) were upregulated and 2,905 (30.98%) were
downregulated. Hierarchical clustering analysis identified
significant changes in the cardiac expression of lncRNAs, miRNAs
and mRNAs in the myocardial I/R group compared with the Sham group
(Fig. 2A-C). The top ten up- and
downregulated lncRNAs, miRNAs and mRNA are presented in Tables SII–SIV, respectively.
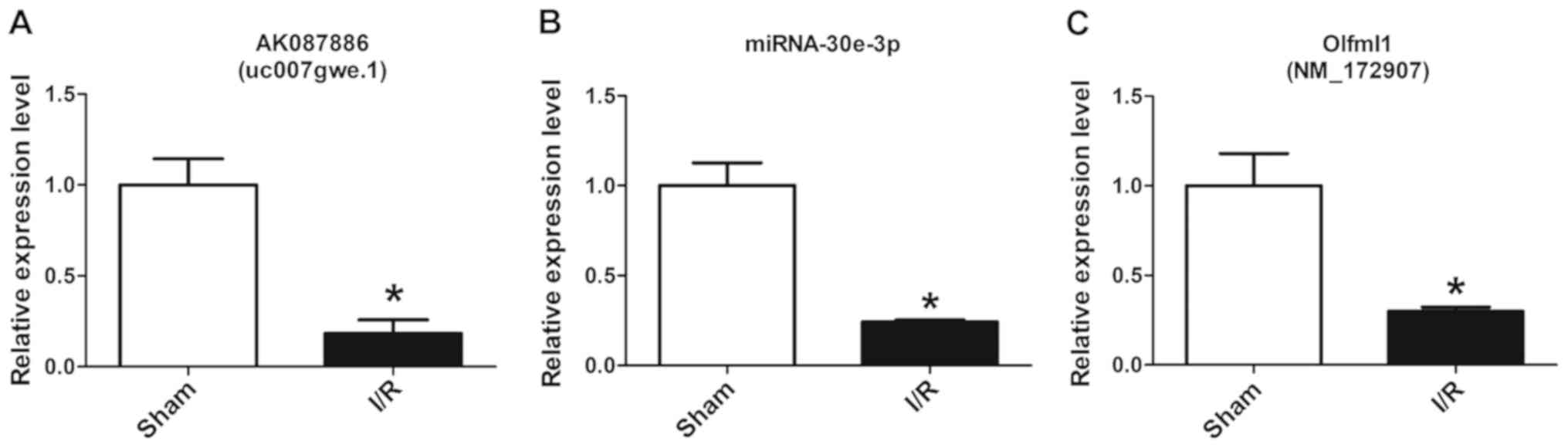
The microarray data was assessed by randomly
detecting the gene expression levels of lncRNA-AK087886,
miRNA-30e-3p and mRNA-Olfml1 using RT-qPCR, which were in the top
ten up- and downregulated lncRNAs, miRNAs and mRNAs, and the
results demonstrated that the expression levels in the I/R group
were significantly lower compared with those of the Sham group
(Fig. 3).
GO and pathway analysis
The up- and downregulated genes were analyzed
individually according to GO analysis, including ‘Biological
Process’, ‘Molecular Function’ and ‘Cellular Component’. The top
ten molecular functions associated with up- and downregulated genes
according to the enrichment score [-log10 (P-value)] are listed in
Table SV. The upregulated genes
were mainly involved in ‘guanosine diphosphate binding’, ‘RNA
polymerase II carboxy-terminal domain kinase activity’,
‘TATA-binding protein-class protein binding’, ‘NAD+
binding’ and ‘protein phosphatase type 2A regulator activity’. The
downregulated genes were mainly involved in ‘proline-rich region
binding’, ‘sodium ion binding’, ‘armadillo repeat domain binding’,
‘alkali metal ion binding’ and ‘peroxisome proliferator activated
receptor binding’.
Pathway analysis was performed to identify the
potential module function of the differentially expressed genes in
myocardial I/R injury, according to KEGG. The pathway enrichment
analysis demonstrated that these upregulated genes were involved in
104 pathways and downregulated genes were involved in 29 pathways.
