Introduction
Lung cancer has been widely acknowledged to have the
highest mortality of all cancer types worldwide, and is classified
pathologically as non-small cell lung cancer (NSCLC) and small cell
lung cancer (SCLC) (1). Lung
adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are
identified as the most common subtypes of NSCLC (2). Surgery is the most general treatment
for early-stage NSCLC, and the combination of cytotoxic and
platinum drugs has become the standard for NSCLC chemotherapy
(3). However, the long-term
survival for postoperative patients with NSCLC remains poor.
Previous studies have indicated that the five-year survival rates
of patients with NSCLC with stage Ia, Ib, IIa and IIb are 70, 60,
55 and 40%, respectively (4,5).
Postoperative adjuvant chemotherapy has a limited effect on
improving the survival rate of patients with NSCLC (6). The biomarker cluster of
differentiation 24, has been recently identified for use in the
prediction of NSCLC disease-free survival (7). However, there are other promising
biomarkers with confirmed prognostic value in NSCLC, including
matrix metallopeptidase 9, aurora kinase A and enhancer of zeste 2
polycomb repressive complex 2 subunit (8).
Non-SMC condensin I complex subunit H (NCAPH) is a
member of the condensin I complex, which is a recently identified
superfamily of proteins termed kleisins (9). Condensin I complex consists of three
non-SMC subunits; NCAPH, chromosome-associated protein (CAP)D2 and
CAPG (10). Condensin serves an
indispensable role in chromosome-wide gene regulation and therefore
controls the architecture and segregation of sister chromatids
(11). In the presence of type I
topoisomerase, the condensin complex may introduce positive
supercoils into relaxed DNA, while type II topoisomerase is
involved in nicked DNA transformation into a positive knotted form
(12). Biallelic mutations in
CAPD2, NCAPH or CAPD3 are essential to ensure accurate mitotic
chromosome condensation in neuron stem cells, ultimately affecting
neuron pool and cortex size (13).
The authors of the present study previously demonstrated that NCAPH
is highly expressed in colon cancer, the knockdown of which
inhibits cell proliferation, migration, and xenograft tumor
formation abilities (14).
However, patients with colon cancer and the overexpression of NCAPH
have a markedly improved prognosis compared with patients who
demonstrate low-expression of NCAPH (14). Other studies note that highly
expressed condensin I complex in non-SMC is associated with the
progression of multiple human cancers: Ryu et al (15) demonstrated that NCAPH is expressed
at a higher level in metastatic melanoma cell lines compared with
less aggressive primary tumor cell lines. Other studies have
revealed that NCAPH can serve as a potential prognostic indicator
for hepatocellular carcinoma and nasopharyngeal cancer (16).
A number of antigens, including carbohydrate antigen
125, are commonly used clinically as auxiliary indicators for the
diagnosis of lung cancer, but they exhibit low sensitivity
(17). Myeloid cell leukemia
sequence 1 (Mcl-1), which is a member of the Bcl-2 family, was
originally considered to be an effective short-term promoter for
cell survival during the differentiation of bone marrow cells
(18–21). Studies have demonstrated that Mcl-1
is a pro-survival protein that is overexpressed in human malignant
tumors (22,23). Mcl-1, which is involved in
chemotherapy resistance and metastasis, is often highly expressed
in NSCLC and its association with other markers can improve
diagnostic sensitivity (24). The
intrinsic apoptotic signaling pathway is closely associated with
NCAPH (GO:0097193: http://www.coexpedia.org/) (25). The dual expression of NCAPH and
Mcl-1 proteins, and their association with clinical and
pathological features in NSCLC remain unknown, with the exception
of the aforementioned studies.
In the present study, the expression of NCAPH and
Mcl-1 was determined in 260 cases of NSCLC, and the association of
NCAPH and Mcl-1 expression with clinical and pathological
characteristics of NSCLC, and its prognosis, was discussed.
Materials and methods
Clinical data
Paraffin-embedded NSCLC tissues from 260 patients
diagnosed with primary NSCLC, none of whom had received any
treatment before the surgery. All patients had complete clinical
and follow-up data. (194 males and 66 females, mean age
60.1±12.76), and 52 control non-cancerous lung disease sections (37
males and 15 females, mean age 57.45±14.56), including
bronchiectasis and pneumatocele, were collected from the Second
Xiangya Hospital between January 2010 and December 2016. The
present study was approved by the Scientific and Research Ethics
Committee of the Second Xiangya Hospital (approval no. S039/2011).
Patients were fully informed about specimen usage and data
retrieval prior to the acquisition of specimens. No information or
images that may expose any relevant patient identification or
violate any individual rights are presented in the present study.
Written informed consent was obtained from each patient. Informed
consent was obtained from a parent and/or legal guardian for
subjects <18 years old. All patients with NSCLC underwent
surgery and none received neoadjuvant radiotherapy or chemotherapy.
Histological diagnosis (26) and
staging (27) were performed for
all patients to confirm NSCLC. Tissue microarray technology was
performed as previously described (28,29).
Immunohistochemistry (IHC) staining
and scores
IHC was performed to detect the expression and
cellular location of NCAPH and Mcl-1. IHC assay was performed as
previously described (16,28). Tissues were fixed in
paraformaldehyde dehydrated in a graded alcohol series, embedded in
paraffin and sectioned at 3 µm. Sections were then deparaffinized,
rehydrated and endogenous peroxidase inactivated using methanol
containing 0.3% H2O2. The slides were
incubated with appropriate pre-immune serum (Normal Goat Serum,
Beyotime Institute of Biotechnology, cat. no. C0269; diluted to 10%
in PBS+0.1% Tween20) for 30 min at room temperature to eliminate
nonspecific staining, and the sections were stained overnight at
4°C with 1:100 dilution of primary antibody to NCAPH
(Rabbit-anti-NCAPH antibody, Fine Test; Wuhan Fine Biotech Co.,
Ltd., cat. no. FNab05579), recombinant anti-Mcl-1 antibody (Abcam,
cat. no. ab32087). Slides were then exposed to an appropriate
secondary antibody (MaxvisionTM2 HRP-Polymer anti-Mouse/Rabbit IHC
kit, Fuzhou Maixin Biotech. Co., Ltd., dilution: 1:1, cat. no.
KIT-5020) for 30 min at 37°C via EnVision™+ Dual Link
System-HRP (Dako; Agilent Technologies, Inc.). DAB and chromogen
solution were used for color reaction enhancement. Hematoxylin was
used to counterstain the slides (37°C for 7 min). A light
microscope (BX53, Olympus Corporation) was used to visualize slides
and the magnification was ×40 (right corner, zoom out) and ×200,
respectively.
