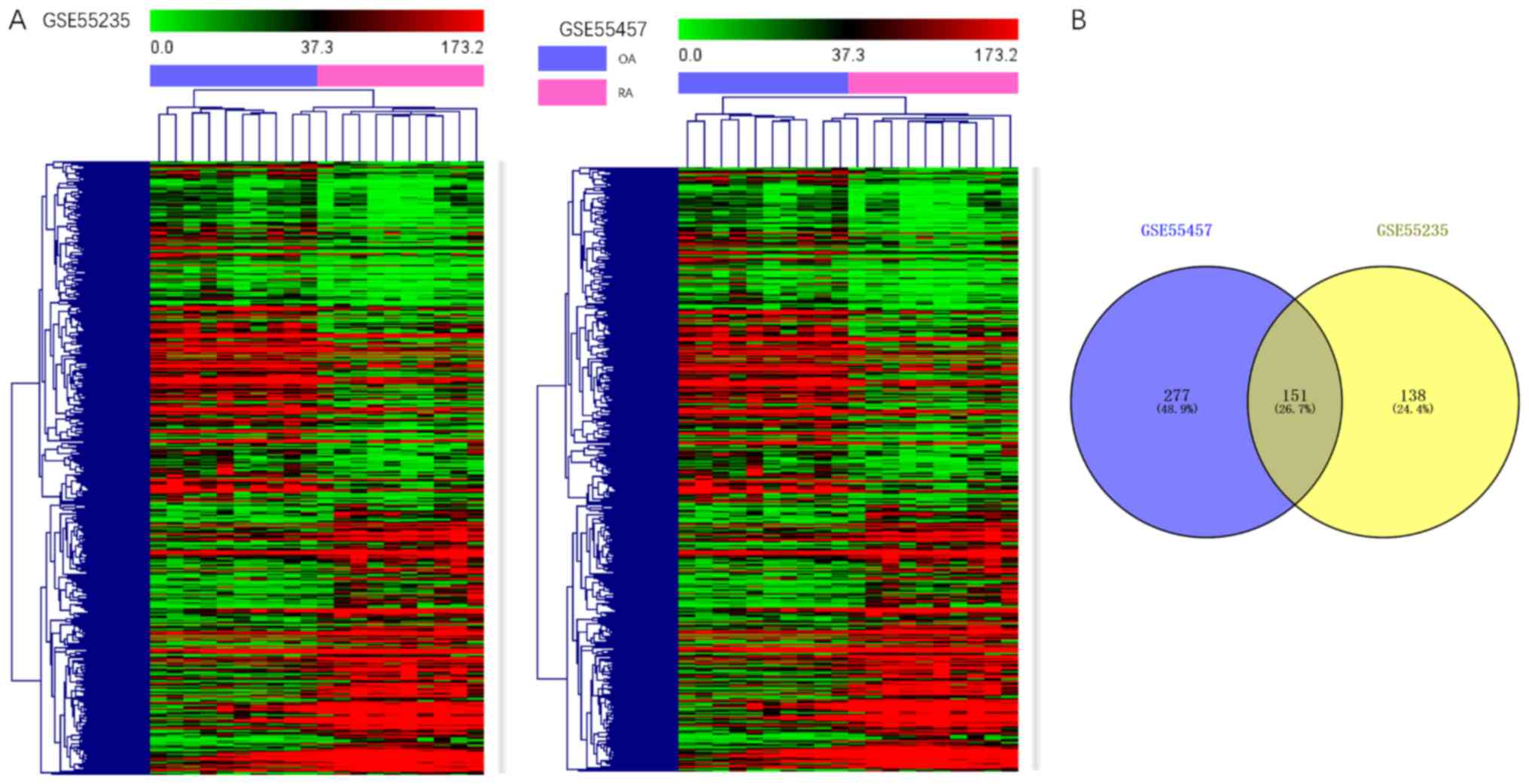
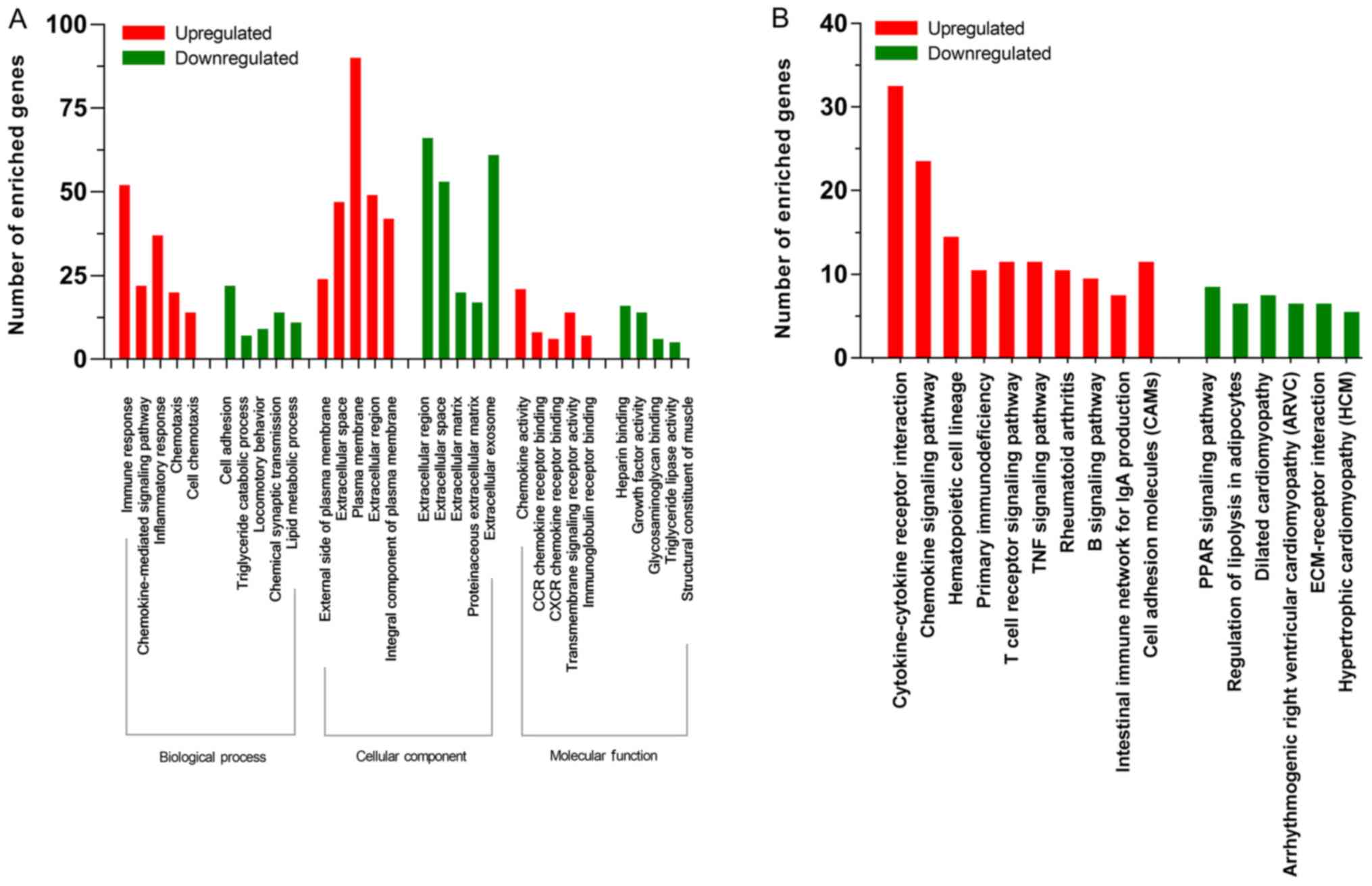
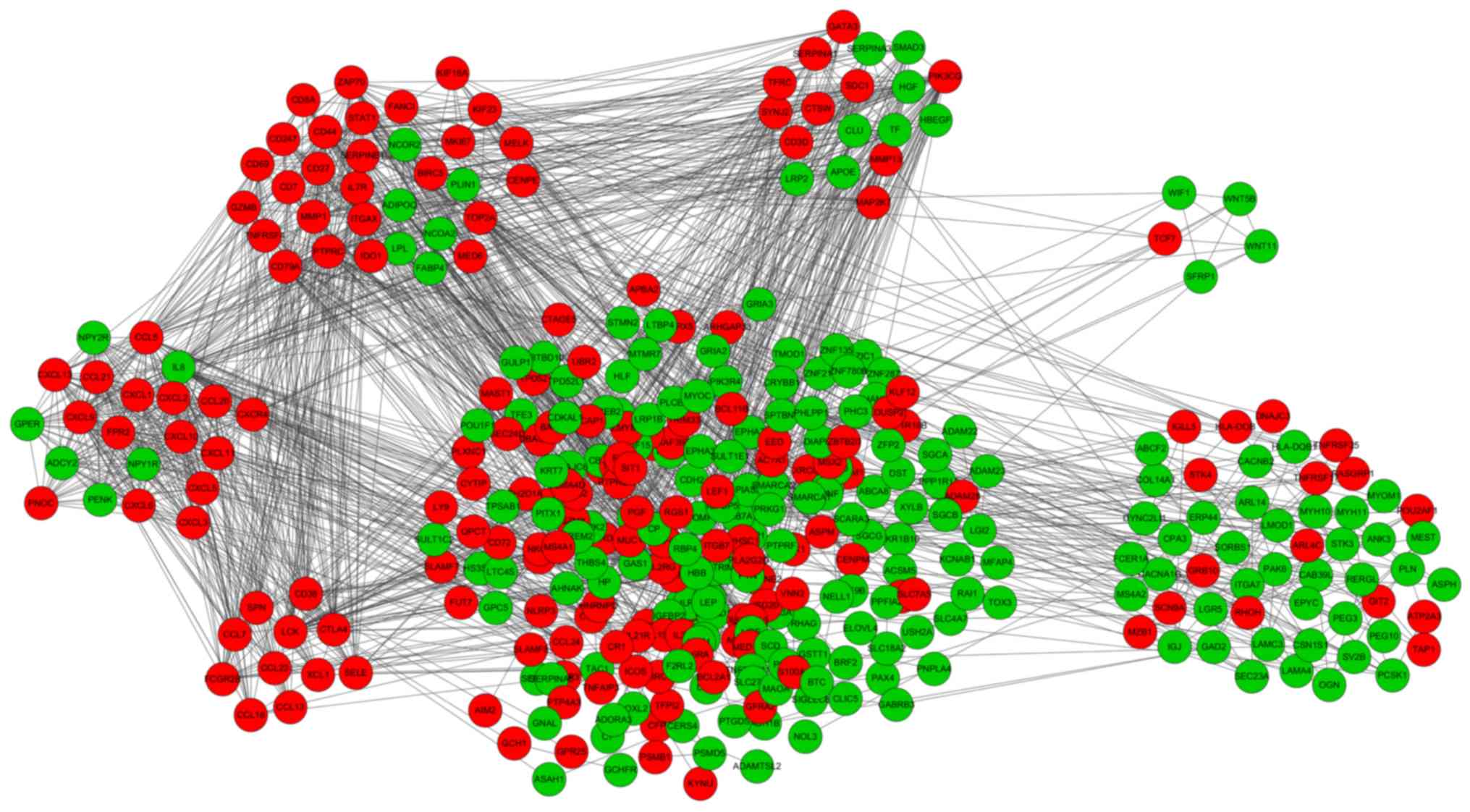
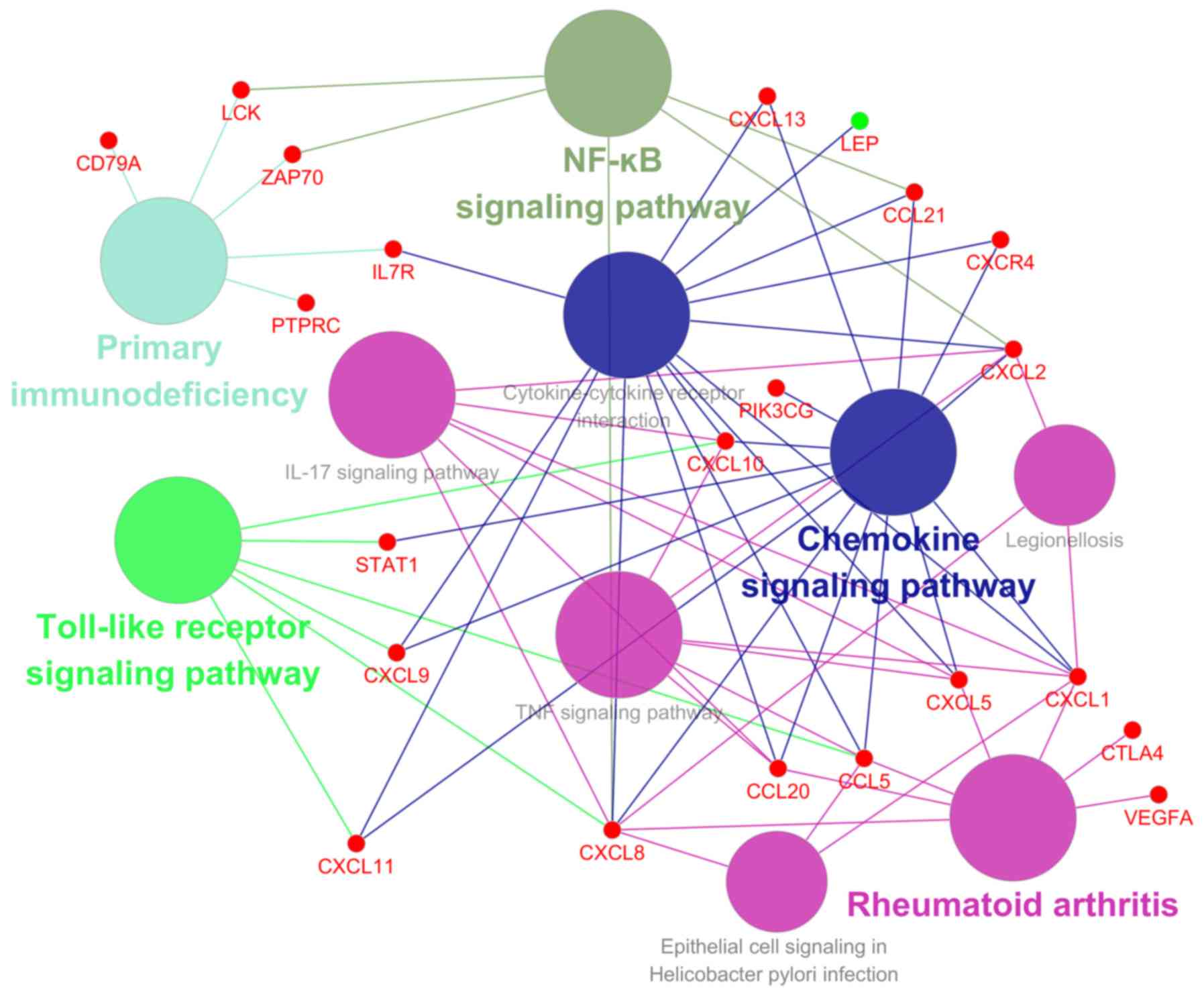
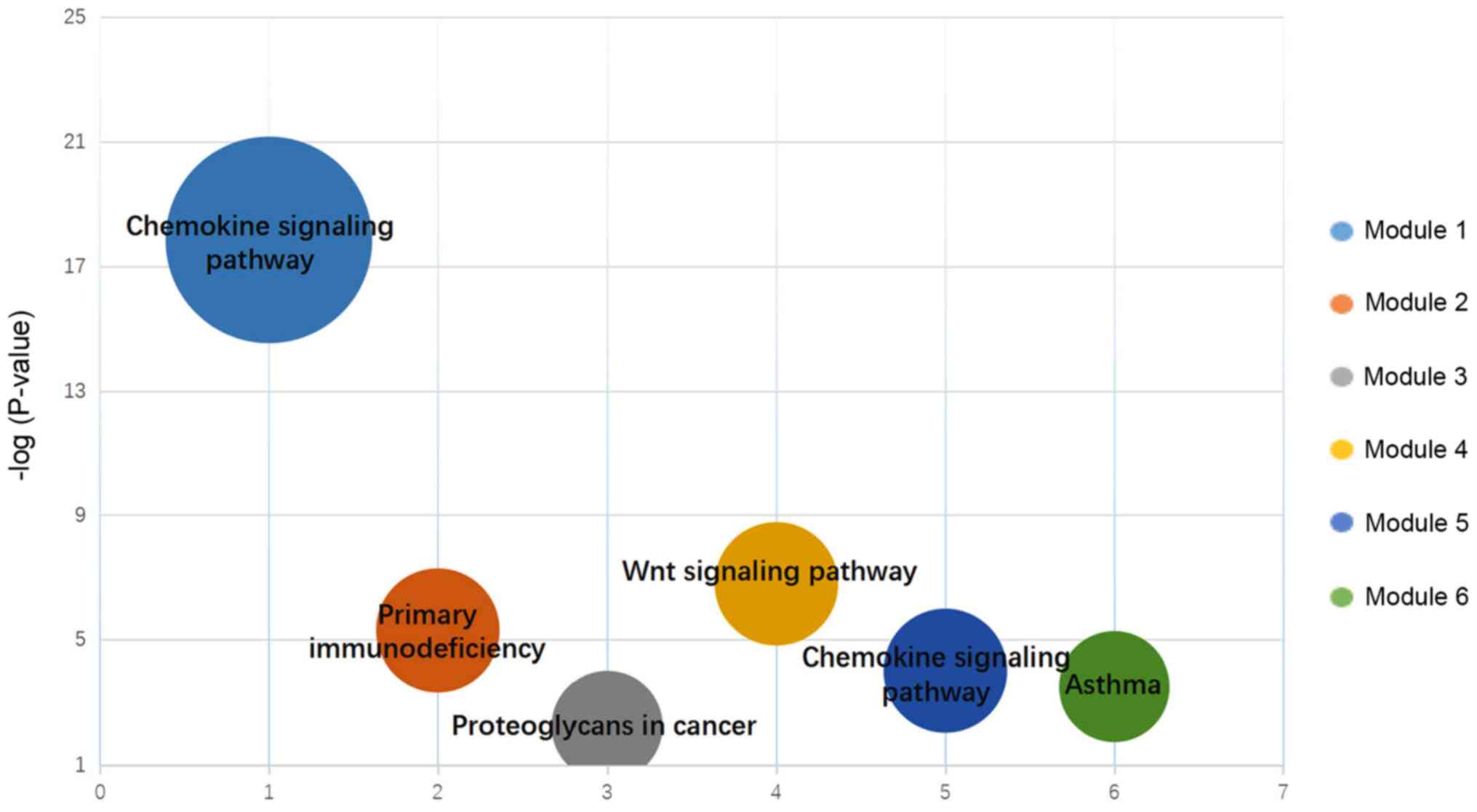