The top ten module functions associated with up- and downregulated
genes according to the enrichment score [-log10 (P-value)] are
listed in Table SVI. Upregulated
genes mainly participated in pathways related to ‘Alzheimer's
disease’, ‘sphingolipid signaling pathway’, ‘protein processing in
endoplasmic reticulum’, ‘non-alcoholic fatty liver disease’ and
‘Parkinson's disease’. Downregulated genes mainly participated in
pathways of ‘endocytosis’, ‘bacterial invasion of epithelial
cells’, ‘type II diabetes mellitus’, ‘oxytocin signaling pathway’
and ‘arrhythmogenic right ventricular cardiomyopathy’.
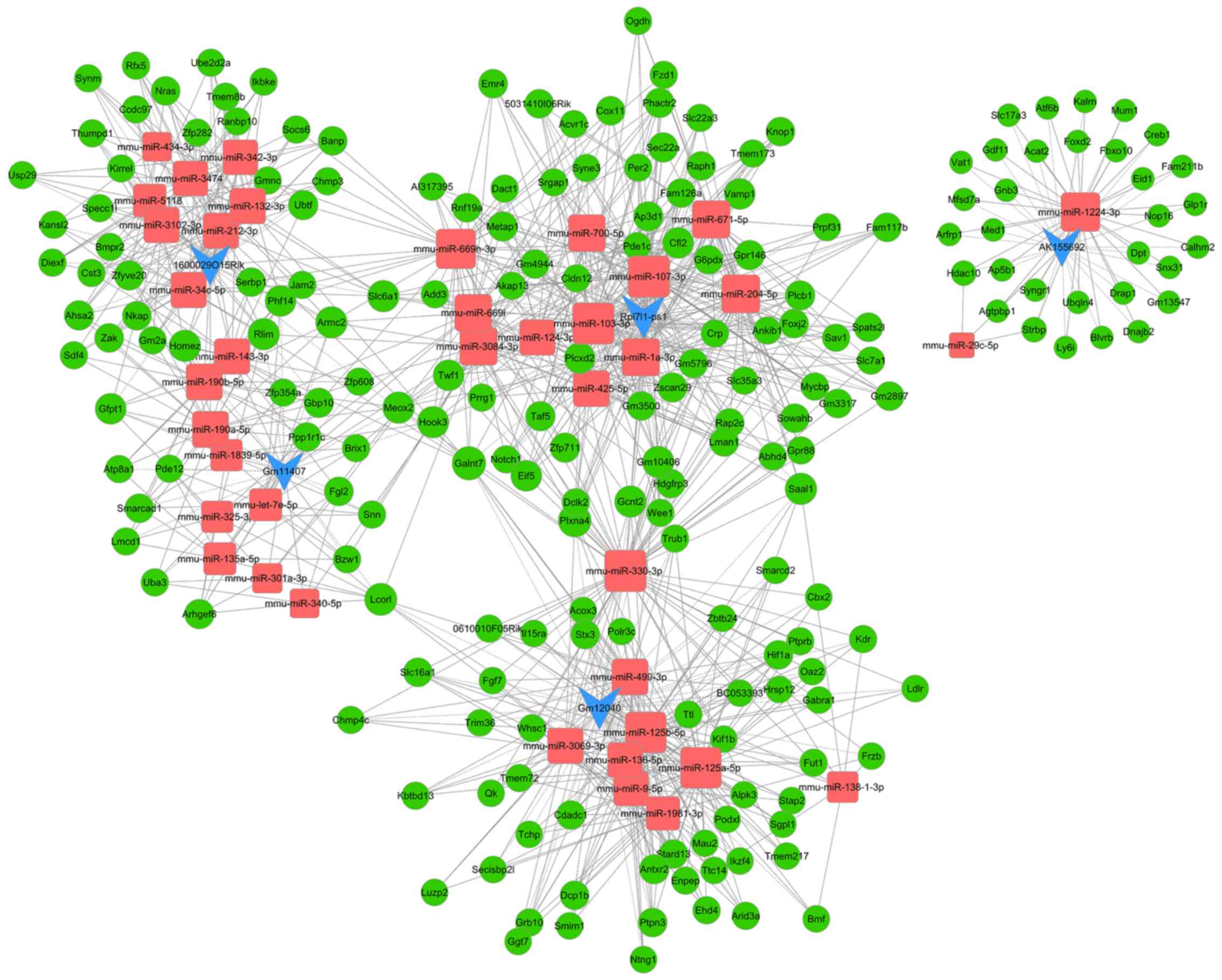
ceRNAs regulatory network
analysis
In total, five lncRNAs, including Gm11407, AK155692,
1600029O15Rik, Rpl7l1-ps1 and Gm12040, were selected from the top
ten up- and downregulated lncRNAs for ceRNA analysis after they
were screened using rigorous standards, including fold change,
P-value, raw intensity, exon sense overlapping and natural
antisense. Then, the lncRNA-miRNA-mRNA ceRNA regulatory network was
constructed, which included five lncRNAs, 38 miRNAs and 196 mRNAs.