NCAPH and Mcl-1 protein expression analysis was
performed using semi-quantitative evaluation (28). The staining intensity for NCAPH and
Mcl-1 was classified on a scale 0–3 based on the observed color
intensity (0=negative, no staining; 1=weak, light brown;
2=moderate, brown and 3=strong, dark brown), and positive rates
were evenly divided into five grades [0 (0%), 1 (1–25%), 2
(26–50%), 3 (51–75%), and 4 (76–100%)]. The final staining score
for NCAPH and Mcl-1 was determined by the formula: Staining
score=positive rates* intensity. The best cut-off score for NCAPH
was determined as 4 (high expression was determined when the score
was ≥4, while scores <4 were considered negative) based on the
survival rate. The optimal cut-off score was determined to be 6 for
Mcl-1. The consistency of the two evaluators (Qiuxia Xiong and
Songqing Fan) was 95% and differences were solved by discussion
between Qiuxia Xiong and Songqing Fan under a two-headed
microscope. A light microscope (BX53; Olympus Corporation) was used
to visualize slides and the magnification was ×40 (right corner,
zoom out) and ×200, respectively.
Statistical analysis
SPSS version 20.0 was used for statistical analysis
(IBM Corp.). The association of NCAPH, Mcl-1, and clinical and
pathological characteristics was analyzed via the χ2
test and verified using The Cancer Genome Atlas (TCGA) database
(v21.0; http://portal.gdc.cancer.gov/). The data were
generated from the database starBase (v2.0; http://starbase.sysu.edu.cn/panGeneCoExp.php).
Overall survival curves were obtained using Kaplan-Meier analysis.
The Cox proportional hazard regression model was used for
multivariate analysis to determine the potential of positive
expression of NCAPH and Mcl-1 as poor prognostic indicators.
One-way ANOVA was used for comparisons between groups, and t-test
was used for further paired comparisons if differences existed.
P<0.05 was considered to indicate a statistically
significant difference.
Results
Protein expression of NCAPH and Mcl-1
in the tissues of LUSC and LUAD
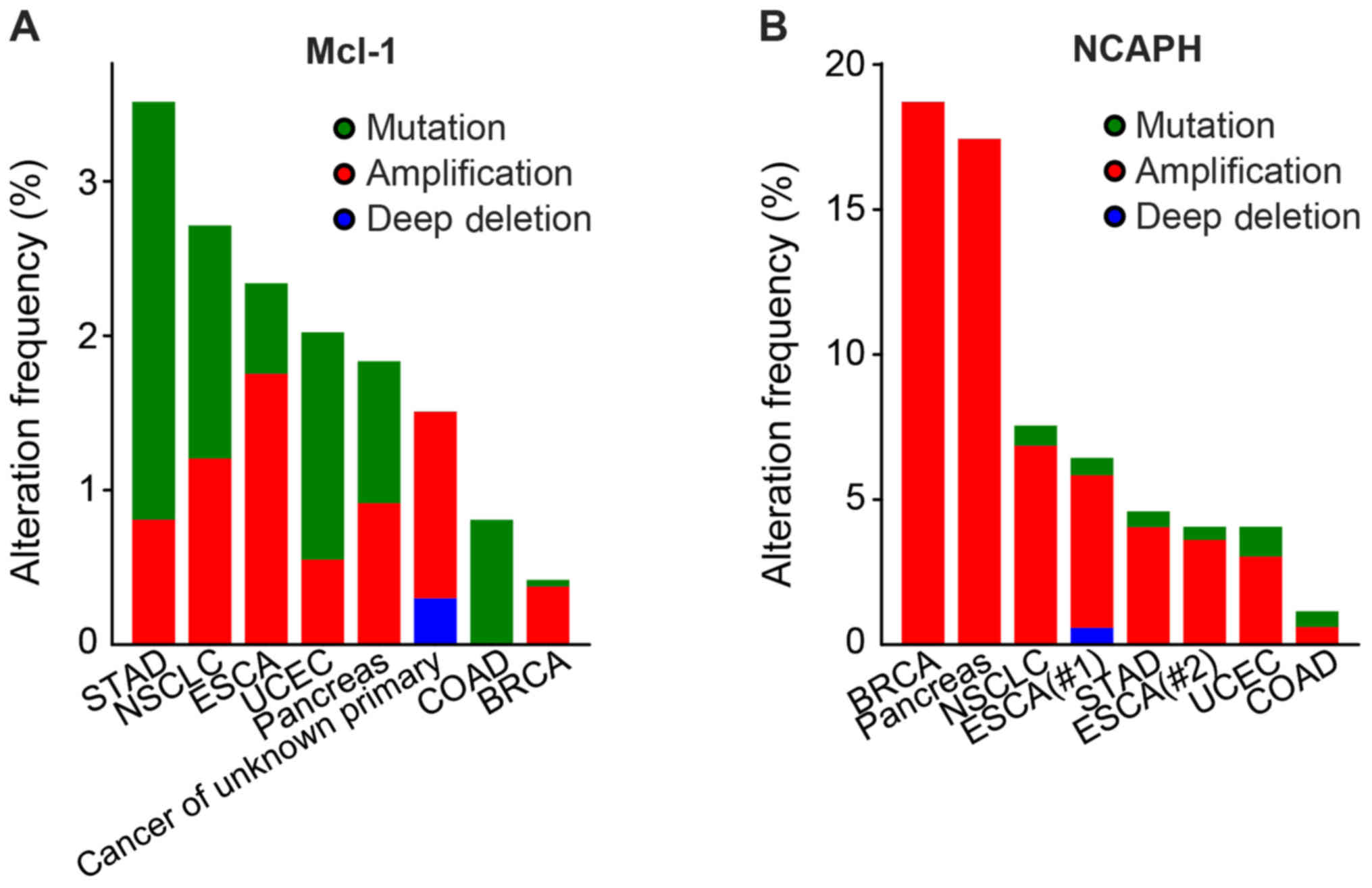
The mutation patterns of NCAPH and Mcl-1 in various
cancers were examined (Fig. 1;
Table I) (30–37).