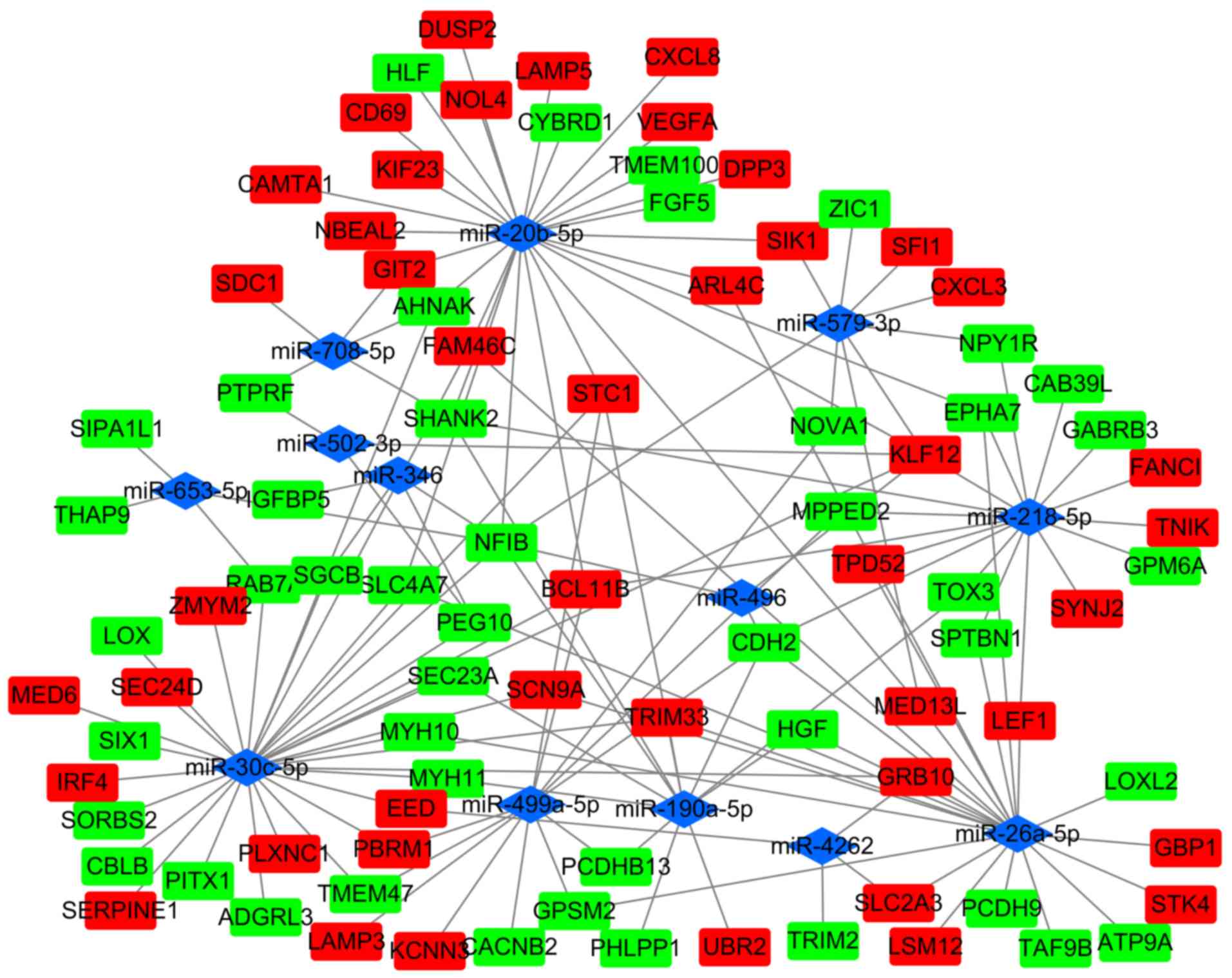
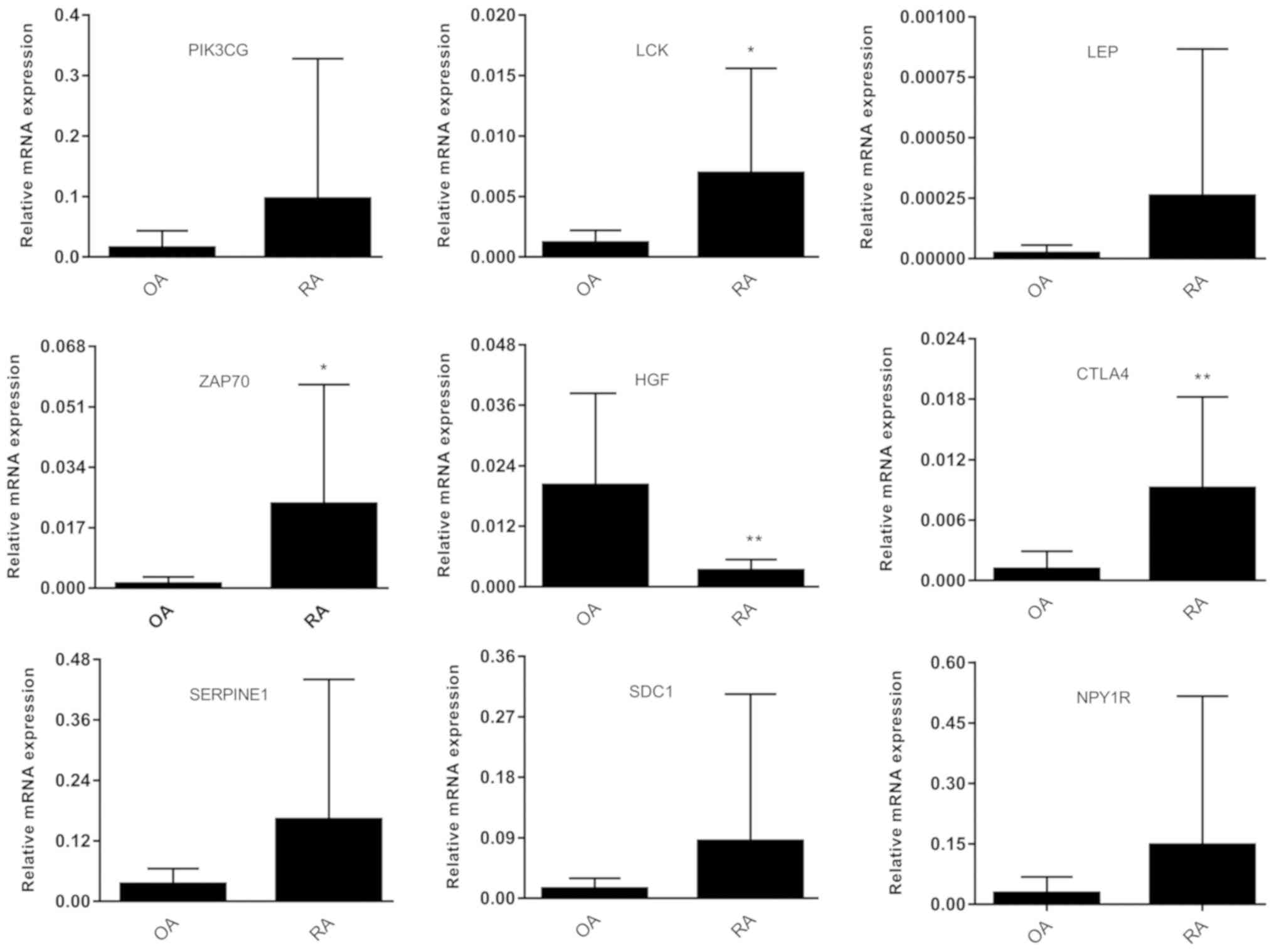
|
1
|
Zhang Q, Dehaini D, Zhang Y, Zhou J, Chen
X and Zhang L, Fang RH, Gao W and Zhang L: Neutrophil
membrane-coated nanoparticles inhibit synovial inflammation and
alleviate joint damage in inflammatory arthritis. Nat Nanotechnol.
13:3513–1190. 2018. View Article : Google Scholar
|
|
2
|
Kikuchi H, Shimada W, Nonaka T, Ueshima S
and Tanaka S: Significance of serine proteinase and matrix
metalloproteinase systems in the destruction of human articular
cartilage. Clin Exp Pharmacol Physiol. 23:885–889. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blits M, Jansen G, Assaraf YG, van de Wiel
MA, Lems WF, Nurmohamed MT, van Schaardenburg D, Voskuyl AE,
Wolbink GJ, Vosslamber S and Verweij CL: Methotrexate normalizes
up-regulated folate pathway genes in rheumatoid arthritis.