The lncRNA-miRNA-mRNA regulatory networks included 239 nodes and
678 edges (Fig. 4). The higher the
number of nodes and edges these RNAs possessed, the increased
significant effects they may have in myocardial I/R injury. Blue,
red and green nodes represented lncRNAs, miRNAs and mRNAs,
respectively. The edges between the nodes represented the
interactions between RNAs. Furthermore, the nodes of the top ten
miRNAs and mRNAs with the highest degree are presented in Table SVII. The nodes with the highest
degrees in lncRNAs, miRNAs and mRNAs were Rpl7l1-ps1 (degree=12,
indicating 12 edges or targets were directly connected to
Rpl7l1-ps1), mmu-miR-330-3p (degree=67) and plexin A4 (Plxna4;
degree=7), respectively. According to the rank scores of ceRNA5
pathways among the top 5 differentially expressed lncRNA and miRNA
and mRNA (33), the following
ceRNA network were chosen: Gm12040-mmu-miR-125a-5p-decapping mRNA
1B (Dcp1b), Rpl7l1-ps1-mmu-miR-124-3p-G protein-coupled receptor
146 (Gpr146), Gm11407-mmu-miR-190a-5p-homeobox and leucine zipper
encoding (HOMEZ), 1600029O15Rik-mmu-miR-132-3p-HOMEZ and
AK155692-mmu-miR-1224-3p-activating transcription factor 6β
(Atf6b).
Discussion
In the present study, the expression profiles of
lncRNAs, miRNAs and mRNAs were investigated in mice hearts after
I/R injury. It was found that, compared with Sham hearts, 14,366
lncRNAs, 151 miRNAs and 9,377 mRNAs were differentially expressed
in the area at risk of I/R hearts. The GO and KEGG pathway analyses
demonstrated the functions and pathways associated with these
differentially expressed genes in myocardial I/R injury. Moreover,
the interactions between lncRNA, miRNA and mRNA were constructed
using ceRNA network analysis, which included five lncRNAs, 38
miRNAs and 196 mRNAs. The lncRNAs, miRNAs and mRNAs with the
highest degree were lncRNA Rpl7l1-ps1, miRNA mmu-miR-330-3p and
mRNA Plxna4. These interactions were indicated by
Gm12040-mmu-miR-125a-5p-Dcp1b, Rpl7l1-ps1-mmu-miR-124-3p-Gpr146,
Gm11407-mmu-miR-190a-5p-HOMEZ, 1600029O15Rik-mmu-miR-132-3p-HOMEZ
and AK155692-mmu-miR-1224-3p-Atf6b. Thus, the present study
provides a novel insight into the regulatory mechanisms and
possible functions of a number of lncRNAs, miRNAs and mRNAs in
myocardial I/R injury.
In the present study, the differently expressed
lncRNAs, miRNAs and mRNAs after myocardial I/R injury in mice were
analyzed, providing valuable information to explore the potential
roles of RNAs in myocardial I/R injury in future studies.
Furthermore, a number of novel lncRNAs, miRNAs and mRNAs, which had
not been reported in previous studies regarding myocardial I/R
injury, were screened, such as Gm11407, AK155692, 1600029O15Rik,
Rpl7l1-ps1, Gm12040, miR-3068, miR-5624, Gm20594, Ndufb6 and Fdft1.