The result showed that the mutation patterns of NCAPH and Mcl-1
existed in various cancers including stomach adenocarcinoma,
non-small cell lung cancer, esophagogastric carcinoma, endometrial
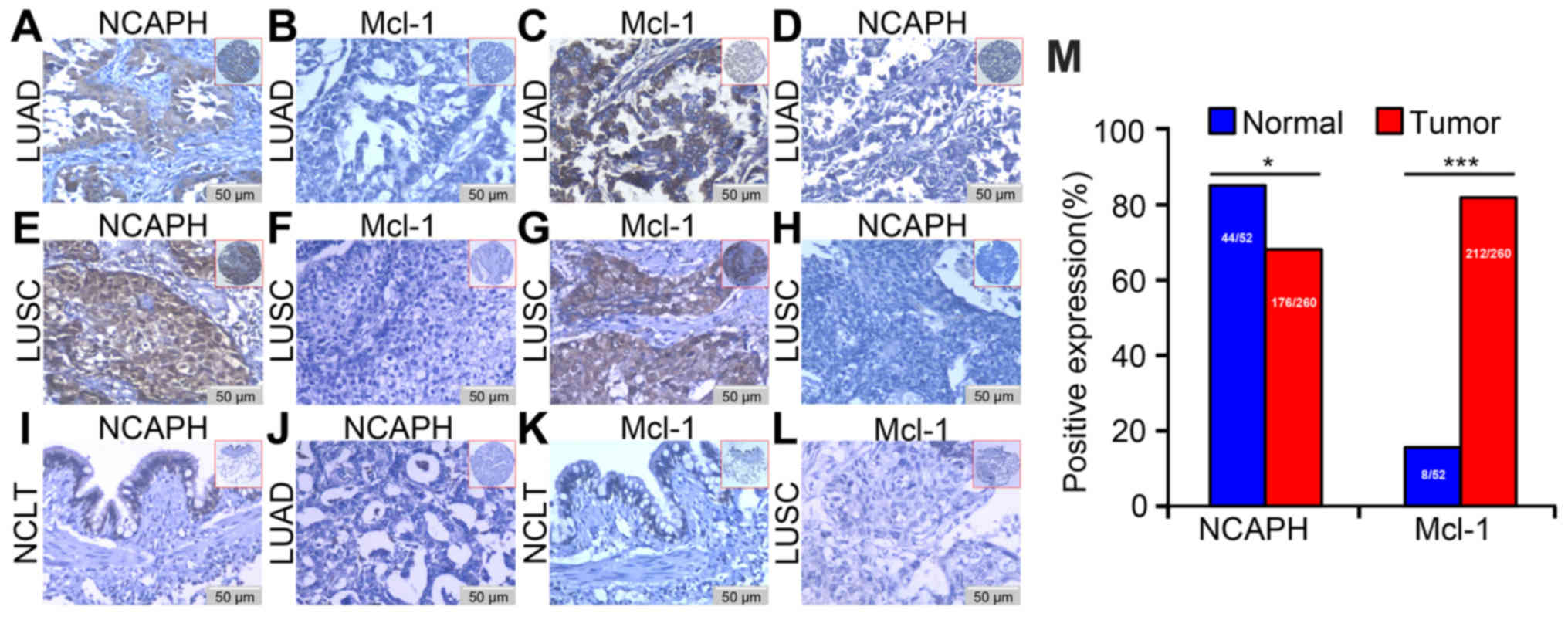
cancer, colorectal cancer and breast cancer. Immunohistochemistry
was conducted to detect the expression and cellular location of
NCAPH and Mcl-1 in LUSC, LUAD and the non-cancerous lung control
tissues to investigate the connection of NCAPH and Mcl-1 in
malignant tissues. The clinicopathological characteristics are
presented in Table II. Expression
of NCAPH and Mcl-1 was high in NSCLC tissues and was identified in
the cell cytoplasm and nuclear tissues and weak positive expression
in the bronchial epithelial cells of non-cancerous normal control
lung tissue. In the LUAD and LUSC tissues, Mcl-1 was negatively
expressed in the matched tissue in which NCAPH was positively
expressed and vice versa (Fig.
2A-H). A weak positive expression of NCAPH was identified in
the control tissues (Fig. 2I). No
positive NCAPH staining was observed in the LUAD tissue sections
when the matched immunoglobulin G isotype antibody was stained as a
negative control (Fig. 2J). Weak
positive expression of Mcl-1 was revealed in the bronchial
epithelial cells of non-cancerous control normal lung tissue
(Fig. 2K). The negative control
demonstrated no expression of Mcl-1 in LUSC (Fig. 2L).
 | Table I.Data sources of the mutation patterns
of NCAPH and Mcl-1 in various types of cancer. |
Table I.
Data sources of the mutation patterns
of NCAPH and Mcl-1 in various types of cancer.
| Author, year | Cancer type | (Refs.) |
|---|
| Taylor et al
(2018) | Stomach
adenocarcinoma | (30) |
| Jordan et al
(2017) | Non-small cell lung
cancer | (31) |
| Janjigian et
al (2018) | Esophagogastric
carcinoma | (32) |
| Soumerai et
al (2018) | Endometrial
cancer | (33) |
| Witkiewicz et
al (2015) | Pancreatic
cancer | (34) |
| Ghandi et al
(2019) | Cancer of unknown
primary | (35) |
| Yaeger et al
(2018) | Colorectal
cancer | (36) |
| Pereira et
al (2016) | Breast cancer | (37) |
 | Table II.Summary of patients with NSCLC and
non-cancerous control lung tissues in the tissue arrays. |
Table II.
Summary of patients with NSCLC and
non-cancerous control lung tissues in the tissue arrays.
| A, NSCLC |
|---|
|
|---|
| Patients
characteristics | No. of patients
(%) |
|---|
| Age (years) |
|
|
<55 | 118 (45.4) |
|
≥55 | 142 (54.6) |
| Sex |
|
|
Male | 194 (74.6) |
|
Female | 66 (25.4) |
| Clinical stage |
|
| Stage I
and II | 125 (48.1) |
| Stage
III and IV | 125 (51.9) |
| Lymph node
status |
|
| No
LNM | 111 (42.7) |
|
LNM | 149 (57.3) |
| Histological
type |
|
|
SCC | 129 (49.6) |
|
ADC | 131 (50.4) |
|
Differentiation |
|
| Well
and moderate | 117 (45.0) |
|
Poor | 143 (55.0) |
| Survival state |
|
|
Living | 160 (61.5) |
|
Mortality | 100 (38.5) |
|
| B, Non-cancerous
lung tissues |
|
| Patients
characteristics | No. of patients
(%) |
|
| Age (years) |
|
|
<55 | 25 (48.1) |
|
≥55 | 27 (51.9) |
| Sex |
|
|
Male | 37 (71.2) |
|
Female | 15 (28.8) |
The expression of NCAPH and Mcl-1 in the
non-cancerous control lung and NSCLC tissues was quantified
(Fig. 2M); the positive percentage
of NCAPH in the non-cancerous lung tissues (84.61%: 44/52) was
higher than that in NSCLC (67.69%; 176/260; P=0.015). The positive
percentage of Mcl-1 in the non-cancerous lung tissues (15.38%;
8/52) was lower compared with that in NSCLC (81.54%; 212/260;
P<0.001). The aforementioned results indicated that the
positive expression percentage of NCAPH was lower in tissues
harvested from patients with patients while the positive
percentages of Mcl-1 were significantly higher compared with
control lung tissue sections.
Association between NCAPH and Mcl-1
and the clinical characteristics in NSCLC
The univariate χ2 test was used to
examine the influence of the altered expression of NCAPH and Mcl-1
in NSCLC tissues on clinical outcomes. The clinicopathological
features investigated included age, clinical stage, sex, lymph node
(LNM) status, pathological grade and survival state. The high
percentage of Mcl-1 expression was significantly associated with
histological type (P<0.0001; Table III). The positive percentages of
NCAPH were significantly increased in the living group (73.8 vs.
58.0%; P=0.008), while the high percentage of Mcl-1 expression was
significantly higher in the mortality group (88.0 vs. 77.5%;
P=0.034). However, no association was observed between the
expression of NCAPH/Mcl-1 and sex, age, LNM stage or pathological
grade.
 | Table III.Analysis of the association between
expression of NCAPH and Mcl-1 proteins and clinicopathological
characteristics of NSCLC. |
Table III.