Arthritis Rheum. 65:2791–2802. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scott DL, Wolfe F and Huizinga TW:
Rheumatoid arthritis. Lancet. 376:1094–1108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xing R, Yang L, Jin Y, Sun L, Li C, Li Z,
Zhao J and Liu X: Interleukin-21 induces proliferation and
proinflammatory cytokine profile of fibroblast-like synoviocytes of
patients with rheumatoid arthritis. Scand J Immunol. 83:64–71.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zaga-Clavellina V, Parra-Covarrubias A,
Ramirez-Peredo J, Vega-Sanchez R and Vadillo-Ortega F: The
potential role of prolactin as a modulator of the secretion of
proinflammatory mediators in chorioamniotic membranes in term human
gestation. Am J Obstet Gynecol. 211:48.e1–e6. 2014. View Article : Google Scholar
|
|
7
|
Mateen S, Zafar A, Moin S, Khan AQ and
Zubair S: Understanding the role of cytokines in the pathogenesis
of rheumatoid arthritis. Clin Chim Acta. 455:161–171. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kurowska W, Kuca-Warnawin EH, Radzikowska
A and Maslinski W: The role of anti-citrullinated protein
antibodies (ACPA) in the pathogenesis of rheumatoid arthritis. Cent
Eur J Immunol. 42:390–398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sokolove J, Bromberg R, Deane KD, Lahey
LJ, Derber LA, Chandra PE, Edison JD, Gilliland WR, Tibshirani RJ,
Norris JM, et al: Autoantibody epitope spreading in the
pre-clinical phase predicts progression to rheumatoid arthritis.
PLoS One. 7:e352962012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang W, Zhong B, Zhang C, Luo C and Zhan
Y: miR-373 regulates inflammatory cytokine-mediated chondrocyte
proliferation in osteoarthritis by targeting the P2X7 receptor.
FEBS Open Bio. 8:325–331. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Scanzello CR and Goldring SR: The role of
synovitis in osteoarthritis pathogenesis. Bone. 51:249–257. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trachana V, Ntoumou E, Anastasopoulou L
and Tsezou A: Studying microRNAs in osteoarthritis: Critical
overview of different analytical approaches. Mech Ageing Dev.
171:15–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Churov AV, Oleinik EK and Knip M:
MicroRNAs in rheumatoid arthritis: Altered expression and
diagnostic potential. Autoimmun Rev. 14:1029–1037. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Evangelatos G, Fragoulis GE, Koulouri V
and Lambrou GI: MicroRNAs in rheumatoid arthritis: From
pathogenesis to clinical impact. Autoimmun Rev. 18:1023912019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin J, Huo R, Xiao L, Zhu X, Xie J, Sun S,
He Y, Zhang J, Sun Y, Zhou Z, et al: A novel p53/microRNA-22/Cyr61
axis in synovial cells regulates inflammation in rheumatoid
arthritis. Arthritis Rheumatol. 66:49–59. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dunaeva M, Blom J, Thurlings R and Pruijn
G: Circulating serum miR-223-3p and miR-16-5p as possible
biomarkers of early rheumatoid arthritis. Clin Exp Immunol.
193:376–385. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Galligan CL, Baig E, Bykerk V, Keystone EC
and Fish EN: Distinctive gene expression signatures in rheumatoid
arthritis synovial tissue fibroblast cells: Correlates with disease
activity. Genes Immun. 8:480–491. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Woetzel D, Huber R, Kupfer P, Pohlers D,
Pfaff M, Driesch D, Häupl T, Koczan D, Stiehl P, Guthke R and Kinne
RW: Identification of rheumatoid arthritis and osteoarthritis
patients by transcriptome-based rule set generation. Arthritis Res
Ther. 16:R842014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saeed AI, Sharov V, White J, Li J, Liang
W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et
al: TM4: A free, open-source system for microarray data management
and analysis. Biotechniques. 34:374–378. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oliveros JC: Venny. An interactive tool
for comparing lists with Venn's diagrams, 2007. https://bioinfogp.cnb.csic.es/tools/venny/index.html
|
|
22
|
Szklarczyk D, Gable Al, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0:
Service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41((Web Server Issue)): W169–W173. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vejnar CE and Zdobnov EM: MiRmap:
Comprehensive prediction of microRNA target repression strength.
Nucleic Acids Res. 40:11673–11683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Loher P and Rigoutsos I: Interactive
exploration of RNA22 microRNA target predictions. Bioinformatics.
28:3322–3323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
32
|
Buckley L, Guyatt G, Fink HA, Cannon M,
Grossman J, Hansen KE, Humphrey MB, Lane NE, Magrey M, Miller M, et
al: 2017 American college of rheumatology guideline for the
prevention and treatment of glucocorticoid-induced osteoporosis.
Arthritis Rheumatol. 69:1521–1537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Elemam NM, Hannawi S and Maghazachi AA:
Role of chemokines and chemokine receptors in rheumatoid arthritis.
