The success rate in the clinical treatment of
neoplastic disease remains highly associated with early detection
of pre-malignant or first stages of malignant tissues. To date,
only few highly specific and sensitive biomarkers are routinely
used in the clinic for early-stage cancer screening or
diagnostics.
Due to mammography screening, which was first
introduced in 2005 in Germany, breast cancer (BC) has been
identified at earlier stages, when treatment options are most
promising and prognosis is most favorable (1). For endometrial cancer (EC) and
ovarian cancer (OC), no standardized screening has yet been
established. Postmenopausal bleeding serves as an early indicator
of EC (2,3) European studies have shown that the
5-year survival rate of endometrial adenocarcinoma is >90% when
detected at stage I compared with a survival rate of ~50% for
advanced stages (II, III, IV) (3,4). OC
remains one of the most challenging types of cancer to detect and
treat. In most cases, tumor progression and metastasis are
unnoticed until the advanced stages (5). According to the Surveillance,
Epidemiology and End Results Program (National Cancer Institute,
USA) database, the 5-year survival rate for localized disease is
>90% in the USA population (6),
however only 20% of ovarian cancer cases are detected at such an
early stage in the USA (5,7).
One possible approach in the identification of novel
potential biomarker candidates is based on expression profiling of
different states, for example comparing malignant and healthy
control expression profiles (7).
In a stepwise filtering process, the discovery, qualification,
verification, potential candidate prioritization and subsequent
validation in adequate cohort sizes demonstrate the applicability
of a biomarker for clinical practice implementation (7). Among a multitude of potential
biomarker types, in previous years one group of nucleic acids has
gained significant attention due to their diverse regulatory
functions (8).
MicroRNAs (miRNAs or miRs) are small non-coding RNA
molecules of ~22 nucleotides in length, which are involved in the
post-transcriptional regulation of gene expression, predominately
via gene silencing. By binding to various mRNA targets,
upregulation of miRNA leads to reduced translation of mRNA or
degradation of its transcript (9).
In cancer, dysregulated miRNA expression plays an important role by
upregulating oncogenes and downregulating tumor-suppressor genes,
thus modulating cell proliferation, differentiation, apoptosis and
stress response (10). The
regulatory influence of miRNAs in breast and gynecological cancer
biology has been demonstrated in a growing number of studies
(8,11–17).
The selection of miRNAs in the present study was based on an
extensive literature search, with the major criterion being
expression changes in the tumor types of BC, EC and OC (Table I), in combination with a proven
detectability of all analyzed miRNA types in in vitro models
as well as in human urine samples (18–20).
miRNA-21 (miR-21) is one of the most common miRNAs
in epithelial cancer, and it generally promotes anti-apoptotic
effects in various malignant tissues and cell lines, including BC,
OC and EC, by downregulating tumor suppressors, such as phosphatase
and tensin homolog (21,22) and programmed cell death protein 4
(23). In patients with BC,
overexpression of miR-21 in the tumor is associated with advanced
tumor stage, lymph node metastasis and poor survival (24). Whereas, in OC cell lines, miR-21
promotes pathways that enhance chemoresistance (25).
In contrast to miR-21, members of the miRNA family
let-7 have most commonly been reported as tumor suppressors by
downregulating Harvey rat sarcoma viral oncogene homolog and
high-mobility group AT-hook 2 (26). However, studies have reported
inconsistent results regarding the individual member let-7b. While
some studies reported that high levels of let-7b in serum and
plasma was associated with a favorable prognosis in cancer
(27,28), a previous meta-analysis
demonstrated reduced survival rates in high-grade serous OC with
high tissue expression of let-7b (29). The tumor suppressing miRNA family
miR-30 has been reported to exhibit pro-apoptotic effects by
silencing ubiquitin-conjugating enzyme 9 and integrin β3 (30). In BC, miR-30a inhibits cell
migration and invasion (31),
whereas expression of miR-30c in tissues is associated with
benefits during endocrine treatment (32) and regulatory effects in
chemotherapy resistance processes (33). Notably, high expression levels of
miR-30c and miR-30e have been observed in OC compared with normal
tissue; however, both miRNAs are associated with an improved
prognosis (34–36).
A more homogenous profiling has been observed for
miR-125b and miR-100. miR-125b and miR-100 mediate the Erb-B2
receptor tyrosine kinase 2 and mechanistic target of rapamycin
pathways, respectively, and downregulation of both miRNAs has been
reported in BC, OC and EC tissue and cell lines (37–41).
The previously described functional implications of the
investigated miRNAs in BC, EC and OC tumor biology are summarized
in Table I.
Due to recent investigations on miRNAs that are
commonly conducted based on different study designs and
environments, the comparison and interpretation of results between
multiple cancer types have become increasingly challenging. The
goal of the present study was to evaluate differences of miRNA
profiling in three of the most common female cancer types: BC, OC
and EC. Instead of solely focusing on individual miRNA types or
families, the present study aimed to investigate the expression
patterns of miRNAs that have great potential to serve as promising
diagnostic tools in the distinction of different tumor types. Based
on three cell types for each type of malignancy, BC, OC and EC, the
detected differences in quantitative expression levels of a set of
25 miRNAs revealed diagnostic biomarker features clustered in
tumor-entity-specific ‘miRNA signatures’. To this end, the in
vitro models used were selected to represent a range of common
subtypes/properties of the respective carcinomas. The data obtained
in this first phase biomarker identification study serve as a basis
to prioritize distinct miRNAs with diagnostic significance that
will be investigated in future studies.