Analysis of the association between
expression of NCAPH and Mcl-1 proteins and clinicopathological
characteristics of NSCLC.
|
| NCAPH | Mcl-1 |
|---|
|
|
|
|
|---|
| Clinicopathological
features | Positive (%) | Negative (%) | P-value | High (%) | Low (%) | P-value |
|---|
| Age (years) |
|
| 0.302 |
|
| 0.567 |
|
<55 | 76 (64.4) | 42 (35.6) |
| 98 (83.1) | 20 (16.9) |
|
|
≥55 | 100 (70.4) | 42 (29.6) |
| 114 (80.3) | 28 (19.7) |
|
| Sex |
|
| 0.922 |
|
| 0.301 |
|
Male | 131 (67.5) | 64 (32.5) |
| 161 (83.0) | 33 (17.0) |
|
|
Female | 45 (68.2) | 21 (31.8) |
| 51 (77.3) | 15 (22.7) |
|
| Histological
type |
|
| 0.858 |
|
| <0.001 |
|
ADC | 88 (68.2) | 41 (31.8) |
| 118 (91.51) | 11 (8.5) |
|
|
SCC | 88 (67.2) | 43 (32.8) |
| 94 (71.8) | 37 (28.2) |
|
| Clinical
stages |
|
| 0.369 |
|
| 0.768 |
| Stage I
and II | 88 (70.4) | 37 (29.6) |
| 101 (80.8) | 24 (19.2) |
|
| Stage
III and V | 88 (65.2) | 47 (34.8) |
| 111 (82.2) | 24 (17.8) |
|
| LNM status |
|
| 0.618 |
|
| 0.421 |
| No
LNM | 77 (69.4) | 34 (39.6) |
| 93 (83.8) | 18 (16.2) |
|
|
LNM | 99 (66.4) | 50 (33.4) |
| 119 (79.9) | 30 (20.1) |
|
| Pathological
grade |
|
| 0.201 |
|
| 0.247 |
| Well
and moderate | 84 (71.8) | 33 (28.2) |
| 99 (84.6) | 18 (15.4) |
|
|
Poor | 92 (64.3) | 51 (35.7) |
| 113 (79.0) | 30 (21.0) |
|
| Survival
status |
|
| 0.008a |
|
| 0.034a |
|
Alive | 118 (73.8) | 42 (26.2) |
| 124 (77.5) | 36 (22.5) |
|
|
Succumbed | 58 (58.0) | 42 (42.0) |
| 88 (88.0) | 12 (12.0) |
|
The pairwise association between NCAPH
and Mcl-1 proteins in NSCLC
Pairwise association (Table IV) demonstrated that positive
expression of NCAPH protein was associated with negative expression
of Mcl-1 protein (r=−0.185, P=0.003), while the positive expression
of Mcl-1 protein was associated with negative expression of NCAPH
protein (r=−0.185, P=0.003). Therefore, the overexpression of NCAPH
protein was negatively associated with the expression of Mcl-1. To
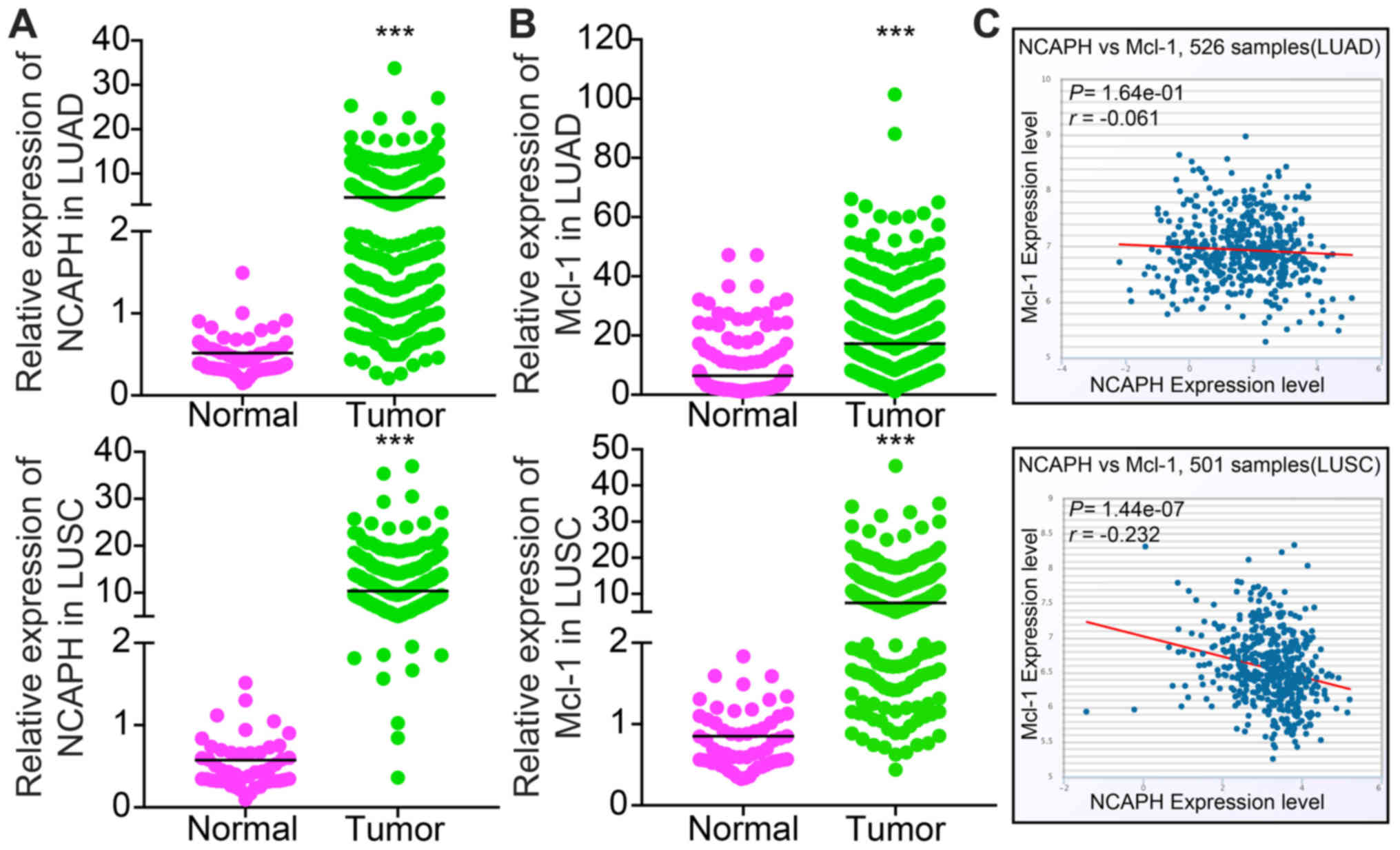
evaluate the expression profiles of NCAPH and Mcl-1 in LUAD and
LUSC, The Cancer Genome Atlas (TCGA) database was analyzed. The
results showed that NCAPH is highly expressed in LUAD and LUSC
(Fig. 3A), and Mcl-1 is highly
expressed in LUAD and LUSC (Fig.