Immunotargets Ther. 9:43–56. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu S, Wu F and Jiang Z: Identification of
hub genes, key miRNAs and potential molecular mechanisms of
colorectal cancer. Oncol Rep. 38:2043–2050. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Orange DE, Agius P, DiCarlo EF, Robine N,
Geiger H, Szymonifka J, McNamara M, Cummings R, Andersen KM, Mirza
S, et al: Identification of three rheumatoid arthritis disease
subtypes by machine learning integration of synovial histologic
features and RNA sequencing data. Arthritis Rheumatol. 70:690–701.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wigerblad G, Bas DB, Fernades-Cerqueira C,
Krishnamurthy A, Nandakumar KS, Rogoz K, Kato J, Sandor K, Su J,
Jimenez-Andrade JM, et al: Autoantibodies to citrullinated proteins
induce joint pain independent of inflammation via a
chemokine-dependent mechanism. Ann Rheum Dis. 75:730–738. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Firestein GS and Mcinnes IB:
Immunopathogenesis of rheumatoid arthritis. Immunity. 46:183–196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kuwabara T, Ishikawa F, Kondo M and
Kakiuchi T: The role of IL-17 and related cytokines in inflammatory
autoimmune diseases. Mediators Inflamm. 2017:39080612017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Siebert S, Tsoukas A, Robertson J and
Mcinnes I: Cytokines as therapeutic targets in rheumatoid arthritis
and other inflammatory diseases. Pharmacol Rev. 67:280–309. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kwon EJ, Park EJ, Choi S, Kim SR, Cho M
and Kim J: PPARγ agonist rosiglitazone inhibits migration and
invasion by downregulating Cyr61 in rheumatoid arthritis
fibroblast-like synoviocytes. Int J Rheum Dis. 20:1499–1509. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fahmi H, Pelletier JP, Di Battista JA,
Cheung HS, Fernandes JC and Martel-Pelletier J: Peroxisome
proliferator-activated receptor gamma activators inhibit MMP-1
production in human synovial fibroblasts likely by reducing the
binding of the activator protein 1. Osteoarthr Cartilage.
10:100–108. 2002. View Article : Google Scholar
|
|
43
|
Li XF, Sun YY, Bao J, Chen X, Li YH, Yang
Y, Zhang L, Huang C, Wu BM, Meng XM and Li J: Functional role of
PPAR-γ on the proliferation and migration of fibroblast-like
synoviocytes in rheumatoid arthritis. Sci Rep. 7:126712017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhebrun DA, Totolyan AA, Maslyanskii AL,
Titov AG, Patrukhin AP, Kostareva AA and Gol'tseva IS: Synthesis of
some CC chemokines and their receptors in the synovium in
rheumatoid arthritis. Bull Exp Biol Med. 158:192–196. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee AY and Körner H: CCR6 and CCL20:
Emerging players in the pathogenesis of rheumatoid arthritis.
Immunol Cell Biol. 92:354–358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Su CM, Hsu CJ, Tsai CH, Huang CY, Wang SW
and Tang CH: Resistin promotes angiogenesis in endothelial
progenitor cells through inhibition of MicroRNA206: Potential
implications for rheumatoid arthritis. Stem Cells. 33:2243–2255.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kraan MC, Patel DD, Haringman JJ, Smith
MD, Weedon H, Ahern MJ, Breedveld FC and Tak PP: The development of
clinical signs of rheumatoid synovial inflammation is associated
with increased synthesis of the chemokine CXCL8 (interleukin-8).
Arthritis Res. 3:65–71. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tanida S, Yoshitomi H, Nishitani K,
Ishikawa M, Kitaori T, Ito H and Nakamura T: CCL20 produced in the
cytokine network of rheumatoid arthritis recruits CCR6+ mononuclear
cells and enhances the production of IL-6. Cytokine. 47:112–118.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lisignoli G, Piacentini A, Cristino S,
Grassi F, Cavallo C, Cattini L, Tonnarelli B, Manferdini C and
Facchini A: CCL20 chemokine induces both osteoblast proliferation
and osteoclast differentiation: Increased levels of CCL20 are
expressed in subchondral bone tissue of rheumatoid arthritis
patients. J Cell Physiol. 210:798–806. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kawashiri SY, Kawakami A, Iwamoto N,
Fujikawa K, Aramaki T, Tamai M, Arima K, Kamachi M, Yamasaki S,
Nakamura H, et al: Proinflammatory cytokines synergistically
enhance the production of chemokine ligand 20 (CCL20) from
rheumatoid fibroblast-like synovial cells in vitro and serum CCL20
is reduced in vivo by biologic disease-modifying antirheumatic
drugs. J Rheumatol. 36:2397–2402. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Luterek-Puszyńska K, Malinowski D,
Paradowska-Gorycka A, Safranow K and Pawlik A: CD28, CTLA-4 and
CCL5 gene polymorphisms in patients with rheumatoid arthritis. Clin
Rheumatol. 36:1129–1135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Agere SA, Akhtar N, Watson JM and Ahmed S:
RANTES/CCL5 induces collagen degradation by activating MMP-1 and
MMP-13 expression in human rheumatoid arthritis synovial
fibroblasts. Front Immunol. 8:13412017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Toyoda Y, Tabata S, Kishi J, Kuramoto T,
Mitsuhashi A, Saijo A, Kawano H, Goto H, Aono Y, Hanibuchi M, et
al: Thymidine phosphorylase regulates the expression of CXCL10 in
rheumatoid arthritis fibroblast-like synoviocytes. Arthritis
Rheumatol. 66:560–568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Laragione T, Brenner M, Sherry B and Gulko
PS: CXCL10 and its receptor CXCR3 regulate synovial fibroblast
invasion in rheumatoid arthritis. Arthritis Rheum. 63:3274–3283.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kuan WP, Tam LS, Wong CK, Ko FW, Li T, Zhu
T and Li EK: CXCL 9 and CXCL 10 as Sensitive markers of disease
activity in patients with rheumatoid arthritis. J Rheumatol.
37:257–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sasaki T, Irie-Sasaki J, Jones RG,
Oliveira-dos-Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A,
Bouchard D, Kozieradzki I, et al: Function of PI3Kgamma in
thymocyte development, T cell activation, and neutrophil migration.
