Introduction
Aristolochic acid (AA) is found in plants of the
genera Aristolochia and Asarum (1). Plants of the Aristolochia
genus are commonly used as herbal medicines in Asia, Central
America and other countries (2).
DNA damage and/or carcinogenicity is caused by AA derived-DNA
adducts after AA exposure, and death or apoptosis of renal tubular
cells is caused by AA-induced DNA damage (3,4). AA
can cause aristolochic acid nephropathy (AAN), leading to acute
kidney injury and/or chronic kidney disease (CKD), accompanied by
tubulointerstitial fibrosis (5,6).
Additionally, AA is a carcinogen associated with urothelial
tumorigenesis following sufficient long-term exposure, as
carcinogenicity is caused by AA derived-DNA adducts after AA
exposure (6).
In acute AAN, AA induces renal injury and
tubulointerstitial fibrosis (5).
Fibrosis is one common final outcome of several kidney diseases
(7). Renal tubulointerstitial
fibrosis is a complex biological process involved in multiple
cellular and signaling pathways. Moreover, fibrosis shares common
pathways in several different organs (8). The progression of renal fibrosis
primarily includes renal injury, recruitment and activation of
immune cells, activation of fibroblasts, epithelial-to-mesenchymal
transition (EMT), extracellular matrix (ECM) production and
deposition, and tubular injury and atrophy (7). Pro-fibrosis factors, such as
transforming growth factor-β 1 (TGF-β1), are key molecular
mediators of renal fibrosis (9,10).
MicroRNAs (miRNAs/miRs) are a class of small,
endogenous, non-coding RNAs that silence target mRNAs (11). miRNAs bind to their target mRNAs
through complementary nucleotide sequences, and the target gene is
suppressed by a miRNA through degradation and/or translation
inhibition of its target mRNA (12). It is suggested that miRNAs serve a
key role in renal fibrosis (13).
For example, miRNAs regulate the effects of pro-fibrosis factors,
EMT and ECM production (14).
To the best of our knowledge, the expression
profiling of miRNAs and mRNAs, and their regulatory pairs or
networks has not been reported in acute AAN. To identify the
expression profiles and regulatory networks of miRNAs and mRNAs in
acute AAN, the present study examined renal genome-wide
differentially expressed (DE) miRNAs and DE mRNAs using deep
sequencing in mouse kidneys induced by a short-term exposure to
AA.
Materials and methods
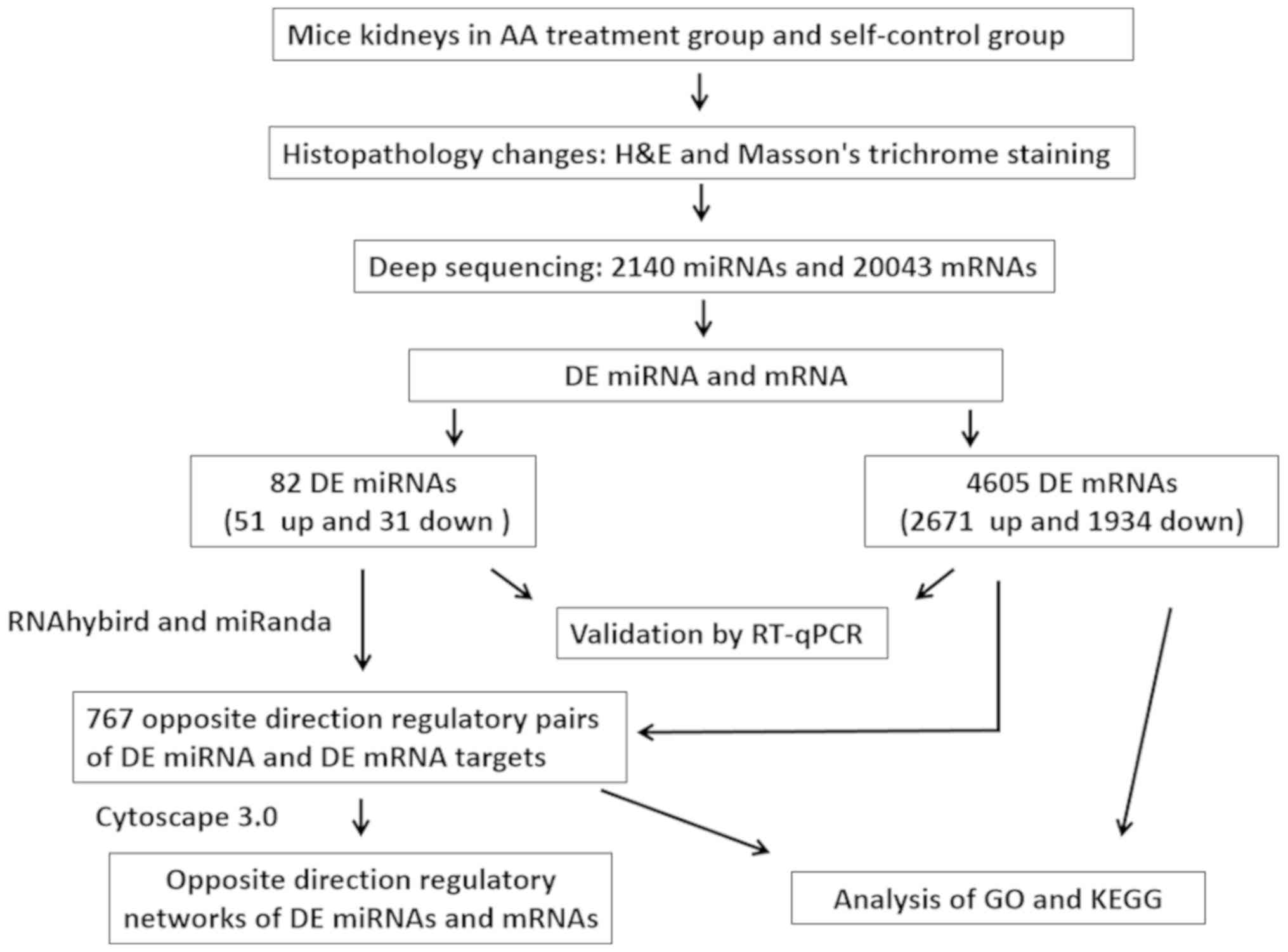
The flow chart of the experimental design of the
present study is shown in Fig.
1.
Animal treatment
A total of four kidney samples were used as the
self-control group, and were obtained through single nephrectomy
from four 8-week-old male C57BL/6 mice (specific pathogen free;
weight, ~20 g; Experimental Animal Center of Soochow University).
The mice were raised in a specific pathogen free animal house with
a constant temperature (24°C), humidity (50%), specific pathogen
free filtered atmosphere and 12 h light/dark cycles. All the mice
had free access and were fed with radiation sterilization food and
water sterilized with high temperature and high pressure. The mice
were anesthetized using chloral hydrate (400 mg/kg intraperitoneal
injection) before single nephrectomy. After 4 weeks, these four
mice were treated with an intraperitoneal injection of AA I (cat.
no. A5512; Sigma-Aldrich; Merck KGaA) at a dosage of 5 mg/kg/day
for 5 days. A total of 100 mg AA I was diluted with 5 ml DMSO
(final concentration, 5%) and 95 ml saline. The final concentration
of the injected AA I was 1 mg/ml. All mice were euthanized by
cervical dislocation. The death of mice was verified by the
stopping of both breathing and the heartbeat. The remaining four
kidney samples were obtained when these four mice were sacrificed
on the 6th day. All kidney samples were divided into two parts, one
of which was fixed in 10% formalin solution for 5 days at room
temperature, while the other was frozen at −80°C in
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The present study was approved by the Ethics Committee of
the Children's Hospital of Soochow University (Suzhou, China;
approval no. 2020-CHSU-012).
Histological analysis
After fixing in 10% formalin solution for ≥3 days at
26°C, all kidney samples were dehydrated and transparent as
follows: 70% alcohol for 1 h, 80% ethanol for 1 h, 90% ethanol for
1 h, 95% ethanol for 1 h, anhydrous ethanol twice for 1 h,
anhydrous ethanol and xylene solution (1:1) for 1 h and xylene
twice for 1 h. The kidney samples were embedded in paraffin and
sectioned into 5-µm-thick slices. The slices were placed in at 60°C
for 30 min, dewaxed after being immersed in xylene twice for 10 min
each time and then immersed in ethanol (100, 95, 80 and 70%) and
deionized water for 5 min. Sections of the kidney samples were
stained with hematoxylin and eosin (HE) or Masson's trichrome. HE
staining were performed as follow: Hematoxylin dye for 10 min at
26°C, rinsed with water and 0.7% hydrochloric acid ethanol for 5
sec. Cells were then washed with water until the cell nuclei were
blue, along with 95% ethanol for 30 sec, alcoholic eosin dye for 30
sec at 26°C, 95% ethanol twice for 30 sec, 100% ethanol twice for 5
min, carbolic Acid and xylene (1:4) for 5 min, xylene twice for 5
min and the slices were sealed with neutral balsam.