3B). In addition, NCAPH expression is negative associated with
Mcl-1, as validated in LUAD and LUSC by utilizing the TCGA dataset
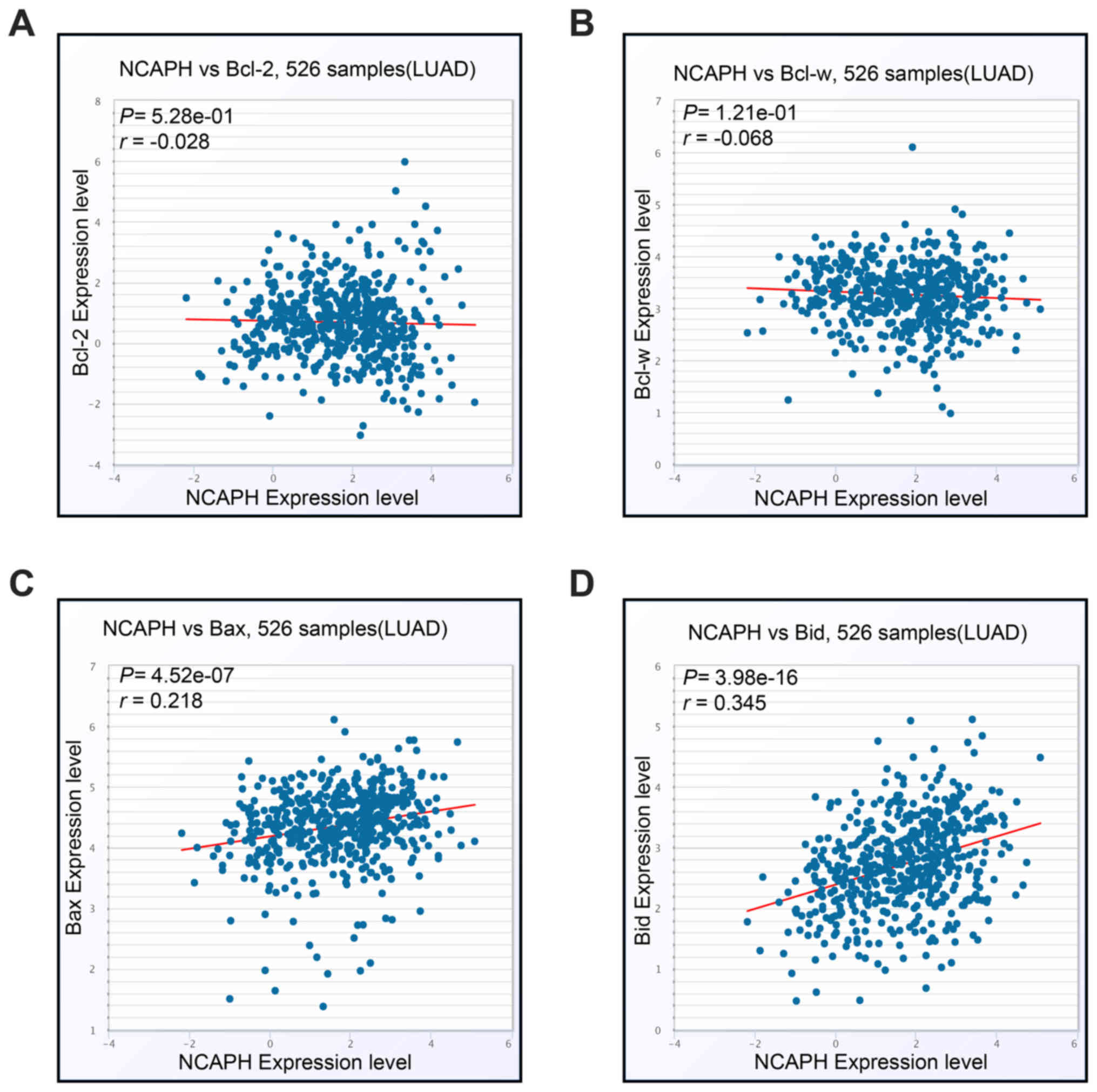
(Fig. 3C) (38). In addition, NCAPH was revealed to
be negatively associated with pro-survival members including Bcl-2
and Bcl-w, while being positively associated with pro-apoptotic
members Bax and Bid (Fig.
4A-D).
 | Table IV.The pairwise correlation between
expression of NCAPH and Mcl-1 proteins in 260 cases of NSCLC. |
Table IV.
The pairwise correlation between
expression of NCAPH and Mcl-1 proteins in 260 cases of NSCLC.
| Protein | NCAPH | Mcl-1 |
|---|
| NCAPH |
|
|
|
Spearman's correlation
coincident | 1 | −0.185 |
|
Significance (2-tailed) | – |
0.003a |
| Mcl-1 |
|
|
|
Spearman's correlation
coincident | −0.185 | 1 |
|
Significance (2-tailed) |
0.003a | – |
Impact of NCAPH and Mcl-1 expression
levels on the prognosis in NSCLC
To serve as a control, no significant difference was
detected in the overall survival (OS) between patients with LUAD
and LUSC from tissue microarrays used in the current study
(Fig. 5A; P=0.554). The results of
Kaplan-Meier curves indicated that the overall survival rates for
patients with lymph node metastasis were lower than for
metastasis-free patients (P=0.002; Fig. 5B). Meanwhile, higher NCAPH
expression in NSCLC was associated with higher OS rates (P=0.003;
Fig. 5C), while patients with high
Mcl-1 expression in NSCLC exhibited significantly worse OS rates
(P=0.032; Fig. 5D).