In addition, the role of numerous RNAs screened by the present
study have not been comprehensively examined in myocardial I/R
injury. Among them, miR-33, as the top upregulated miRNA in the
present study, has been revealed to suppress cardiac remodeling via
regulation of adaptive fibrotic response in patients with heart
failure (34). However, whether
miR-33 has a role in inhibiting cardiac remodeling following
myocardial I/R injury requires further investigation. It was also
previously reported that the expression of miR-378a, which was
indicated to be downregulated in the present study, is altered at
an earlier stage in kidney I/R injury compared with kidney injury
molecule-1 (35). As biomarkers
for the diagnosis of diseases, RNAs are important in early
detection compared with protein molecules (36). However, whether miR-378a is useful
for the early diagnosis of acute myocardial infarction is yet to be
elucidated. Pyruvate dehydrogenase kinase 4 (Pdk4), the mRNA that
was significantly increased in the present study, has been shown to
promote metabolic remodeling of cardiomyocytes in late pregnancy
(37), but, there is currently no
evidence that Pdk4 can improve energy metabolism after myocardial
I/R injury. It has been revealed that farnesyl-diphosphate
farnesyltransferase 1 (Fdft1), one of the top ten downregulated
mRNAs, is closely associated with lipid metabolism in
obesity-related type 2 diabetes and cardiovascular disease
(38,39). However, the relationship between
Fdft1 and lipid metabolism disorder in myocardial I/R injury has
not been previously reported. Therefore, the expression profiles of
differentially expressed lncRNAs, miRNAs and mRNAs identified in
the present study may provide potential targets for future research
regarding myocardial I/R injury.
To the best of our knowledge, the present study was
the first to predict the lncRNA-miRNA-mRNA ceRNA interactions in
myocardial I/R injury. It was found that Rpl7l1-ps1, mmu-miR-330-3p
and Plxna4 were the lncRNA, miRNA and mRNA with the highest degree
nodes, respectively, indicating their potential role in myocardial
I/R injury. Currently, there is no report on the role of Rpl7l1-ps1
in any disease. Previous studies on miR-330-3p have shown that it
is involved in tumors, such as lung cancer, gastric cancer and
pancreatic cancer, with the exception of one study, which revealed
that miR-330-3p enhanced hypertrophic response of cardiomyocytes
(40–43). Combined with the present results,
these findings indicate that miR-330-3p may participate in
myocardial I/R injury by interacting with lncRNAs and mRNAs.
Plxnax4 has been reported to serve a role in Alzheimer's disease
and modulation of tau phosphorylation, however it has not been
examined in myocardial I/R injury or other cardiac diseases, and
thus further investigation is required (44). The lncRNA-miRNA-mRNA ceRNA
interactions were demonstrated by Gm12040-mmu-miR-125a-5p-Dcp1b,
Rpl7l1-ps1-mmu-miR-124-3p-Gpr146, Gm11407-mmu-miR-190a-5p-HOMEZ,
1600029O15Rik-mmu-miR-132-3p-HOMEZ and
AK155692-mmu-miR-1224-3p-Atf6b in the present study. Furthermore, a
previous study suggested that Gpr146 was involved in constructing
the C-peptide signaling complex in microvascular diseases of
diabetes, which could attenuate cardiac contractile dysfunction in
myocardial I/R injury (45,46).
Thus, in line with the present result, lncRNA Rpl7l1 may regulate
Gpr146 via mmu-miR-124-3p and participate in the cardiac protective
mechanisms in myocardial I/R injury. It has been shown that Atf6b
can regulate the pressure overload-induced cardiac hypertrophic
response, which indicates the possible role of
AK155692-mmu-miR-1224-3p-Atf6b in myocardial I/R injury (47). However, to the best of our
knowledge, there are currently no reports on Dcp1b and HOMEZ, and
additional studies are required to predict the functions of
Gm12040-mmu-miR-125a-5p-Dcp1b, Gm11407-mmu-miR-190a-5p-HOMEZ or
1600029O15Rik-mmu-miR-132-3p-HOMEZ.