Masson's trichrome staining was performed as follow:
Staining with Regaud's hematoxylin dye for 10 min at 26°C, rinsed
with unstained color with water, 0.7% hydrochloric acid ethanol for
5 sec, washed with water for 8 min, ponceaux and acid fuchsin (0.7%
ponceaux, 0.3% acid fuchsin and 1% acetic acid) for 8 min, 2%
acetic acid solution for 45 sec, 1% phosphomolybdic acid for 4 min,
aniline blue for 5 min and 0.2% acetic acid until no obvious blue
color escaped from the slices. Subsequently, slices were rinsed
with 95% ethanol twice for 30 sec, 100% ethanol twice for 5 min and
xylene twice for 5 min, and then sealed with neutral balsam. The
slices were observed and captured at ×400 magnification under a
light microscope.
RNA extraction
Total RNA was extracted from the kidney samples
using TRIzol® reagent according to the manufacturer's
protocol. Total RNA was qualified and quantified using NanoDrop and
an Agilent 2100 bioanalyzer (both from Thermo Fisher Scientific,
Inc.).
Sequencing of miRNAs
Small RNAs from the total RNA were obtained from the
18–30 nt region following 15% urea polyacrylamide gel
electrophoresis (urea-PAGE) gel electrophoresis. After the addition
of 3′ and 5′ adaptors (MGIEasy Small RNA Library Prep Kit V2.0; BGI
Genomics), the cDNA of small RNAs was synthesized and enriched by
reverse transcription and PCR amplification. The reverse
transcription was performed using the First Strand Master Mix and
Superscript II Reverse Transcriptase (SuperScript™ First-Strand
Synthesis System; Invitrogen; Thermo Fisher Scientific, Inc.) at
42°C for 1 h and 70°C for 15 min. PCR was performed using PCR
Primer Cocktail and PCR Master Mix (MGIEasy Small RNA Library Prep
Kit V2.0; BGI Genomics) to enrich the cDNA fragments, under the
following conditions: Initial denaturation at 95°C for 3 min,
followed by 18 cycles at 98°C for 20 sec, 56°C for 15 sec and 72°C
for 15 sec, with a final extension at 72°C for 10 min. The library
of small RNAs was constructed from the PCR products in the 100–120
bp region, separated by agarose gel electrophoresis, eliminating
primer-dimers and other potential byproducts. The library of small
RNAs was denatured by heat (94°C), and the single-stranded DNA was
cyclized. The library of cyclized small RNAs was sequenced using a
BGISEQ-500 platform (BGI Genomics). The clean data was filtered
from the raw data. The clean reads of miRNAs were mapped to the
mouse genome and other sRNAs database with annotation using the
software Anchor Alignment-Based Small RNA Annotation version 1.0
(University of Nevada) (15),
except Rfam version 12.0, which was performed using cmsearch
version 1.1 (16).
Sequencing of mRNAs
mRNA was selected using poly-T oligo-attached
magnetic beads, and ribosomal RNA was depleted. mRNA was fragmented
and reverse transcribed into double-stranded cDNA using N6 random
primers. The reverse transcription was performed using the First
Strand Master Mix, N6 random primers and Superscript II Reverse
Transcriptase (SuperScript™ First-Strand Synthesis System;
Invitrogen; Thermo Fisher Scientific, Inc.) at 42°C for 1 h and
70°C for 15 min. The dscDNA was subjected to end-repair and
3′-adenylation. A bubble adapter was ligated to the double ends of
the dscDNA. After purifying, the ligated products were amplified by
PCR. PCR was performed using PCR Primer Cocktail and PCR Master Mix
(MGIEasy RNA Library Prep Kit V3.0; BGI Genomics) to enrich the
cDNA fragments, under the following conditions: Initial
denaturation at 95°C for 3 min, followed by 18 cycles at 98°C for
20 sec, 56°C for 15 sec and 72°C for 15 sec; with a final extension
at 72°C for 10 min. The PCR products were denatured by heat (94°C)
and the single-stranded DNA was cyclized using splint oligo and DNA
ligase. The cyclized mRNA was sequenced using the BGISEQ-500
platform. The clean mRNA data were filtered from the raw data and
mapped to the mouse genome with annotation using HISAT version
2.0.4 (Center for Computational Biology; Johns Hopkins University)
(17). A detailed explanation of
the methods of miRNA and mRNA sequencing and analysis is provided
in Data S1.
Functional analysis of the DE miRNAs
and DE mRNAs
Hierarchical clustering analysis was performed
between all 8 kidney samples using function hclust of R software
version 3.5.2 (R Core Team). Principal component analysis (PCA) was
performed with all 8 kidney samples using function princomp of R
software version 3.5.2 (R Core Team). Between the AA-treated group
and the self-control group, DE miRNAs were defined using a cut-off
with read count >50, adjusted P-values <0.001 and
log2 fold changes (AA/control) >1.5 or <-1.5. DE
mRNAs were defined using a cut-off with read count >50, adjusted
P-values <0.001 and log2 fold changes (AA/control)
>1.0 or <-1.0. These cut-off points were used to assist in
identifying the key DE miRNAs and DE mRNAs enriched in biological
processes and pathways, which may be involved in the
pathophysiology of acute AAN.
Target genes of miRNAs were identified based on the
similarity of the sequences with target genes, predicted using
RNAhybrid version 2.2 (18) and
miRanda (version released in 2010) (19). The functions and pathways analysis
of both DE mRNAs and the target genes of DE miRNAs were performed
using Gene Ontology (GO) (20) and
the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases
(21). Networks of DE miRNAs and
DE mRNAs were drawn using Cytoscape version 3.0 (National Institute
of General Medical Sciences).
Reverse transcription-quantitative
(RT-q) PCR
To validate the DE miRNA and DE mRNA results,
RT-qPCR was performed on reference DE miRNAs and DE mRNAs from the
total RNA of all 8 kidney samples extracted as aforementioned.
RT-qPCR assays were performed using a Light Cycler 480 II real-time
PCR system (Roche Diagnostics). U6 and GAPDH were used as the
internal controls for miRNA and mRNA, respectively. The M-MLV
Reverse Transcriptase kit (Promega Corp.) was used for the
synthesis of cDNA, using the following conditions: 42°C for 1 h and
70°C for 10 min. The cDNA products of mRNA were amplified using
SYBR Premix Ex Taq kit (Takara Bio, Inc.) using the following
thermocycling conditions: 95°C for 30 sec; followed by 40 cycles of
95°C for 5 sec and 60°C for 30 sec, and a final cycle of 95°C for
15 sec, 60°C for 30 sec and 95°C for 15 sec. All the primers
(Shanghai GeneChem Co., Ltd.) for detection of mRNAs are listed in
Table I. The RT-qPCR reactions for
all miRNAs were performed using a miDETECTA Track™ miRNA qRT-PCR
Starter kit (Guangzhou RiboBio Co., Ltd.) according to the
manufacturer's protocol. RT-qPCR was performed in triplicate. The
expression levels of the selected miRNAs and mRNAs were normalized
to those of U6 and GAPDH, respectively. The changes in expression
of the assessed miRNAs and mRNAs between the AA treatment group and
the self-control group were calculated using the 2−ΔΔCq
method (22).