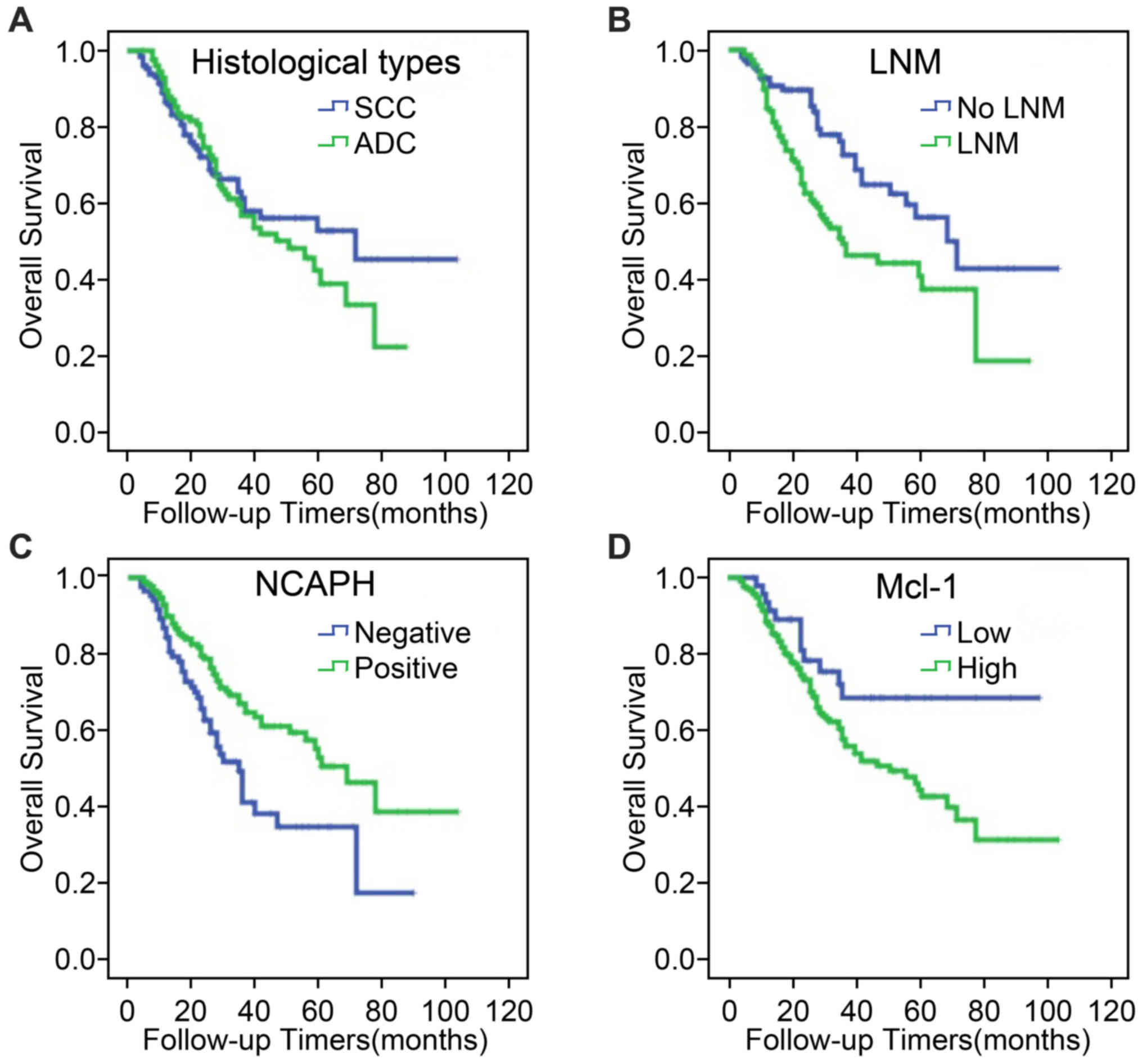 | Figure 5.Kaplan-Meier curves were used to
compare the overall survival curves of patients with NSCLC
according to clinicopathological characteristics and expression of
NCAPH and Mcl-1 proteins. (A) Kaplan-Meier analysis of the patients
with LUAD (green) and LUSC (blue) from tissue microarrays used in
the current study, P=0.554. (B) Kaplan-Meier analysis of the
patients with lymph node metastasis (green) and no lymph node
metastasis (blue) from tissue microarrays used in the current
study, P=0.002. (C) Kaplan-Meier analysis of the association
between NCAPH expression and the overall survival (blue, negative
expression of NCAPH; green, positive expression of NCAPH), P=0.003.
(D) Kaplan-Meier analysis of the association between Mcl-1
expression and the overall survival (blue, low expression of Mcl-1;
green, high expression of Mcl-1), P=0.032. NSCLC, non-small cell
lung cancer; NCAPH, non-SMC condensin I complex subunit H; Mcl-1,
myeloid cell leukemia sequence 1; LUSC, lung squamous cell
carcinoma; LUAD, lung adenocarcinoma; LNM, lymph node metastasis;
SCC, lung squamous carcinoma; ADC, lung adenocarcinoma. |
Multivariate Cox proportional hazard regression
analysis was performed on 260 NSCLC cases. Pathological grades
(P=0.017) and clinical stages (P=0.003) were revealed to serve as
prognostic factors (Table V).
Additionally, the expression of NCAPH (P=0.003) and Mcl-1 (P=0.038)
could be considered as distinct prognostic factors in patients with
NSCLC. No clinical effect based on age, sex, histological types,
LNM, or treatment strategy was detected.
 | Table V.Summary of multivariate analysis of
Cox proportional hazard regression for overall survival in 260
cases of patients with NSCLC. |
Table V.
Summary of multivariate analysis of
Cox proportional hazard regression for overall survival in 260
cases of patients with NSCLC.
|
|
|
|
| 95.0% CI for Exp
(B) |
|---|
|
|
|
|
|
|
|---|
| Variable | Wald | Sig. | Exp (B) | Lower | Upper |
|---|
| Age | 0.001 | 0.976 | 1.006 | 0.667 | 1.517 |
| Sex | 2.442 | 0.118 | 0.673 | 0.409 | 1.106 |
| Histological
types | 3.285 | 0.070 | 1.516 | 0.967 | 2.376 |
| Pathological
grades | 5.746 | 0.017a | 1.689 | 1.100 | 2.593 |
| LNM | 3.819 | 0.051 | 1.595 | 0.999 | 2.547 |
| Clinical
stages | 8.982 | 0.003a | 2.055 | 1.283 | 3.291 |
| Treatment
strategy | 0.087 | 0.768 | 0.918 | 0.519 | 1.624 |
| NCAPH | 8.539 | 0.003a | 0.544 | 0.361 | 0.818 |
| Mcl-1 | 4.324 | 0.038a | 1.955 | 1.039 | 3.678 |
Discussion
NCAPH is one of the members of the barr gene family
and is located on chromosome 2q11.2 (39). A previous study has indicated that
NCAPH is one of the essential factors for maintaining cell survival
and is indispensable in mitotic chromosome cohesion and separation
(40). Mcl-1 is homologous to
Bcl-2 and possesses an anti-apoptotic effect in regulating cell
survival (41). Compared with
healthy lungs, the overexpression of Mcl-1 in NSCLC lines is
associated with poor patient prognosis. Mcl-1 belongs to the
pro-apoptotic Bcl-2 family, maintains its inactive monomeric state
and antagonizes the signaling of cellular apoptosis, particularly
in Mcl-1-overexpressing NSCLC cell lines (23). In addition, the depletion of Mcl-1
results in increased sensitivity to radiotherapy and chemotherapy
in NSCLC cells (42). Huang et
al (43) revealed that Mcl-1
promotes the migration of lung cancer cells through a mechanism
involving Ca2+-dependent reactive oxygen species
production. Therefore, Mcl-1 has been demonstrated to serve an
important role in NSCLC cell survival by limiting apoptotic
signaling.
In the present study, the protein expression of
NCAPH and Mcl-1 were examined by IHC in tissues of 260 cases of
NSCLC. The data demonstrated that NCAPH and Mcl-1 were highly
expressed in NSCLC cancerous tissues; with positive expression in
the cytoplasm and nucleus and weak positive expression in the
bronchial epithelial cells of non-cancerous normal control lung
tissue. The expression of NCAPH and Mcl-1 was negatively associated
in the matched tissues of LUAD and LUSC. Pairwise association also
demonstrated that NCAPH and Mcl-1 overexpression were inversely
associated.