Previous studies have investigated the expression
profiles of lncRNAs, mRNAs and miRNAs in myocardial I/R injury, the
majority of which have focused on the interactions between two
types of RNA. For example, Liu et al (18) identified 151 differentially
expressed lncRNAs and 110 mRNAs in the infarct region after
myocardial I/R injury in mice, and function analysis revealed that
these differentially expressed transcripts were associated with
‘immune response’, ‘spermine catabolic process’, ‘taxis’,
‘cytokine-cytokine receptor interaction’, ‘the chemokine signaling
pathway’ and ‘nucleotide oligomerization domain-like receptor
signaling pathway’. Moreover, Wu et al (48) reported that 168 lncRNAs and 126
mRNAs were differentially expressed after myocardial infarction in
mice, and these differentially expressed genes were enriched in 41
signaling pathways, including ‘complement and coagulation
cascades’, ‘cytokine-cytokine receptor interaction’ and ‘chemokine
signaling pathways’. Gao et al (49) also demonstrated that 1,197 lncRNAs
and 2,066 mRNAs were upregulated, whereas 1,403 lncRNAs and 2,871
mRNAs were downregulated after ischemic heart failure in rats,
which were closely related to the ‘mitogen-activated protein
kinase-signaling pathway’, ‘T cell receptor signaling pathway’,
‘prion diseases’ and ‘cell adhesion molecules’. In addition, Liu
et al (50) investigated
the time-course of lncRNA and mRNA expression in the peripheral
blood of patients with acute ST-segment elevation myocardial
infarction and percutaneous coronary intervention. These authors
reported 135 RNAs that were significantly associated with
myocardial I/R injury, and that mRNA SH2 domain containing 3C and
general transcription factor IIH subunit 4 may be the most
responsive transcriptional regulators in the early-phase of
myocardial I/R injury (50). The
present study expanded on these findings by investigating the
interactions between three types of RNA, and in line with previous
studies, provided a potential research area for future studies on
non-coding RNAs in myocardial I/R injury.
The present results provided information on RNAs
that could be used for the diagnosis and prognosis of acute
myocardial infarction. lncRNAs and miRNAs have been identified as
possible biomarkers for diagnosis and prognosis of diseases,
especially in cancer (51,52). A previous study also revealed that
lncRNA HOX transcript antisense RNA acted as a potential biomarker
for the prognosis of patients with squamous cell carcinoma of the
head and neck (53). Moreover,
Zhong et al (54) reported
that lncRNAs, including taurine upregulated 1, SPRY4 intronic
transcript 1 and hepatocellular carcinoma upregulated lncRNA, may
serve as moderate predictors of survival in human cancer. However,
biomarkers for the diagnosis of myocardial infarction are still
mainly focused on protein molecules, including plasma troponin and
CK-MB (55). High-sensitive
cardiac troponin I has been used to improve the sensitivity of the
diagnosis of acute myocardial infarction, but at the cost of lower
specificity (56). Therefore,
identifying novel biomarkers for the accurate diagnosis of
myocardial infarction is of great significance. Currently some
miRNAs, such as miR-1, miR-499 and miR-133, have been revealed as
biomarkers for the diagnosis of acute myocardial infarction
(57). miR-208a, expressed
specifically in cardiomyocytes, also has a high sensitivity and
specificity for myocardial infarction diagnosis (58). However, additional lncRNAs and
miRNAs that could be used as potential biomarkers for the diagnosis
and prognosis of acute myocardial infarction are yet to be
elucidated.
In conclusion, the present results demonstrated the
expression profiles of differentially expressed lncRNAs, miRNAs and
mRNAs, as well as predicted their functions and potential pathways
in myocardial I/R injury. Furthermore, the lncRNA-miRNA-mRNA ceRNA
interaction networks, including five lncRNAs, 38 miRNAs and 196
mRNAs, were predicted in myocardial I/R injury. However, a
limitation of the present study was that intervention experiments
of RNAs were not included, and further verifications on the
potential functions of these RNAs in myocardial I/R injury were not
performed.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key
Research and Development Program of China (grant nos.
2017YFC0908700 and 2017YFC0908703), National Natural Science
Foundation of China (grant nos. 81772036, 81671952, 81873950,
81873953, 81570401 and 81571934), National Science and Technology
Fundamental Resources Investigation Project (grant nos.
2018FY100600 and 2018FY100602), Taishan Pandeng Scholar Program of
Shandong Province (grant no. tspd20181220), Taishan Young Scholar
Program of Shandong Province (grant nos. tsqn20161065 and
tsqn201812129), Key Research and Development Program of Shandong
Province (grant no. 2018GSF118003) and the Fundamental Research
Funds of Shandong University (grant no. 2018JC011).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC, PG, RZ, JW and BL contributed to the conception
and design. RZ, BL, WW, XF and BZ performed the experiments. RZ,
BL, JW, QY, MX and FX contributed to the acquisition, analysis and
interpretation of data, drafted the article or revised it
critically for important intellectual content. All authors reviewed
and approved the final version.