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| mRNAs | Forward
(5′-3′) | Reverse
(3′-5′) |
|---|
| GAPDH |
TGGTGAAGGTCGGTGTGAAC |
GCTCCTGGAAGATGGTGATGG |
| TGF-β1 |
AGCCTGCCTCTTGAGTCCCT |
CTCCCAAGGAAAGGTAGGTGAT |
| IL-11 |
GCTGGGACATTGGGATCTTTG |
GAGCTGTAAACGGCGGAGTAG |
| Timp1 |
CCCCAGAAATCAACGAGACC |
GTACGCCAGGGAACCAAGAA |
| Serpine1 |
GACACCCTCAGCATGTTCATC |
TTGGTCGGAAAGACTTGTGAA |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| miR-21a-5p |
GUACUUAUCAGACUGAUGUUGA |
UCAACAUCAGUCUGAUAAGUUA |
| miR-124-3p |
TCTTTAAGGCACGCGGTG |
TATGGTTTTGACGACTGTGTGAT |
| miR-132-3p |
GCGCGCGTAACAGTCTACAGG |
GTCGTATCCAGTGCAGGGTCC |
| miR-1968-5P |
TGCAGCTGTTAAGGATGGTGGACT |
GCGAGCACAGAATTAATACGAC |
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Statistical analysis was performed using GraphPad Prism
version 8.0 (GraphPad Software, Inc.). Statistical differences were
calculated using a paired Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Renal histopathological alterations
induced by AA
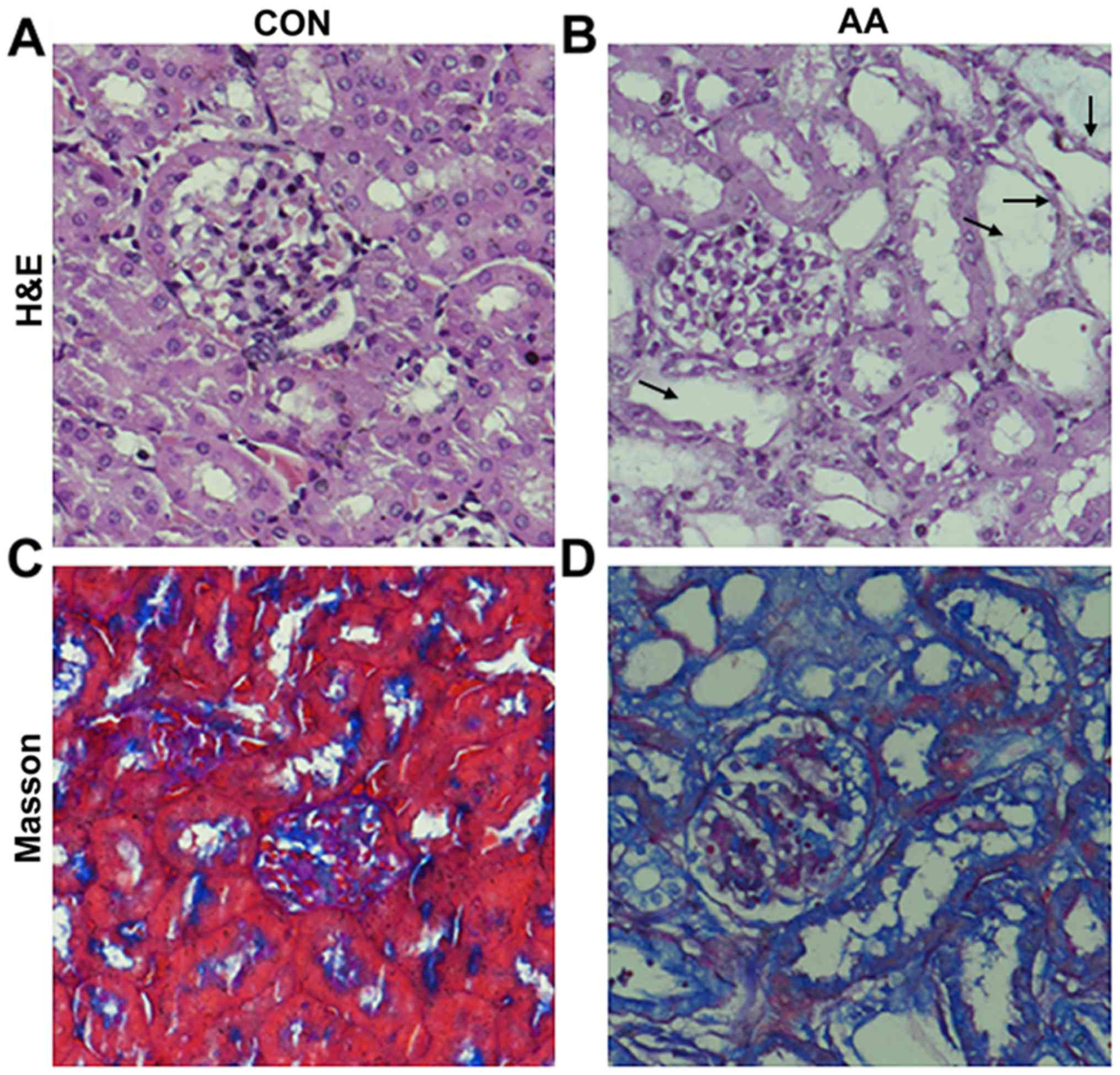
Histopathological changes in the kidney samples were
detected using H&E and Masson's trichrome staining. There were
markedly widened tubular lumens and flattened tubular cells in the
AA-treated group compared with in the self-control group (Fig. 2A-D). Tubulointerstitial fibrosis
was induced by AA treatment, as shown by the observable increase in
collagen based on the Masson's trichrome staining in the AA-treated
group compared with in the self-control group (Fig. 2C and D).
Renal DE mRNAs induced by AA
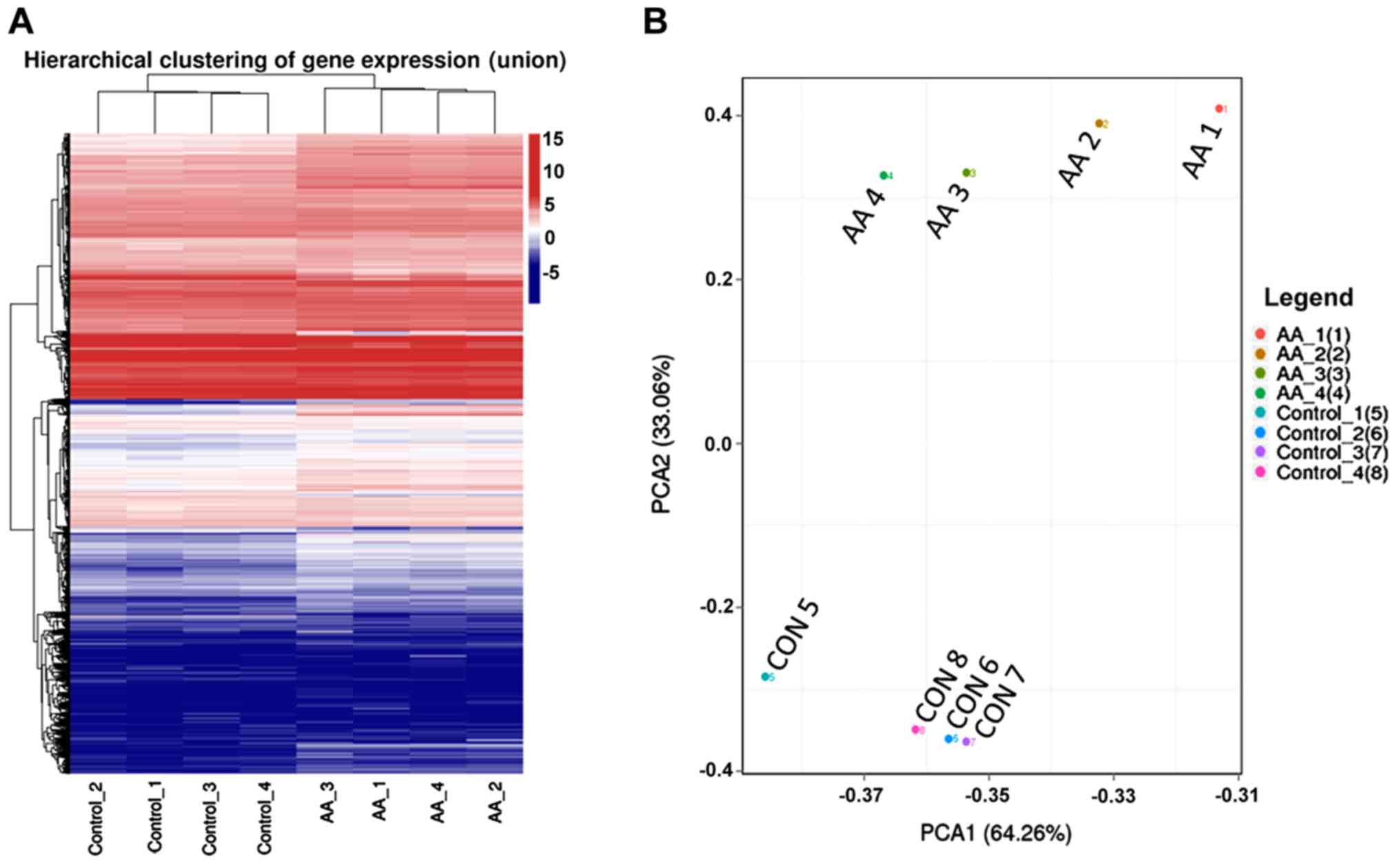
A total of 20,043 mRNAs were detected using deep
sequencing in the 8 kidney samples. The mRNA expression patterns
were significantly different between the AA-treated group and the
self-control group based on hierarchical clustering analysis and
PCA (Fig. 3).
Among the total detected mRNAs, 4,605 were
considered to be significantly DE between the AA-treated group and
the self-control group. Of these, 2,671 DE mRNAs were upregulated
and 1,934 were downregulated.