Science. 287:1040–1046. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hayer S, Pundt N, Peters MA, Wunrau C,
Kühnel I, Neugebauer K, Strietholt S, Zwerina J, Korb A, Penninger
J, et al: PI3Kgamma regulates cartilage damage in chronic
inflammatory arthritis. FASEB J. 23:4288–4298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Camps M, Rückle T, Ji H, Ardissone V,
Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Françon B,
et al: Blockade of PI3Kgamma suppresses joint inflammation and
damage in mouse models of rheumatoid arthritis. Nat Med.
11:936–943. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kumar Singh P, Kashyap A and Silakari O:
Exploration of the therapeutic aspects of Lck: A kinase target in
inflammatory mediated pathological conditions. Biomed Pharmacother.
108:1565–1571. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Farag AK, Elkamhawy A, Londhe AM, Lee KT,
Pae AN and Roh EJ: Novel LCK/FMS inhibitors based on
phenoxypyrimidine scaffold as potential treatment for inflammatory
disorders. Eur J Med Chem. 141:657–675. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xiong Y, Mi BB, Liu MF, Xue H, Wu QP and
Liu GH: Bioinformatics analysis and identification of genes and
molecular pathways involved in synovial inflammation in rheumatoid
arthritis. Med Sci Monitor. 25:2246–2256. 2019. View Article : Google Scholar
|
|
62
|
Procaccini C, Pucino V, Mantzoros CS and
Matarese G: Leptin in autoimmune diseases. Metabolism. 64:92–104.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
La Cava A: Leptin in inflammation and
autoimmunity. Cytokine. 98:51–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Batún-Garrido JAJ, Salas-Magaña M,
Juárez-Rojop IE, Hernández-Núñez E and Olán F: Relationship between
leptin concentrations and disease activity in patients with
rheumatoid arthritis. Med Clin (Barc). 150:341–344. 2018.(In
English, Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tian G, Liang JN, Wang ZY and Zhou D:
Emerging role of leptin in rheumatoid arthritis. Clin Exp Immunol.
177:557–570. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Toussirot É, Michel F, Binda D and
Dumoulin G: The role of leptin in the pathophysiology of rheumatoid
arthritis. Life Sci. 140:29–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Toussirot E, Grandclément E, Gaugler B,
Michel F, Wendling D, Saas P and Dumoulin G; CBT-506: Serum
adipokines and adipose tissue distribution in rheumatoid arthritis
and ankylosing spondylitis. A comparative study. Front Immunol.
4:4532013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gremese E, Tolusso B, Fedele AL, Canestri
S, Alivernini S and Ferraccioli G: ZAP-70+ B cell subset influences
response to B cell depletion therapy and early repopulation in
rheumatoid arthritis. J Rheumatol. 39:2276–2285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kugyelka R, Prenek L, Olasz K, Kohl Z,
Botz B, Glant TT, Berki T and Boldizsár F: ZAP-70 regulates
autoimmune arthritis via alterations in T cell activation and
apoptosis. Cells. 8(pii): E5042019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Romo-Tena J, Gómez-Martín D and
Alcocer-Varela J: CTLA-4 and autoimmunity: New insights into the
dual regulator of tolerance. Autoimmun Rev. 12:1171–1176. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Molnarfi N, Benkhoucha M, Funakoshi H,
Nakamura T and Lalive PH: Hepatocyte growth factor: A regulator of
inflammation and autoimmunity. Autoimmun Rev. 14:293–303. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pavón MA, Arroyo-Solera I, Céspedes MV,
Casanova I, León X and Mangues R: uPA/uPAR and SERPINE1 in head and
neck cancer: Role in tumor resistance, metastasis, prognosis and
therapy. Oncotarget. 7:57351–57366. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Deng W, Feng X, Li X, Wang D and Sun L:
Hypoxia-inducible factor 1 in autoimmune diseases. Cell Immunol.
303:7–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Teng YH, Aquino RS and Park PW: Molecular
functions of syndecan-1 in disease. Matrix Biol. 31:3–16. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jaiswal AK, Sadasivam M and Hamad ARA:
Unexpected alliance between syndecan-1 and innate-like T cells to
protect host from autoimmune effects of interleukin-17. World J
Diabetes. 9:220–225. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bedoui S, Miyake S, Lin Y, Miyamoto K, Oki
S, Kawamura N, Beck-Sickinger A, von Hörsten S and Yamamura T:
Neuropeptide Y (NPY) suppresses experimental autoimmune
encephalomyelitis: NPY1 receptor-specific inhibition of
autoreactive Th1 responses in vivo. J Immunol. 171:3451–3458. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kang X, Qian Z, Liu J, Feng D, Li H, Zhang
Z, Jin X, Ma Z, Xu M, Li F, et al: Neuropeptide Y acts directly on
cartilage homeostasis and exacerbates progression of osteoarthritis
through NPY2R. J Bone Miner Res. Feb 26–2020.(Epub ahead of print).
View Article : Google Scholar
|
|
78
|
Liu L, Xu Q, Cheng L, Ma C, Xiao L, Xu D,
Gao Y, Wang J and Song H: NPY1R is a novel peripheral blood marker
predictive of metastasis and prognosis in breast cancer patients.
Oncol Lett. 9:891–896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kara F, Yildirim A, Gumusdere M, Karatay
S, Yildirim K and Bakan E: Association between hepatocyte growth
factor (HGF) gene polymorphisms and serum HGF levels in patients
with rheumatoid arthritis. Eurasian J Med. 46:176–185. 2014.
View Article : Google Scholar : PubMed/NCBI
|