Ethics approval and consent to
participate
All animal procedures were approved by the Animal
Use and Care Committee of Shandong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reed GW, Rossi JE and Cannon CP: Acute
myocardial infarction. Lancet. 389:197–210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Virani SS, Alonso A, Benjamin EJ,
Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR,
Cheng S, Delling FN, et al American Heart Association Council on
Epidemiology and Prevention Statistics Committee and Stroke
Statistics Subcommittee, : Heart Disease and Stroke Statistics-2020
Update: A Report From the American Heart Association. Circulation.
141:e139–e596. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yellon DM and Hausenloy DJ: Myocardial
reperfusion injury. N Engl J Med. 357:1121–1135. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y: The novel regulatory role of
lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med.
22:5768–5775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma M, Hui J, Zhang QY, Zhu Y, He Y and Liu
XJ: Long non-coding RNA nuclear-enriched abundant transcript 1
inhibition blunts myocardial ischemia reperfusion injury via
autophagic flux arrest and apoptosis in streptozotocin-induced
diabetic rats. Atherosclerosis. 277:113–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang W, Li Y and Wang P: Long non-coding
RNA-ROR aggravates myocardial ischemia/reperfusion injury. Braz J
Med Biol Res. 51:e65552018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren L, Chen S, Liu W, Hou P, Sun W and Yan
H: Downregulation of long non-coding RNA nuclear enriched abundant
transcript 1 promotes cell proliferation and inhibits cell
apoptosis by targeting miR-193a in myocardial ischemia/reperfusion
injury. BMC Cardiovasc Disord. 19:1922019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Archer K, Broskova Z, Bayoumi AS, Teoh JP,
Davila A, Tang Y, Su H and Kim IM: Long Non-Coding RNAs as Master
Regulators in Cardiovascular Diseases. Int J Mol Sci.
16:23651–23667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei C, Luo T, Zou S, Zhou X, Shen W, Ji X,
Li Q and Wu A: Differentially expressed lncRNAs and miRNAs with
associated ceRNA networks in aged mice with postoperative cognitive
dysfunction. Oncotarget. 8:55901–55914. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, He XN, Li C, Gong L and Liu M:
Identification of Candidate Genes and MicroRNAs for Acute
Myocardial Infarction by Weighted Gene Coexpression Network
Analysis. BioMed Res Int. 2019:57426082019.PubMed/NCBI
|
|
12
|
Blythe AJ, Fox AH and Bond CS: The ins and
outs of lncRNA structure: How, why and what comes next? Biochim
Biophys Acta. 1859:46–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li S, Chen X, Liu X, Yu Y, Pan H, Haak R,
Schmidt J, Ziebolz D and Schmalz G: Complex integrated analysis of
lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol.
73:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li N, Ponnusamy M, Li MP, Wang K and Li
PF: The Role of MicroRNA and LncRNA-MicroRNA Interactions in
Regulating Ischemic Heart Disease. J Cardiovasc Pharmacol Ther.
22:105–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang K, Sun T, Li N, Wang Y, Wang JX, Zhou
LY, Long B, Liu CY, Liu F and Li PF: MDRL lncRNA regulates the
processing of miR-484 primary transcript by targeting miR-361. PLoS
Genet. 10:e10044672014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals, . Guide for the Care and Use of Laboratory Animals. The
National Academies Collection: Reports funded by National
Institutes of Health. National Academies Press. (Washington, DC).
2011."ref-label" rowspan="1" colspan="1">
17
|
Xue L, Yang F, Han Z, Cui S, Dai S, Xu F,
Zhang C, Wang X, Pang J, Pan C, et al: ALDH2 mediates the
dose-response protection of chronic ethanol against endothelial
senescence through SIRT1/p53 pathway. Biochem Biophys Res Commun.