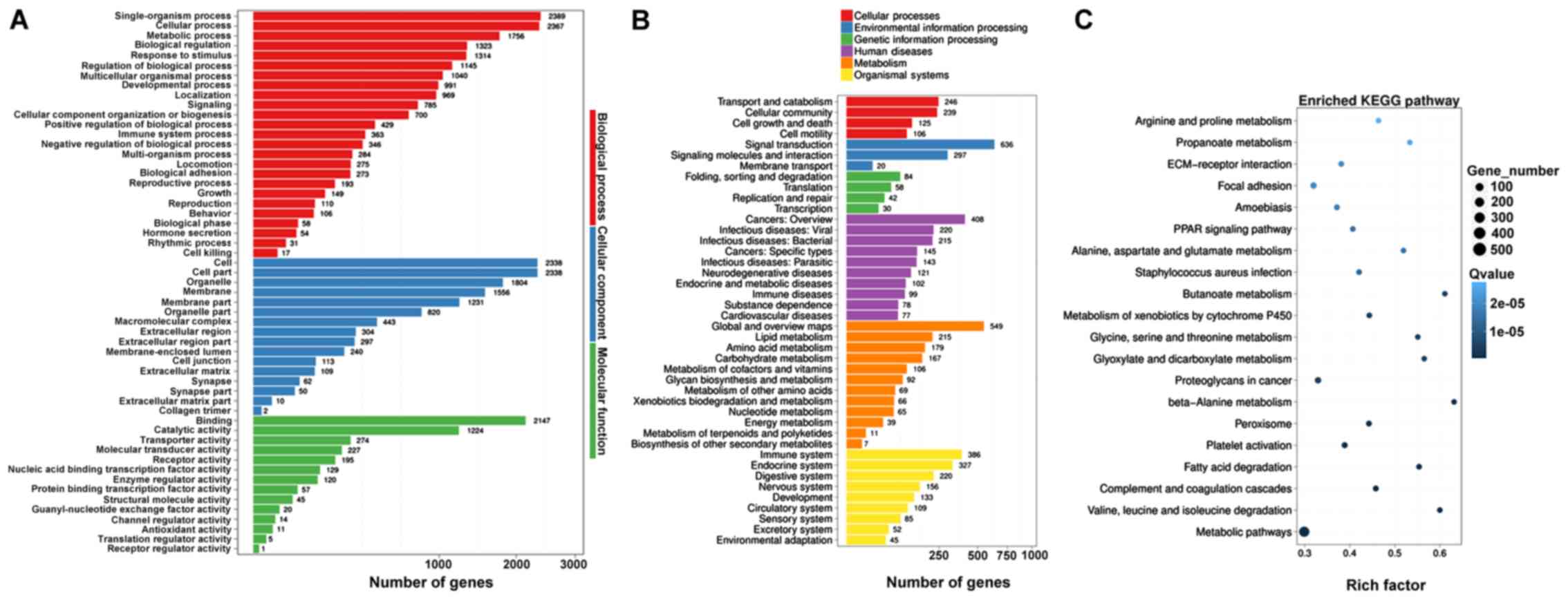
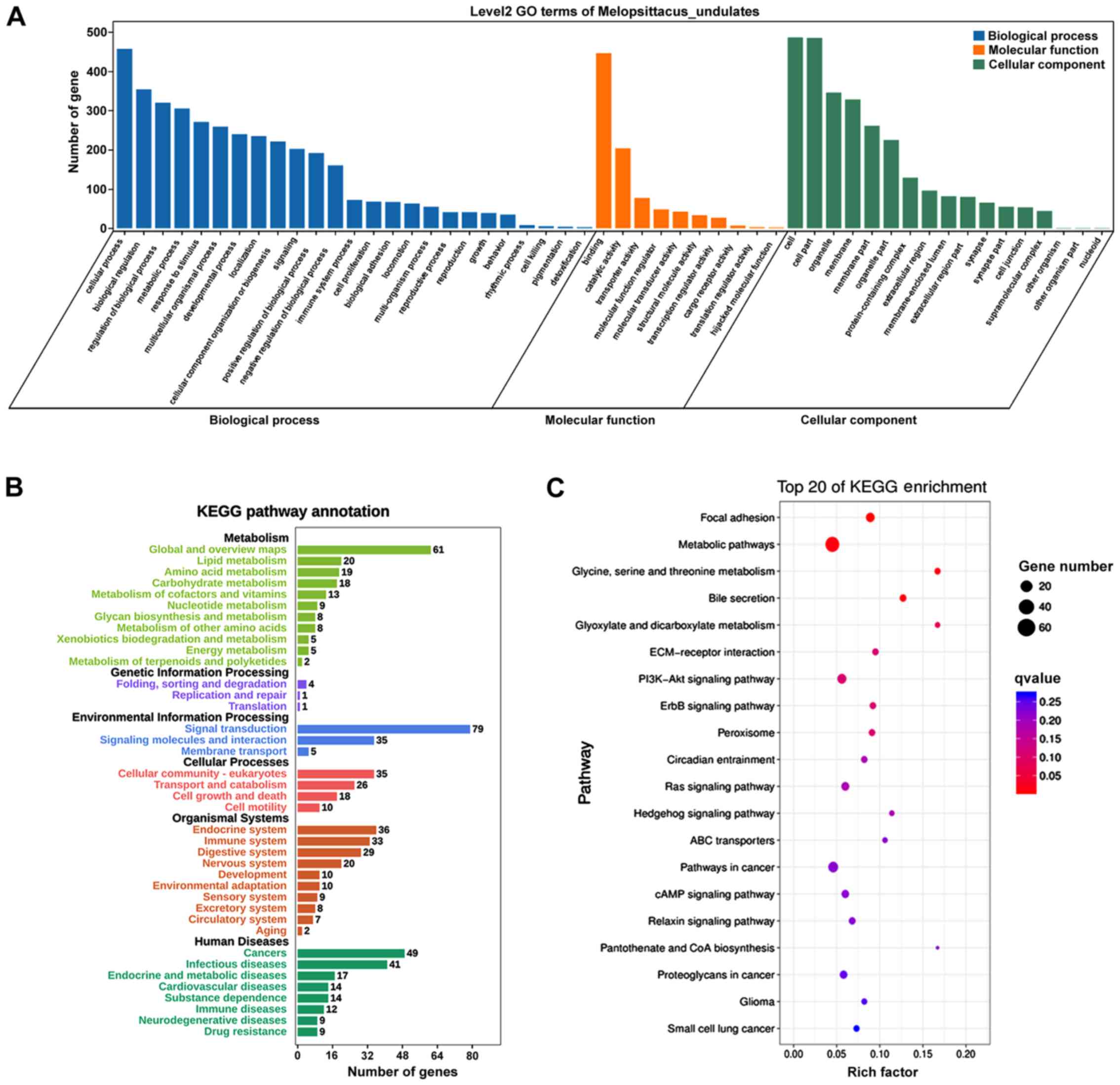
AA treatment induced extensive alterations in
cellular function and processes based on the results of GO and KEGG
pathway analysis on DE mRNAs. GO analysis of DE mRNAs identified
‘single-organism process’ and ‘cellular process’ as the most
altered biological process, ‘cell’ and ‘cell part’ as the most
altered cellular component, and ‘binding’ and ‘catalytic activity’
as the most altered molecular function (Fig. 4A). KEGG analysis identified ‘signal
transduction’, ‘global and overview maps’, ‘cancers: Overview’,
‘immune system’ and ‘endocrine system’ as the most altered pathways
(Fig. 4B). Furthermore, ‘amino
acid metabolism’, ‘fatty acid degradation’, ‘complement and
coagulation cascades’ and ‘ECM-receptor interaction’ were the most
enriched pathways (Fig. 4C). The
‘amino acid metabolism’ included all types of amino acid
metabolism, such as ‘Arginine and proline metabolism’, ‘Alanine,
Aspartic acid and glutamate metabolism’, ‘glycine, serine and
threonine metabolism’ and ‘Valine, leucine and isoleucine
degradation’.
Renal DE miRNAs induced by AA
A total of 2,140 miRNAs were detected by deep
sequencing. Among these, 82 DE miRNAs were identified between the
AA-treated and the self-control group (Table SI). Of these, 51 DE miRNAs were
upregulated in the AA treatment group compared with in the
self-control group, and 31 DE miRNAs were downregulated.
The top 10 upregulated miRNAs in this study were
miR-124-5p, miR-122-3p, miR-124-3p, miR-344d-3p, miR-409-5p,
miR-129-2-3p, miR-496a-3p, miR-147-3p, miR-5128 and miR-7649-3p
(Table SI). Among the 51
upregulated DE miRNAs, miRNAs with the highest expression levels in
the AA treatment group were miR-21a-5p, miR-31-5p, miR-21a-3p,
miR-146b-5p, miR-212-3p, miR-34b-3p, miR-132-3p, miR-34c-3p and
miR-34c-5p. The 10 most downregulated miRNAs were miR-3073a-5p,
miR-3073b-3p, miR-3073b-5p, miR-1948-5p, miR-92a-2-5p,
miR-3073a-3p, miR-669c-5p, miR-7085-5p, miR-1968-5p and miR-1948-3p
(Table SI). Among the 31
downregulated DE miRNAs, the highest expressed miRNAs in the
self-control group were miR-187-3p, miR-190a-5p, miR-122-5p,
miR-486a-5p and let-7a-1-3p.
The profibrotic miR-21, miR-433 and miR-132
families, and the oncogenic miR-34 family were significantly
increased, while the anti-fibrotic miR-122-5p and let-7a-1-3p were
significantly decreased following AA treatment.
Validation of DE miRNAs and DE
mRNAs
To validate the DE miRNAs and DE mRNAs, RT-qPCR was
performed on miR-21a-5p, miR-324-3p, miR-132-3p, miR-1968-5p, TIMP
metallopeptidase inhibitor 1 (Timp1), serpin family E member 1
(Serpine1), interleukin (IL)-11 and TGF-β1. These miRNA and mRNA
were chosen as they were among the most altered mRNAs and miRNAs,
which also had high expression levels (high read counts). Moreover,
the chosen mRNAs are pro-fibrosis factors (23,24).