Precise biomarkers are essential for providing a
reference for NSCLC prognosis in the tumor-node-metastasis staging
system; one of the most effective prognostic methods for operable
NSCLC (42). Therefore, it is
important to identify novel biomarkers that can help to determine
the risks of tumor occurrence and progression. According to the
association analysis data between NCAPH and Mcl-1, the high
expression percentage of Mcl-1 was positively associated with
histological type in NSCLC. In addition, the positive percentage
expression of NCAPH was significantly elevated in the living group,
while the percentage expression of Mcl-1 was increased in the
mortality group. Positive NCAPH expression was associated with
higher OS rates, whereas, high Mcl-1 expression was inversely
associated with OS rates. These results indicated that the
expression of NCAPH and Mcl-1 may be associated with the
development and progression of NSCLC and can be considered to be
independent prognostic factors. Multiple regulatory mechanisms
including ubiquitination and subsequent proteasomal degradation
have been shown to control Mcl-1 functions by regulating its
expression in response to different stimuli (44). Recent studies validate the
non-apoptotic functions of Mcl-1 in autophagy, mitochondrial
homeostasis and protein kinase cascade signaling (45). Mcl-1 physically interacts with a
number of cell cycle regulators in the nucleus (including PCNA and
Chk1), thus regulating the checkpoint response (46,47).
This leads to the hypothesis that NCAPH could also form a complex
with, and negatively regulate, Mcl-1 expression during NSCLC
progression. This hypothesis should be investigated in future
studies due to the lack of current proteasome data from the NCAPH
complex. In conclusion, the expression of NCAPH protein was
associated with the expression of Mcl-1 in patients with NSCLC, and
the positive expression of NCAPH may serve as one of the
independent prognostic factors in surgically resected patients with
this disease.
Acknowledgements
The authors would like to thank Dr Nigel W. Fraser,
Dept of Microbiology, Pereleman School of Medicine, University of
Pennsylvania, USA, for his instructive comments on the writing of
the manuscript.
Funding
The present study was supported by Yunnan Provincial
Department of science and Technology-Kunming Medical University
applied basic research joint project in 2019 (grant no. 2019FE001
(−042)) and Scientific research projects of research institutions
in medical and health units of Yunnan Province in 2018 (grant no.
2018NS0111).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QX, YD, SF, LD and XC conceived and designed the
experiments. YD, QX, SF, BL, XJ, CX, QT, HZ and JW performed the
experiments. QX, SF, CX, CC and JW analyzed the data. YD, QX, SF,
XC, JW, HZ, YD and QT contributed reagents, materials or analytical
tools. YD, QX, SF, LD, QT and CX wrote the manuscript. QX, SF, XC
and BL accessed the full-text articles. XJ generated the revised
Fig. 4. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Scientific and
Research Ethics Committee of the Second Xiangya Hospital (approval
no. S039/2011). Patients were fully informed about specimen usage
and data retrieval prior to the acquisition of specimens. Written
informed consent was obtained from each patient. Informed consent
was obtained from a parent and/or legal guardian for subjects
<18 years old.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:2916–27. 2016. View Article : Google Scholar
|
|
2
|
Chen M, Liu X, Du J, Wang XJ and Xia L:
Differentiated regulation of immune-response related genes between
LUAD and LUSC subtypes of lung cancers. Oncotarget. 8:133–144.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yoshioka H, Shimokawa M, Seto T, Morita S,
Yatabe Y, Okamoto I, Tsurutani J, Satouchi M, Hirashima T, Atagi S,
et al: Final overall survival results of WJTOG3405, a randomized
phase III trial comparing gefitinib versus cisplatin with docetaxel
as the first-line treatment for patients with stage IIIB/IV or
postoperative recurrent EGFR mutation-positive non-small-cell lung
cancer. Ann Oncol. 30:1978–1984. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bobbili P, Ryan K, Duh MS, Dua A,
Fernandes AW, Pavilack M and Gomez JE: Treatment patterns and
overall survival among patients with unresectable, stage III
non-small-cell lung cancer. Future Oncol. 15:3381–3393. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
León-Atance P, Moreno-Mata N,
González-Aragoneses F, Cañizares-Carretero MA, García-Jiménez MD,
Genovés-Crespo M, Honguero-Martínez AF, Rombolá CA, Simón-Adiego CM
and Peñalver-Pascual R: Multicenter analysis of survival and
prognostic factors in pathologic stage I non-small-cell lung cancer
according to the new 2009 TNM Classification. Arch Bronconeumol.
47:441–446. 2011.(In English, Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Criss SD, Mooradian MJ, Watson TR, Gainor
JF, Reynolds KL and Kong CY: Cost-effectiveness of atezolizumab
combination therapy for first-line treatment of metastatic
nonsquamous non-small cell lung cancer in the United States. JAMA
Netw Open. 2:e19119522019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosser CJ and Goodison S: CD24, a
promising biomarker in NSCLC. Biomark Med. 4:4952010. View Article : Google Scholar
|
|
8
|
Zhan SJ, Liu B and Linghu H: Identifying
genes as potential prognostic indicators in patients with serous
ovarian cancer resistant to carboplatin using integrated
bioinformatics analysis. Oncol Rep. 39:2653–2663. 2018.PubMed/NCBI
|
|
9
|
Sun C, Huang S, Wang H, Xie R, Zhang L,
Zhou Q, He X and Ju W: Non-SMC condensin I complex subunit H
enhances proliferation, migration, and invasion of hepatocellular
carcinoma. Mol Carcinog. 58:2266–2275. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hirano T: Condensins: Universal organizers
of chromosomes with diverse functions. Genes Dev. 26:1659–1678.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wood AJ, Severson AF and Meyer BJ:
Condensin and cohesin complexity: The expanding repertoire of
functions. Nat Rev Genet. 11:391–404. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kimura K, Cuvier O and Hirano T:
Chromosome condensation by a human condensin complex in xenopus egg
extracts. J Biol Chem. 276:5417–5420. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin CA, Murray JE, Carroll P, Leitch A,
Mackenzie KJ, Halachev M, Fetit AE, Keith C, Bicknell LS, Fluteau
A, et al: Mutations in genes encoding condensin complex proteins
cause microcephaly through decatenation failure at mitosis. Genes
Dev. 30:2158–2172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin L, Jiang LP, Shen QS, Xiong QX, Zhuo
X, Zhang LL, Yu HJ, Guo X, Luo Y, Dong J, et al: NCAPH plays
important roles in human colon cancer. Cell Death Dis. 8:e26802017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ryu B, Kim DS, Deluca AM and Alani RM:
Comprehensive expression profiling of tumor cell lines identifies
molecular signatures of melanoma progression. PLoS One. 2:e5942007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu L, Jiang Y, Zheng J, Xie G, Li J, Shi L
and Fan S: Aberrant expression of β-catenin and e-cadherin is
correlated with poor prognosis of nasopharyngeal cancer. Hum
Pathol. 44:1357–1364. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Granville CA and Dennis PA: An overview of
lung cancer genomics and proteomics. Am J Respir Cell Mol Biol.