504:777–783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu Y, Li G, Lu H, Li W, Li X, Liu H, Li
X, Li T and Yu B: Expression profiling and ontology analysis of
long noncoding RNAs in post-ischemic heart and their implied roles
in ischemia/reperfusion injury. Gene. 543:15–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shimizu T, Suzuki S, Sato A, Nakamura Y,
Ikeda K, Saitoh S, Misaka S, Shishido T, Kubota I and Takeishi Y:
Cardio-protective effects of pentraxin 3 produced from bone
marrow-derived cells against ischemia/reperfusion injury. J Mol
Cell Cardiol. 89B:B306–B313. 2015. View Article : Google Scholar
|
|
20
|
Imbeaud S, Graudens E, Boulanger V, Barlet
X, Zaborski P, Eveno E, Mueller O, Schroeder A and Auffray C:
Towards standardization of RNA quality assessment using
user-independent classifiers of microcapillary electrophoresis
traces. Nucleic Acids Res. 33:e562005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
O'Leary NA, Wright MW, Brister JR, Ciufo
S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B,
Ako-Adjei D, et al: Reference sequence (RefSeq) database at NCBI:
Current status, taxonomic expansion, and functional annotation.
Nucleic Acids Res 44D. D733–D745. 2016. View Article : Google Scholar
|
|
22
|
Singh A, Shannon CP, Kim YW, Yang CX,
Balshaw R, Cohen Freue GV, Gauvreau GM, FitzGerald JM, Boulet LP,
O'Byrne PM, et al: Novel Blood-based Transcriptional Biomarker
Panels Predict the Late-Phase Asthmatic Response. Am J Respir Crit
Care Med. 197:450–462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Haas BJ, Papanicolaou A, Yassour M,
Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber
M, et al: De novo transcript sequence reconstruction from RNA-seq
using the Trinity platform for reference generation and analysis.
Nat Protoc. 8:1494–1512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and
ER− breast cancer cell lines. J Cell Mol Med.
19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guzzi PH, Tradigo G and Veltri P: Using
miRNA-Analyzer for the Analysis of miRNA Data. Microarrays (Basel).
5:52016.
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bo H, Liu Z, Tang R, Gong G, Wang X, Zhang
H, Zhu F, Zhou D, Zhu W, Tan Y, et al: Testicular biopsies
microarray analysis reveals circRNAs are involved in the
pathogenesis of non-obstructive azoospermia. Aging (Albany NY).
12:2610–2625. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
31
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila.
Genome Biol. 5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res 42D. D92–D97. 2014. View Article : Google Scholar
|
|
33
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X and Fernández-Hernando C: miR-33
Regulation of Adaptive Fibrotic Response in Cardiac Remodeling.
Circ Res. 120:753–755. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zou YF, Wen D, Zhao Q, Shen PY, Shi H,
Zhao Q, Chen YX and Zhang W: Urinary MicroRNA-30c-5p and
MicroRNA-192-5p as potential biomarkers of
ischemia-reperfusion-induced kidney injury. Exp Biol Med (Maywood).
242:657–667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xi X, Li T, Huang Y, Sun J, Zhu Y, Yang Y
and Lu ZJ: RNA Biomarkers: Frontier of Precision Medicine for
Cancer. Noncoding RNA. 3:32017.
|
|
37
|
Liu LX, Rowe GC, Yang S, Li J, Damilano F,
Chan MC, Lu W, Jang C, Wada S, Morley M, et al: PDK4 Inhibits
Cardiac Pyruvate Oxidation in Late Pregnancy. Circ Res.
121:1370–1378. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ding J, Reynolds LM, Zeller T, Müller C,
Lohman K, Nicklas BJ, Kritchevsky SB, Huang Z, de la Fuente A,
Soranzo N, et al: Alterations of a Cellular Cholesterol Metabolism
Network Are a Molecular Feature of Obesity-Related Type 2 Diabetes
and Cardiovascular Disease. Diabetes. 64:3464–3474. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ligthart S, Vaez A, Hsu YH, Stolk R,
Uitterlinden AG, Hofman A, Alizadeh BZ, Franco OH and Dehghan A;
Inflammation Working Group of the CHARGE Consortium; PMI-WG-XCP;
LifeLines Cohort Study, : Bivariate genome-wide association study
identifies novel pleiotropic loci for lipids and inflammation. BMC
Genomics. 17:4432016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen T, Yang Z, Liu C, Wang L, Yang J,
Chen L and Li W: Circ_0078767 suppresses non-small-cell lung cancer
by protecting RASSF1A expression via sponging miR-330-3p. Cell
Prolif. 52:e125482019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Z, Qu H, Gong W and Liu A:
Up-regulation and tumor-promoting role of SPHK1 were attenuated by
miR-330-3p in gastric cancer. IUBMB Life. 70:1164–1176. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiong X, Shi Q, Yang X, Wang W and Tao J:
LINC00052 functions as a tumor suppressor through negatively
modulating miR-330-3p in pancreatic cancer. J Cell Physiol.