As shown in Fig. 5, changes in
expression levels of the assessed miRNAs and mRNAs based on RT-qPCR
were consistent with the results of deep sequencing (Table SI). Relative expression levels of
miR-21a-5p (3.90±0.90 vs. 1.00±0.08), miR-324-3p (2.87±0.62 vs.
1.00±0.13) and miR-132-3p (3.81±0.77 vs. 1.00±0.25) were
significantly increased in the AA-treated group compared with that
in the self-control group, respectively, and the expression level
of miR-1968-5p (0.23±0.02 vs. 1.00±0.07) was significantly
decreased (Fig. 5A-D). Relative
mRNA levels of Timp1 (304.24±88.65 vs. 1.00±0.56), Serpine1
(92.07±44.17 vs. 1.00±0.56), IL-11 (23.25±1.48 vs. 1.00±0.47) and
transforming growth factor (TGF)-β1 (3.85±0.62 vs. 1.00±0.16) were
significantly increased in the AA-treated group compared with that
in the self-control group (Fig.
5E-H).
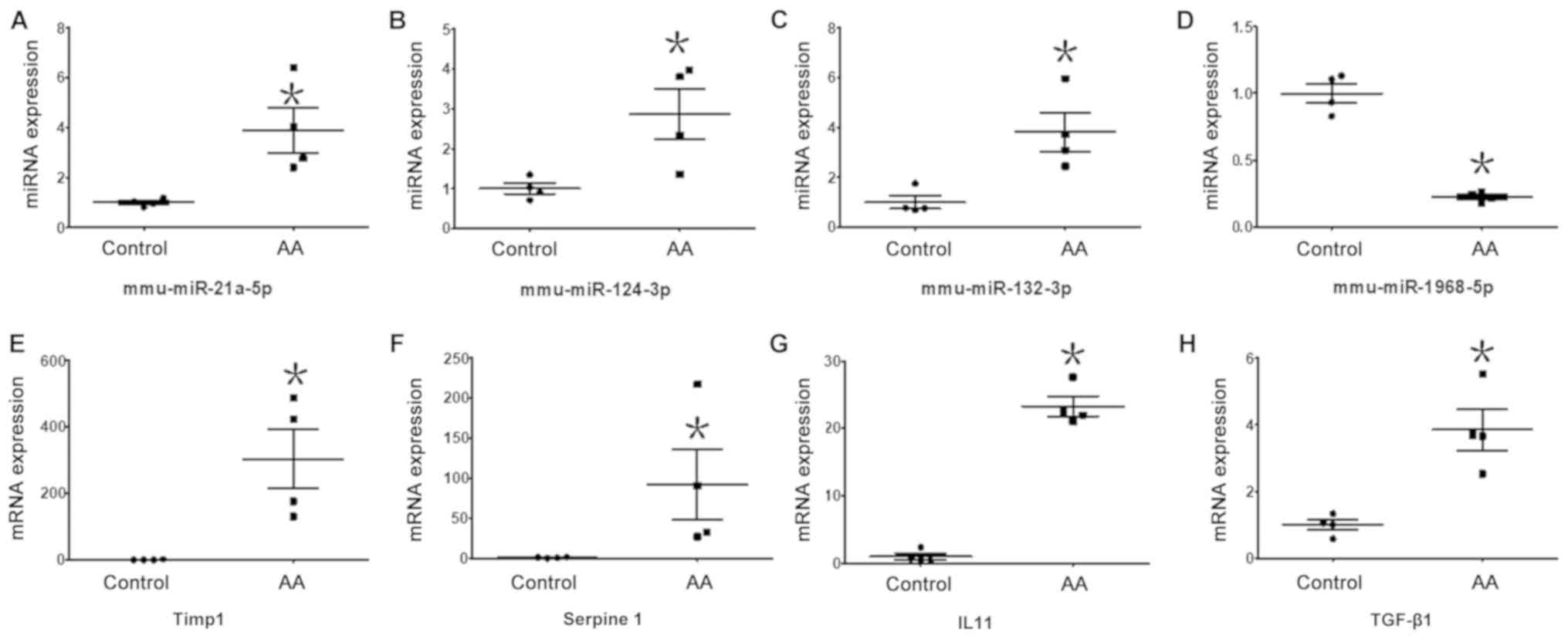 | Figure 5.Expression levels of DE miRNAs and DE
mRNAs validated by RT-qPCR. Expression levels of 4 DE miRNAs and 4
DE mRNAs were measured using RT-qPCR and normalized to U6 or GAPDH,
respectively. Relative expression levels of (A) miR-124-3p
(P=0.0398), (B) miR-21a-5p (P=0.0391) and (C) miR-132-3p (P=0.0399)
were significantly increased in the AA group compared with in the
self-control group. Expression levels of (D) miR-1968-5p (P=0.0014)
were significantly decreased in the AA group compared with in the
self-control group. Relative mRNA levels of (E) Timp1 (P=0.0416),
(F) Serpine1 (P=0.0475), (G) IL11 (P=0.0038) and (H) TGF-β1
(P=0.0146) were significantly increased in the AA group compared
with in the self-control group. *P<0.05. AA, aristolochic acid;
DE, differentially expressed; miRNA/miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; AA, aristolochic acid; IL11,
interleukin 11; TGF-β1, transforming growth factor β1; mmu, Mus
musculus. |
Integrated analysis of DE miRNA-DE
mRNA target pairs
Targets of DE miRNAs were predicted using both
RNAhybrid and miRanda. A total of 8,230 pairs of DE miRNA-mRNA
targets were found. Compared with the DE mRNA data in the present
study, 1,569 pairs of DE miRNA-DE mRNA targets were found. Among
these, there was a total of 767 opposite direction regulatory pairs
of DE miRNA-DE mRNA targets, including 416 pairs of upregulated DE
miRNAs and downregulated DE mRNAs, and 351 pairs of downregulated
DE miRNAs and upregulated DE mRNAs (Data S2).
These 767 opposite direction regulatory pairs of DE
miRNAs and DE mRNAs included 82 DE miRNAs and 624 DE mRNAs. Using
GO analysis on the 624 DE mRNAs, it was identified that ‘cellular
process’, ‘biological regulation’ and ‘regulation of biological
process’ were the most notably altered biological processes,
‘binding’ was the most changed molecular function, and ‘cell’ and
‘cell part’ were the most highly altered cellular components
(Fig. 6A).
KEGG analysis on the aforementioned 624 DE mRNAs
identified ‘signal transduction’, ‘global and overview maps’,
‘cancers’, ‘endocrine system’, ‘cellular community-eukaryotes’ and
‘immune system’ as the main altered pathways (Fig. 6B). The results were similar to that
of total DE mRNAs. The most enriched KEGG pathways were identified
as signaling pathways, ECM-associated pathways, ‘metabolic
pathways’ and ‘pathways in cancer’ (Fig. 6C). Signal transduction was the most
changed pathway, and signaling pathways were the most enriched
pathways. A total of 40 signaling pathways and 6 ECM-associated
pathways were identified by KEGG analysis. The most altered
signaling pathways included ‘Hedgehog signaling pathway’, ‘ABC
transporters’, ‘ErbB signaling pathway’, ‘cAMP signaling pathway’,
‘Relaxin signaling pathway’, ‘Ras signaling pathway’ and ‘PI3K-Akt
signaling pathway’ (Fig. 6C). The
most altered ECM-associated pathways included ‘ECM-receptor
interaction’ and ‘focal adhesion’ (Fig. 6C).