32:169–176. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kozopas KM, Yang T, Buchan HL, Zhou P and
Craig RW: MCL1, a gene expressed in programmed myeloid cell
differentiation, has sequence similarity to BCL2. Proc Natl Acad
Sci USA. 90:3516–3520. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han Y, Wu N, Jiang M, Chu Y, Wang Z, Liu
H, Cao J, Liu H, Xu B and Xie X: Long non-coding RNA MYOSLID
functions as a competing endogenous RNA to regulate MCL-1
expression by sponging miR-29c-3p in gastric cancer. Cell Prolif.
52:e126782019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kour S, Rana S, Contreras JI, King HM,
Robb CM, Sonawane YA, Bendjennat M, Crawford AJ, Barger CJ, Kizhake
S, et al: CDK5 inhibitor downregulates Mcl-1 and sensitizes
pancreatic cancer cell lines to navitoclax. Mol Pharmacol.
96:419–429. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meister MT, Boedicker C, Linder B, Kogel
D, Klingebiel T and Fulda S: Concomitant targeting of Hedgehog
signaling and MCL-1 synergistically induces cell death in
Hedgehog-driven cancer cells. Cancer Lett. 465:1–11. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Akgul C: Mcl-1 is a potential therapeutic
target in multiple types of cancer. Cell Mol Life Sci.
66:1326–1336. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Guttikonda S, Roberts L, Uziel T,
Semizarov D, Elmore SW, Leverson JD and Lam LT: Mcl-1 is critical
for survival in a subgroup of non-small-cell lung cancer cell
lines. Oncogene. 30:1963–1968. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Whitsett TG, Mathews IT, Cardone MH, Lena
RJ, Pierceall WE, Bittner M, Sima C, LoBello J, Weiss GJ and Tran
NL: Mcl-1 mediates TWEAK/Fn14-induced non-small cell lung cancer
survival and therapeutic response. Mol Cancer Res. 12:550–559.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang S, Kim CY, Hwang S, Kim E, Kim H,
Shim H and Lee I: COEXPEDIA: Exploring biomedical hypotheses via
co-expressions associated with medical subject headings (MeSH).
Nucleic Acids Res. 45(D1):D389–D396. 2017. View Article : Google Scholar
|
|
26
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: WHO Classification of tumours of the lung,
pleura, thymus and heart. WHO Classification of Tumours. 7. 4th.
IARC; Lyon: 2015
|
|
27
|
Boffa DJ and Greene FL: Reacting to
changes in staging designations in the 7th edition of the AJCC
staging manual. Ann Surg Oncol. 18:1–3. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wen Q, Wang W, Luo J, Chu S, Chen L, Xu L,
Zang H, Alnemah MM, Ma J and Fan S: CGP57380 enhances efficacy of
RAD001 in non-small cell lung cancer through abrogating mTOR
inhibition-induced phosphorylation of eIF4E and activating
mitochondrial apoptotic pathway. Oncotarget. 7:27787–27801. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wen Q, Wang W, Chu S, Luo J, Chen L, Xie
G, Xu L, Li M and Fan S: Flot-2 expression correlates with EGFR
levels and poor prognosis in surgically resected non-small cell
lung cancer. PLoS One. 10:e01321902015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Taylor AM, Shih J, Ha G, Gao GF, Zhang X,
Berger AC, Schumacher SE, Wang C, Hu H, Liu J, et al: Genomic and
functional approaches to understanding cancer aneuploidy. Cancer
Cell. 33:676–689.e673. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jordan EJ, Kim HR, Arcila ME, Barron D,
Chakravarty D, Gao J, Chang MT, Ni A, Kundra R, Jonsson P, et al:
Prospective comprehensive molecular characterization of lung
adenocarcinomas for efficient patient matching to approved and
emerging therapies. Cancer Discov. 7:596–609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Janjigian YY, Sanchez-Vega F, Jonsson P,
Chatila WK, Hechtman JF, Ku GY, Riches JC, Tuvy Y, Kundra R,
Bouvier N, et al: Genetic predictors of response to systemic
therapy in esophagogastric cancer. Cancer Discov. 8:49–58. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Soumerai TE, Donoghue MTA, Bandlamudi C,
Srinivasan P, Chang MT, Zamarin D, Cadoo KA, Grisham RN,
O'Cearbhaill RE, Tew WP, et al: Clinical utility of prospective
molecular characterization in advanced endometrial cancer. Clin
Cancer Res. 24:5939–5947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Witkiewicz AK, McMillan EA, Balaji U, Baek
G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et
al: Whole-exome sequencing of pancreatic cancer defines genetic
diversity and therapeutic targets. Nat Commun. 6:67442015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ghandi M, Huang FW, Jané-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yaeger R, Chatila WK, Lipsyc MD, Hechtman
JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A,
Donoghue MTA, et al: Clinical sequencing defines the genomic
landscape of metastatic colorectal cancer. Cancer Cell.
33:125–136.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Neuwald AF and Hirano T: HEAT repeats
associated with condensins, cohesins, and other complexes involved
in chromosome-related functions. Genome Res. 10:1445–1452. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lawrimore J and Bloom K: The regulation of
chromosome segregation via centromere loops. Crit Rev Biochem Mol
Biol. 54:352–370. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Senichkin VV, Streletskaia AY, Zhivotovsky
B and Kopeina GS: Molecular comprehension of Mcl-1: From gene
structure to cancer therapy. Trends Cell Biol. 29:549–562. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song L, Coppola D, Livingston S, Cress D
and Haura EB: Mcl-1 regulates survival and sensitivity to diverse
apoptotic stimuli in human non-small cell lung cancer cells. Cancer
Biol Ther. 4:267–276. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang H, Shah K, Bradbury NA, Li C and
White C: Mcl-1 promotes lung cancer cell migration by directly
interacting with VDAC to increase mitochondrial Ca2+ uptake and
reactive oxygen species generation. Cell Death Dis. 5:e14822014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mojsa B, Lassot I and Desagher S: Mcl-1
ubiquitination: Unique regulation of an essential survival protein.
Cells. 3:418–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Young AI, Timpson P, Gallego-Ortega D,
Ormandy CJ and Oakes SR: Myeloid cell leukemia 1 (MCL-1), an
unexpected modulator of protein kinase signaling during invasion.
Cell Adh Migr. 12:513–523. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fujise K, Zhang D, Liu J and Yeh ET:
Regulation of apoptosis and cell cycle progression by MCL1:
Differential role of proliferating cell nuclear antigen. J Biol
Chem. 275:39458–39465. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jamil S, Mojtabavi S, Hojabrpour P, Cheah
S and Duronio V: An essential role for MCL-1 in ATR-mediated CHK1
phosphorylation. Mol Biol Cell. 19:3212–3220. 2008. View Article : Google Scholar : PubMed/NCBI
|