234:15619–15626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen Y, Liu X, Chen L, Chen W, Zhang Y,
Chen J, Wu X, Zhao Y, Wu X and Sun G: The long noncoding RNA XIST
protects cardiomyocyte hypertrophy by targeting miR-330-3p. Biochem
Biophys Res Commun. 505:807–815. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jun G, Asai H, Zeldich E, Drapeau E, Chen
C, Chung J, Park JH, Kim S, Haroutunian V, Foroud T, et al: PLXNA4
is associated with Alzheimer disease and modulates tau
phosphorylation. Ann Neurol. 76:379–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yosten GL, Kolar GR, Redlinger LJ and
Samson WK: Evidence for an interaction between proinsulin C-peptide
and GPR146. J Endocrinol. 218:B1–B8. 2013. View Article : Google Scholar
|
|
46
|
Young LH, Ikeda Y, Scalia R and Lefer AM:
C-peptide exerts cardioprotective effects in myocardial
ischemia-reperfusion. Am J Physiol Heart Circ Physiol.
279:H1453–H1459. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Correll RN, Grimes KM, Prasad V, Lynch JM,
Khalil H and Molkentin JD: Overlapping and differential functions
of ATF6α versus ATF6β in the mouse heart. Sci Rep. 9:20592019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wu T, Wu HD, Xu ZX, Han F, Zhang BQ, Sun J
and Hu SJ: Abnormal expression of long non-coding RNAs in
myocardial infarction. Heart Vessels. 32:1253–1261. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gao W, Wang ZM, Zhu M, Lian XQ, Zhao H,
Zhao D, Yang ZJ, Lu X and Wang LS: Altered long noncoding RNA
expression profiles in the myocardium of rats with ischemic heart
failure. J Cardiovasc Med (Hagerstown). 16:473–479. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu H, Xu D, Zhong X, Xu D, Chen G, Ge J
and Li H: LncRNA-mRNA competing endogenous RNA network depicts
transcriptional regulation in ischaemia reperfusion injury. J Cell
Mol Med. 23:2272–2276. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fu LL, Li CJ, Xu Y, Li LY, Zhou X, Li DD,
Chen SX, Wang FG, Zhang XY and Zheng LW: Role of lncRNAs as Novel
Biomarkers and Therapeutic Targets in Ovarian Cancer. Crit Rev
Eukaryot Gene Expr. 27:183–195. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
White NM and Maher CA: The potential use
of lncRNAs found in the 8q24 region as biomarkers for colon cancer.
Ann Oncol. 28:1688–1689. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Troiano G, Caponio VCA, Boldrup L, Gu X,
Muzio LL, Sgaramella N, Wang L and Nylander K: Expression of the
long non-coding RNA HOTAIR as a prognostic factor in squamous cell
carcinoma of the head and neck: A systematic review and
meta-analysis. Oncotarget. 8:73029–73036. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhong Y, Chen Z, Guo S, Liao X, Xie H,
Zheng Y, Cai B, Huang P, Liu Y, Zhou Q, et al: TUG1, SPRY4-IT1, and
HULC as valuable prognostic biomarkers of survival in cancer: A
PRISMA-compliant meta-analysis. Medicine (Baltimore). 96:e85832017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kehl DW, Iqbal N, Fard A, Kipper BA, De La
Parra Landa A and Maisel AS: Biomarkers in acute myocardial injury.
Transl Res. 159:252–264. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Keller T, Zeller T, Peetz D, Tzikas S,
Roth A, Czyz E, Bickel C, Baldus S, Warnholtz A, Fröhlich M, et al:
Sensitive troponin I assay in early diagnosis of acute myocardial
infarction. N Engl J Med. 361:868–877. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang C and Jing Q: Non-coding RNAs as
biomarkers for acute myocardial infarction. Acta Pharmacol Sin.
39:1110–1119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He
J, Qin YW and Jing Q: Circulating microRNA: A novel potential
biomarker for early diagnosis of acute myocardial infarction in
humans. Eur Heart J. 31:659–666. 2010. View Article : Google Scholar : PubMed/NCBI
|