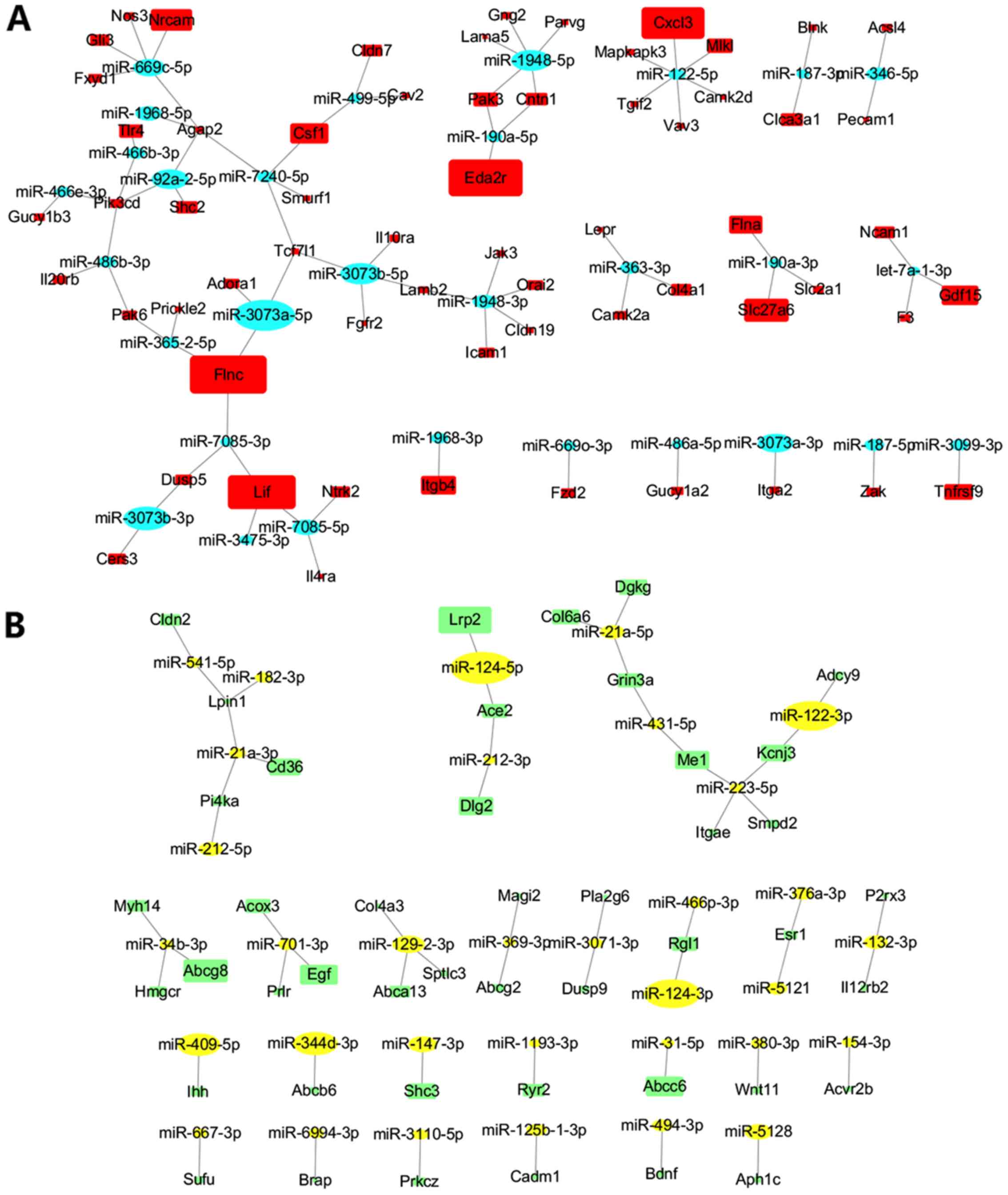
Regulatory networks of DE miRNAs on DE
mRNAs involved in signaling pathways and ECM-associated pathways
following AA treatment
Signaling pathways were the most enriched KEGG
pathways. Additionally, ECM-associated pathways were enriched
pathways and closely associated with renal fibrosis. A total of 63
DE miRNAs and their 107 DE mRNA targets were found from the
aforementioned 40 signaling pathways and 6 ECM-associated pathways.
As shown in Fig. 7, opposing
direction regulatory networks of DE miRNAs-DE mRNAs included 134 DE
miRNA-DE mRNA pairs (Table SII).
Regulatory networks included 54 pairs of increased DE miRNAs and
concurrent decreased DE mRNAs (Fig.
7B), and 80 pairs of decreased DE miRNAs and concurrent
increased DE mRNAs (Fig. 7A).
Among these 63 DE miRNAs, 44 miRNAs had >1 DE mRNA target. Among
them, the increased DE miRNAs included the miR-21 family
(miR-21a-5p and miR-21a-3p), miR-124 family (miR-124-5p and
miR-124-3p), miR-129-2-3p, miR-34b-3p, and so on; the decreased DE
miRNAs included the miR-3073 family (miR-3073a-5p, miR-3073a-3p and
miR-3073b-5p), miR-1948 family (miR-1948-5p and miR-1948-3p),
miR-122-5p, miR-669c-5p and let7a-1-3p (Fig. 7; Table SII).
Discussion
In the present study, short-term exposure (5 days)
to aristolochic acid (AA) induced renal injury and
tubulointerstitial fibrosis in acute aristolochic acid nephropathy
(AAN) in mice. To the best of our knowledge, the present study is
the first to report the changes in genome-wide DE miRNAs and DE
mRNAs, and their regulatory pairs and networks in acute AAN.
There was notable dysregulation of the miRNA
expression profile following treatment with AA. Specifically, 82 DE
miRNAs were identified following AA treatment, the most notable
alterations of which occurred in the profibrotic miR-21, miR-433
and miR-132 families, and the oncogenic miR-34 family, which were
significantly increased, and in the anti-fibrotic miR-122-5p and
let-7a-1-3p, which were significantly decreased. Similar increases
in the miR-21 and miR-34 families following long-term exposure to a
carcinogenic dose (10 mg/kg/day for 12 weeks) of AA in rats have
been previously reported in rats (25,26).
Activity of the TGF-β/Smad3 signaling pathway is increased in renal
fibrosis by miR-21 (25,26) via Smad7, PTEN, miR-433 (27) and Azin1. Silencing of miR-132 was
reported to reduce renal fibrosis (28,29).
Significant downregulation of let-7a was reported to be associated
with renal fibrosis in chronic kidney disease (30).
Global mRNA profiling was notably altered following
AA treatment. Amino acid metabolism, fatty acid metabolism,
‘complement and coagulation cascades’, ‘ECM-receptor interaction’
and ‘focal adhesion’ were the most significantly enriched pathways,
based on KEGG analysis of DE mRNAs. The pathways of ‘complement and
coagulation cascades’, ‘ECM-receptor interaction’ and ‘focal
adhesion’ directly participate in the process of tubulointerstitial
fibrosis (23,31). Amino acid metabolism pathways are
involved in the production of matrix collagen during fibrosis,
which is enriched in glycine, hydroxyproline, and hydroxylysine,
and other amino acid-derived compounds (32).
Additionally, GO and KEGG analysis were used to
analyze the 624 DE mRNAs regulated by DE miRNAs. The renal tubular
injury was largely the result of death or apoptosis of tubular
epithelial cells (33,34). The following tubulointerstitial
fibrosis was induced by infiltration of immune cells, production
and secretion of pro-fibrosis factors, downregulation and
inhibition of anti-fibrosis factors, myofibroblasts activation,
EMT, ECM production, tubular atrophy and others (7). In this process, the most altered
biological process in GO analysis may serve an important role,
which included ‘biological regulation’, ‘metabolic process’ and
‘response to stimulus’ in the biological process category,
‘binding’ in the molecular function category, and ‘organelle’ and
‘membrane part’ in the cellular component category. This process
was also affected by the most altered pathways in KEGG analysis,
which included ‘signal transduction’, metabolism, ‘endocrine
system’, ‘cellular community - eukaryotes’ and ‘immune system’.
Additionally, ‘cancers’ were one of the top changed pathways, as AA
is also a carcinogen, and EMT in fibrosis is an important pathway
in cancer.
In KEGG, similar to the results obtained from
analysis of the total DE miRNAs, ECM-associated pathways, metabolic
pathways and cancer-associated pathways were the most enriched KEGG
pathways. However, unlike the results of total DE miRNAs, signaling
pathways were identified as the most enriched KEGG pathways
regulated by DE miRNAs. A total of 40 signaling pathways were
identified. In the most significantly altered signaling pathway in
this study, fibrosis was reported in previous studies to be
involved in the ‘Hedgehog signaling pathway’ (35–37),
‘ABC transporters’ (38), ‘ErbB
signaling pathway’ (39,40), ‘cAMP signaling pathway’ (41,42),
‘Relaxin signaling pathway’ (43,44),
‘Ras signaling pathway’ (45,46)
and ‘PI3K-Akt signaling pathway’ (47,48).
Additionally, six ECM-associated pathways were directly associated
with renal fibrosis.
It has been suggested that DE miRNAs serve an
important role in renal injury and tubulointerstitial fibrosis by
regulating the DE mRNAs enriched in the signaling pathways and
ECM-associated pathways. In the aforementioned 40 enriched
signaling pathways and 6 ECM-associated pathways, the opposite
direction regulatory networks of 63 DE miRNAs and their 107 DE mRNA
targets were drawn. A total of 134 opposite direction regulatory
pairs of DE miRNAs and DE mRNAs were included in these regulatory
networks. The majority of the 63 DE miRNAs had >1 DE mRNA
target. For example, the increased DE miRNAs with the largest
numbers of targets included the miR-21 family (miR-21a-5p and
miR-21a-3p), the miR-124 family (miR-124-5p and miR-124-3p),
miR-129-2-3p and miR-34b-3p. The decreased DE miRNAs with the
largest numbers of targets included miR-122-5p, the miR-3073 family
(miR-3073a-5p, miR-3073a-3p and miR-3073b-5p), the miR-1948 family
(miR-1948-5p and miR-1948-3p), miR-669c-5p and let7a-1-3p. Among
these, fibrosis was reported to be regulated by the increase in
expression of the miR-21 family (49–54),
miR-124 family (55,56) and miR-34b-3p (57,58),
and the decrease in miR-122-5p (59–61)
and let7a-1-3p (62). The present
study was the first to report that AAN was associated with an
increase in miR-129-2-3p expression and a decrease in the
expression levels of the miR-3073 family (miR-3073a-5p,
miR-3073a-3p and miR-3073b-5p), miR-1948 family (miR-1948-5p and
miR-1948-3p) and miR-669c-5p.
In the present self-controlled study, mice were
treated with AA, following unilateral nephrectomy. This
self-control design may help increase the sensitivity and accuracy
of results by decreasing the interference of individual variations.
However, there may be other influencing factors, including
anesthesia, hyper-perfusion and residual renal function following
unilateral nephrectomy. These factors were likely less influencing
than the AA-induced acute renal injury. Although these factors
often had a notable effect under the background of hypertension,
diabetes, genetic defect, toxic drugs or other harmful conditions,
no significant changes were reported in the control mice group
given only unilateral nephrectomy (63–65).
The nature and severity of the renal response to unilateral
nephrectomy were reported to be strain-dependent in mice (66). It has been reported that the
unilateral nephrectomy did not induce sclerosis or fibrosis in the
glomerulus, tubulointerstitium or vasculature after 8 weeks in the
C57 strain mice (66).
Acute AAN in humans results in AA-induced acute
kidney injury and may progress to chronic kidney disease (CKD, as
necroinflammation, an auto amplification loop between tubular cell
death/apoptosis and interstitial inflammation, results in continued
renal injury and tubulointerstitial fibrosis (67–69).
Halting the progression of CKD following AA exposure by inhibition
or even resolution of AA-induced renal tubular injury and
tubulointerstitial fibrosis should be considered as a potential
therapeutic approach. The present study identified DE miRNAs, their
mRNA targets and the enriched pathways of the mRNA targets, which
may assist in improving the understanding of the underlying
mechanisms. The DE miRNAs and their regulated mRNAs represent
potential targets that may protect the kidney from continued
tubular injury and tubulointerstitial fibrosis following AA
exposure, and may assist in the development of novel clinical
therapies for treatment of CKD following AA exposure.
In conclusion, in the present self-controlled study,
renal injury and tubulointerstitial fibrosis were induced by
short-term treatment with AA in mice, and genome-wide DE miRNAs and
DE mRNAs, and their regulated pairs and networks were identified.
The present results may assist in improving the understanding of
the role of the DE miRNAs and their mRNA targets in the
pathophysiology of renal injury and tubulointerstitial fibrosis in
acute AAN.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from
Jiangsu Commission of Health (grant nos. F201439 and QNRC305) and
Changzhou Commission of Health (grant no. QN201724).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL designed this study. ZZ participated in the
design and writing of the study, the major experiments and data
analysis. XX, YS and YZ participated in the in vivo
experiments. XX, WQ and FW participated in the in vitro
experiments. XZ, CB, HH and SL analyzed the data from the deep
sequencing results of miRNA and mRNA. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Children's Hospital of Soochow University
(approval no. 2020-CHSU-012).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schaneberg BT, Applequist WL and Khan IA:
Determination of aristolochic acid I and II in North American
species of Asarum and Aristolochia. Pharmazie.
57:3367–689. 2002.
|
|
2
|
Michl J, Jennings HM, Kite GC, Ingrouille
MJ, Simmonds MS and Heinrich M: Is aristolochic acid nephropathy a
widespread problem in developing countries? A case study of
Aristolochia indica L. in Bangladesh using an
ethnobotanical-phytochemical approach. J Ethnopharmacol.
149:235–244. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bara T Jr, Gurzu S, Sugimura H, Bara T,
Beleaua MA and Jung I: A systematic review of the possible
carcinogenic role of the aristolochic acid. Rom J Morphol Embryol.
58:41–44. 2017.PubMed/NCBI
|
|
4
|
Michl J, Ingrouille MJ, Simmonds MSJ and
Heinrich M: Naturally occurring aristolochic acid analogues and
their toxicities. Nat Prod Rep. 31:676–693. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang L, Li X and Wang H: Possible
mechanisms explaining the tendency towards interstitial fibrosis in
aristolochic acid-induced acute tubular necrosis. Nephrol Dial
Transplant. 22:445–456. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jadot I, Decleves AE, Nortier J and Caron
N: An integrated view of aristolochic acid nephropathy: Update of
the literature. Int J Mol Sci. 18:2972017. View Article : Google Scholar
|
|
7
|
Humphreys BD: Mechanisms of renal
fibrosis. Annu Rev Physiol. 80:309–326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rockey DC, Bell PD and Hill JA: Fibrosis-a
common pathway to organ injury and failure. N Engl J Med.
372:1138–1149. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yeh YC, Wei WC, Wang YK, Lin SC, Sung JM
and Tang MJ: Transforming growth factor-{beta}1 induces
Smad3-dependent {beta}1 integrin gene expression in
epithelial-to-mesenchymal transition during chronic
tubulointerstitial fibrosis. Am J Pathol. 177:1743–1754. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Y, Feng XJ, Liu XY, Wang LH and Zheng
GP: The effect of transforming growth factor β(1) in the transition
of bone marrow-derived macrophages into myofibroblasts during renal
fibrosis. Zhonghua Nei Ke Za Zhi. 56:610–613. 2017.(In Chinese).
PubMed/NCBI
|
|
11
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bushati N and Cohen SM: MicroRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chung AC and Lan HY: MicroRNAs in renal
fibrosis. Front Physiol. 6:502015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang B and Ricardo S: Role of microRNA
machinery in kidney fibrosis. Clin Exp Pharmacol Physiol.
41:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang C, Xie Y and Yan W: 1 AASRA: An
anchor alignment-based small RNA. bioRxiv. 2017.(Epub ahead for
print).
|
|
16
|
Nawrocki EP and Eddy SR: Infernal 1.1:
100-fold faster RNA homology searches. Bioinformatics.
29:2933–2935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kruger J and Rehmsmeier M: RNAhybrid:
microRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34((Web Server Issue)): W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human microRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mi H, Muruganujan A, Ebert D, Huang X and
Thomas PD: PANTHER version 14: More genomes, a new PANTHER GO-slim
and improvements in enrichment analysis tools. Nucleic Acids Res.
47D:D419–D426. 2019. View Article : Google Scholar
|
|
21
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45D:D353–D361. 2017.
View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Horowitz JC and Thannickal VJ: Mechanisms
for the resolution of organ fibrosis. Physiology (Bethesda).
34:43–55. 2019.PubMed/NCBI
|
|
24
|
Schafer S, Viswanathan S, Widjaja AA, Lim
WW, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E,
et al: IL-11 is a crucial determinant of cardiovascular fibrosis.
Nature. 552:110–115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Z, Qin T, Wang K, Hackenberg M, Yan J,
Gao Y, Yu LR, Shi L, Su Z and Chen T: Integrated microRNA, mRNA,
and protein expression profiling reveals microRNA regulatory
networks in rat kidney treated with a carcinogenic dose of
aristolochic acid. BMC Genomics. 16:3652015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng F, Li Z, Yan J, Manjanatha M, Shelton
S, Yarborough S and Chen T: Tissue-specific microRNA responses in
rats treated with mutagenic and carcinogenic doses of aristolochic
acid. Mutagenesis. 29:357–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li R, Chung AC, Dong Y, Yang W, Zhong X
and Lan HY: The microRNA miR-433 promotes renal fibrosis by
amplifying the TGF-β/Smad3-Azin1 pathway. Kidney Int. 84:1129–1144.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bijkerk R, de Bruin RG, van Solingen C,
van Gils JM, Duijs JM, van der Veer EP, Rabelink TJ, Humphreys BD
and van Zonneveld AJ: Silencing of microRNA-132 reduces renal
fibrosis by selectively inhibiting myofibroblast proliferation.
Kidney Int. 89:1268–1280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou SG, Zhang W, Ma HJ, Guo ZY and Xu Y:
Silencing of lncRNA TCONS_00088786 reduces renal fibrosis through
miR-132. Eur Rev Med Pharmacol Sci. 22:166–173. 2018.PubMed/NCBI
|
|
30
|
Muralidharan J, Ramezani A, Hubal M,
Knoblach S, Shrivastav S, Karandish S, Scott R, Maxwell N, Ozturk
S, Beddhu S, et al: Extracellular microRNA signature in chronic
kidney disease. Am J Physiol Renal Physiol. 312:F982–F991. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y: Cellular and molecular mechanisms
of renal fibrosis. Nat Rev Nephrol. 7:684–696. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gaugg MT, Engler A, Bregy L,
Nussbaumer-Ochsner Y, Eiffert L, Bruderer T, Zenobi R, Sinues P and
Kohler M: Molecular breath analysis supports altered amino acid
metabolism in idiopathic pulmonary fibrosis. Respirology.
24:437–444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park JS, Choi HI, Kim DH, Kim CS, Bae EH,
Ma SK and Kim SW: Alpha-lipoic acid attenuates p-cresyl
sulfate-induced renal tubular injury through suppression of
apoptosis and autophagy in human proximal tubular epithelial cells.
Biomed Pharmacother. 112:1086792019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dong Q, Jie Y, Ma J, Li C, Xin T and Yang
D: Renal tubular cell death and inflammation response are regulated
by the MAPK-ERK-CREB signaling pathway under hypoxia-reoxygenation
injury. J Recept Signal Transduct Res. 39:383–391. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kramann R: Hedgehog Gli signalling in
kidney fibrosis. Nephrol Dial Transplant. 31:1989–1995. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou D, Tan RJ and Liu Y: Sonic hedgehog
signaling in kidney fibrosis: A master communicator. Sci China Life
Sci. 59:920–929. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fabian SL, Penchev RR, St-Jacques B, Rao
AN, Sipila P, West KA, McMahon AP and Humphreys BD: Hedgehog-Gli
pathway activation during kidney fibrosis. Am J Pathol.
180:1441–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kai Y, Yoneyama H, Koyama J, Hamada K,
Kimura H and Matsushima K: Treatment with chondroitinase ABC
alleviates bleomycin-induced pulmonary fibrosis. Med Mol Morphol.
40:128–140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vermeulen Z, Hervent AS, Dugaucquier L,
Vandekerckhove L, Rombouts M, Beyens M, Schrijvers DM, De Meyer
GRY, Maudsley S, De Keulenaer GW and Segers VFM: Inhibitory actions
of the NRG-1/ErbB4 pathway in macrophages during tissue fibrosis in
the heart, skin, and lung. Am J Physiol Heart Circ Physiol.
313:H934–H945. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Scheving LA, Zhang X, Threadgill DW and
Russell WE: Hepatocyte ERBB3 and EGFR are required for maximal
CCl4-induced liver fibrosis. Am J Physiol Gastrointest Liver
Physiol. 311:G807–G816. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Essam RM, Ahmed LA, Abdelsalam RM and
El-Khatib AS: Phosphodiestrase-1 and 4 inhibitors ameliorate liver
fibrosis in rats: Modulation of cAMP/CREB/TLR4 inflammatory and
fibrogenic pathways. Life Sci. 222:245–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Han K, Zhang Y and Yang Z: Cilostazol
protects rats against alcohol-induced hepatic fibrosis via
suppression of TGF-β1/CTGF activation and the cAMP/Epac1 pathway.
Exp Ther Med. 17:2381–2388. 2019.PubMed/NCBI
|
|
43
|
Kanai AJ, Konieczko EM, Bennett RG, Samuel
CS and Royce SG: Relaxin and fibrosis: Emerging targets,
challenges, and future directions. Mol Cell Endocrinol. 487:66–74.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng G, Cai J, Chen X, Chen L, Ge W, Zhou
X and Zhou H: Relaxin ameliorates renal fibrosis and expression of
endothelial cell transition markers in rats of
isoproterenol-induced heart failure. Biol Pharm Bull. 40:960–966.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Newbury LJ, Wang JH, Hung G, Hendry BM and
Sharpe CC: Inhibition of Kirsten-Ras reduces fibrosis and protects
against renal dysfunction in a mouse model of chronic folic acid
nephropathy. Sci Rep. 9:140102019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou G, Li J, Zeng T, Yang P and Li A: The
regulation effect of WNT-RAS signaling in hypothalamic
paraventricular nucleus on renal fibrosis. J Nephrol. 33:289–297.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang J, Zhu H, Huang L, Zhu X, Sha J, Li
G, Ma G, Zhang W, Gu M and Guo Y: Nrf2 signaling attenuates
epithelial-to-mesenchymal transition and renal interstitial
fibrosis via PI3K/Akt signaling pathways. Exp Mol Pathol.
111:1042962019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dou F, Liu Y, Liu L, Wang J, Sun T, Mu F,
Guo Q, Guo C, Jia N, Liu W, et al: Aloe-Emodin ameliorates renal
fibrosis via inhibiting PI3K/Akt/mTOR signaling pathway in vivo and
in vitro. Rejuvenation Res. 22:218–229. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun L, Xu T, Chen Y, Qu W, Sun D, Song X,
Yuan Q and Yao L: Pioglitazone attenuates kidney fibrosis via
miR-21-5p modulation. Life Sci. 232:1166092019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tian C, Wang Y, Chang H, Li J and La X:
Spleen-kidney supplementing formula alleviates renal fibrosis in
diabetic rats via TGF-β1-miR-21-PTEN signaling pathway. Evid Based
Complement Alternat Med. 2018:38243572018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tang CR, Luo SG, Lin X, Wang J and Liu Y:
Silenced miR-21 inhibits renal interstitial fibrosis via targeting
ERK1/2 signaling pathway in mice. Eur Rev Med Pharmacol Sci. 23 (3
Suppl):S110–S116. 2019.
|
|
52
|
McClelland AD, Herman-Edelstein M, Komers
R, Jha JC, Winbanks CE, Hagiwara S, Gregorevic P, Kantharidis P and
Cooper ME: miR-21 promotes renal fibrosis in diabetic nephropathy
by targeting PTEN and SMAD7. Clin Sci (Lond). 129:1237–1249. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang J, Gao Y, Ma M, Li M, Zou D, Yang J,
Zhu Z and Zhao X: Effect of miR-21 on renal fibrosis by regulating
MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem
Biophys. 67:537–546. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhong X, Chung AC, Chen HY, Meng XM and
Lan HY: Smad3-mediated upregulation of miR-21 promotes renal
fibrosis. J Am Soc Nephrol. 22:1668–1681. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou H, Qiu ZZ, Yu ZH, Gao L, He JM, Zhang
ZW and Zheng J: Paeonol reverses promoting effect of the
HOTAIR/miR-124/Notch1 axis on renal interstitial fibrosis in a rat
model. J Cell Physiol. 234:14351–14363. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhou H, Gao L, Yu ZH, Hong SJ, Zhang ZW
and Qiu ZZ: lncRNA HOTAIR promotes renal interstitial fibrosis by
regulating Notch1 pathway via the modulation of miR-124. Nephrology
(Carlton). 24:472–480. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Disayabutr S, Kim EK, Cha SI, Green G,
Naikawadi RP, Jones KD, Golden JA, Schroeder A, Matthay MA, Kukreja
J, et al: miR-34 miRNAs regulate cellular senescence in type II
alveolar epithelial cells of patients with idiopathic pulmonary
fibrosis. PLoS One. 11:e1583672016. View Article : Google Scholar
|
|
58
|
Li WQ, Chen C, Xu MD, Guo J, Li YM, Xia
QM, Liu HM, He J, Yu HY and Zhu L: The rno-miR-34 family is
upregulated and targets ACSL1 in dimethylnitrosamine-induced
hepatic fibrosis in rats. FEBS J. 278:1522–1532. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sun Y, Wang H, Li Y, Liu S, Chen J and
Ying H: miR-24 and miR-122 negatively regulate the transforming
growth Factor-β/smad signaling pathway in skeletal muscle fibrosis.
Mol Ther Nucleic Acids. 11:528–537. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lou G, Yang Y, Liu F, Ye B, Chen Z, Zheng
M and Liu Y: miR-122 modification enhances the therapeutic efficacy
of adipose tissue-derived mesenchymal stem cells against liver
fibrosis. J Cell Mol Med. 21:2963–2973. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Halasz T, Horvath G, Par G, Werling K,
Kiss A, Schaff Z and Lendvai G: miR-122 negatively correlates with
liver fibrosis as detected by histology and FibroScan. World J
Gastroenterol. 21:7814–7823. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li N, Wang LJ, Xu WL, Liu S and Yu JY:
MicroRNA3795p suppresses renal fibrosis by regulating the
LIN28/let7 axis in diabetic nephropathy. Int J Mol Med.
44:1619–1628. 2019.PubMed/NCBI
|
|
63
|
Cevikbas F, Schaefer L, Uhlig P, Robenek
H, Theilmeier G, Echtermeyer F and Bruckner P: Unilateral
nephrectomy leads to up-regulation of syndecan-2- and
TGF-beta-mediated glomerulosclerosis in syndecan-4 deficient male
mice. Matrix Biol. 27:42–52. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xia Y, Jin X, Yan J, Entman ML and Wang Y:
CXCR6 plays a critical role in angiotensin II-induced renal injury
and fibrosis. Arterioscler Thromb Vasc Biol. 34:1422–1428. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Uil M, Scantlebery AML, Butter LM, Larsen
PWB, de Boer OJ, Leemans JC, Florquin S and Roelofs JJTH: Combining
streptozotocin and unilateral nephrectomy is an effective method
for inducing experimental diabetic nephropathy in the ‘resistant’
C57Bl/6J mouse strain. Sci Rep. 8:55422018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Esposito C, He CJ, Striker GE, Zalups RK
and Striker LJ: Nature and severity of the glomerular response to
nephron reduction is strain-dependent in mice. Am J Pathol.
154:891–897. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu BC, Tang TT and Lv LL: How tubular
epithelial cell injury contributes to renal fibrosis. Adv Exp Med
Biol. 1165:233–252. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu BC, Tang TT, Lv LL and Lan HY: Renal
tubule injury: A driving force toward chronic kidney disease.
Kidney Int. 93:568–579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Youl ENH, Husson C, El Khattabi C, El Mere
S, Decleves AE, Pochet S, Nortier J and Antoine MH:
Characterization of cytotoxic effects of aristolochic acids on the
vascular endothelium. Toxicol In Vitro. 65:1048112020. View Article : Google Scholar : PubMed/NCBI
|