The BC cell lines BT-20 (cat. no. 300130; CLS Cell
Lines Service GmbH), BT-474 (cat. no. 00131; CLS Cell Lines Service
GmbH) and SK-BR-3 (cat. no. 300333; CLS Cell Lines Service GmbH),
the EC cell lines Ishikawa (cat. no. 99040201; Sigma-Aldrich; Merck
KGaA), EFE-184 (cat. no. ACC 230; Leibniz Institute DSMZ-German
Collection of Microorganisms and Cell Cultures GmbH) and AN3CA
(cat. no. 300119; CLS Cell Lines Service GmbH), and the OC cell
lines SK-OV-3 (cat. no. 300342; CLS Cell Lines Service GmbH),
EFO-27 (cat. no. ACC 191; Leibniz Institute DSMZ-German Collection
of Microorganisms and Cell Cultures GmbH) and OAW-42 (cat. no.
300304; CLS Cell Lines Service GmbH) were incubated in a humidified
atmosphere at 37°C and 5% CO2. Ishikawa cells were
cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% newborn calf serum (Gibco; Thermo Fisher
Scientific, Inc.), 1% HEPES buffer (Gibco; Thermo Fisher
Scientific, Inc.) and 100 U/ml Penicillin/Streptomycin
(Sigma-Aldrich; Merck KGaA). The BT-20, SK-BR-2, EFE-184, AN3CA,
SK-OV-3 and EFO-28 cells were cultured in DMEM/F12 (cat. no.
31331-028; Thermo Fisher Scientific, Inc.) supplemented with 10%
newborn calf serum, 1% HEPES buffer and 100 U/ml
Penicillin/Streptomycin. The BT-47 and OAW-42 cells were cultured
in DMEM/F12 supplemented with 2.5% insulin (Insuman
rapid®; Sanofi S.A.).
miRNA from cultured cells was isolated using the
innuPREP Micro RNA kit (Analytik Jena US LLC), according to the
manufacturer's instructions. Isolated RNA was quantitatively
determined using the NanoDrop ND1000 (VWR International GmbH). RNA
samples were stored at −20°C until further processing.
Total RNA was isolated using GeneMATRIX Universal
RNA/miRNA Purification kit (cat. no. E3599; EURx®;
Roboklon GmbH) according to manufacturer's protocol. A total of 1
µg isolated RNA per sample was used for RT. The RT reaction mix
contained 5 µl RT-buffer (5X), 1 µl 2.5 µM poly A adapter primer
(Apara Bioscience GmbH), 0.5 µl 5 mM dNTPs (Jena Bioscience), 0.25
µl Maxima reverse transcriptase (Thermo Fisher Scientific, Inc.),
0.25 µl SUPERase In RNase inhibitor (Thermo Fisher Scientific,
Inc.), 0.5 µl 10 mM ATP (New England Biolabs, Inc.), 0.25 µl poly A
polymerase, and 1 µg RNA sample. The reaction was performed on a
thermal cycler (Eppendorf) at 37°C for 60 min and stopped at 85°C
for 10 min. Processed cDNA was stored at 4°C.
The relative expression levels of specific miRNAs
were assessed by qPCR using the SYBR-Green assay in a duplicate
analysis. A total of 1 µl cDNA per sample with a concentration of 5
ng/µl was mixed with 9 µl Master Mix, containing 1 µl buffer (10X),
0.5 µl 5 mM dNTPs (Jena Bioscience GmbH), 0.5 µl 5 µM primer
(Apara), 0.5 µl SYBR-Green (Roche Diagnostics), 0.05 µl HotStart
Taq (Jena Bioscience GmbH) and 6.45 µl nuclease-free water
(Analytik Jena US LLC). The primer pairs consisted of a universal
reverse primer (30–32) and a specific miRNA sense primer.
The qPCR was performed on a LightCycler® 480 instrument
(Roche Diagnostics) at 95°C for 5 min, followed by 40 cycles at
95°C for 5 sec, 62°C for 15 sec and 72°C for 10 sec. Data were
analyzed with the LightCycler® 480 software (Roche
Molecular Systems, Inc.; Version 1.5.1). The relative expression of
each miRNA was determined using the 2−ΔΔCq method
(42,43) based on the housekeeping genes small
nucleolar RNA, C/D box 48 (RNU48), miR-26b, miR-16 and miR-103,
with the ‘BestKeeper’ software tool (Version 1) (43). The specific primer sequences are
listed in Table II.
The expression levels of all BC, EC and
OC-associated miRNAs were determined as mean ΔCq values
of the miRNA normalized against the geometric mean of the four
housekeeping genes RNU48, miR-16, miR-26b and miR-103. The
expression levels of all miRNA types were separately analyzed using
a linear model with cell line as the independent variable. The
regression coefficients with 95% confidence intervals were
tabulated. This led to color coded heatmaps in which red colors
indicate strong deviations in the positive direction and blue
colors indicate strong deviations in the negative direction from
the expression level in the cell line that served as a reference
(AN3CA, BT-474 and BT-20). Dark colors correspond to a
P<0.00005, and light colors correspond to a P<0.00025. All
other comparisons are presented in gray.
In the present study, the expression levels of 25
BC, EC and OC-associated miRNAs (let-7a, let-7b, let-7d, miR-7, −9,
−15b, −17, −19b, −20a, −20b, −21, −27a, −29a, −30a, −30c, −30e,
−92a, −100, −106b, −125b, −128.1, −200b, −200c, −221, −222) were
quantified in three BC, EC and OC cell lines. The characteristics
of each cell line are presented in Table III.
The statistical analyses demonstrated that comparing
the three different cell types (AN3CA, BT-474 and BT-20) revealed a
range of moderately to highly differentially expressed miRNAs,
which exhibited either marked upregulation or downregulation. By
clustering miRNAs with respect to their differential expression
characteristics, subgroups of miRNAs featuring potential biomarkers
to discriminate between BC, OC and EC cells could be created. The
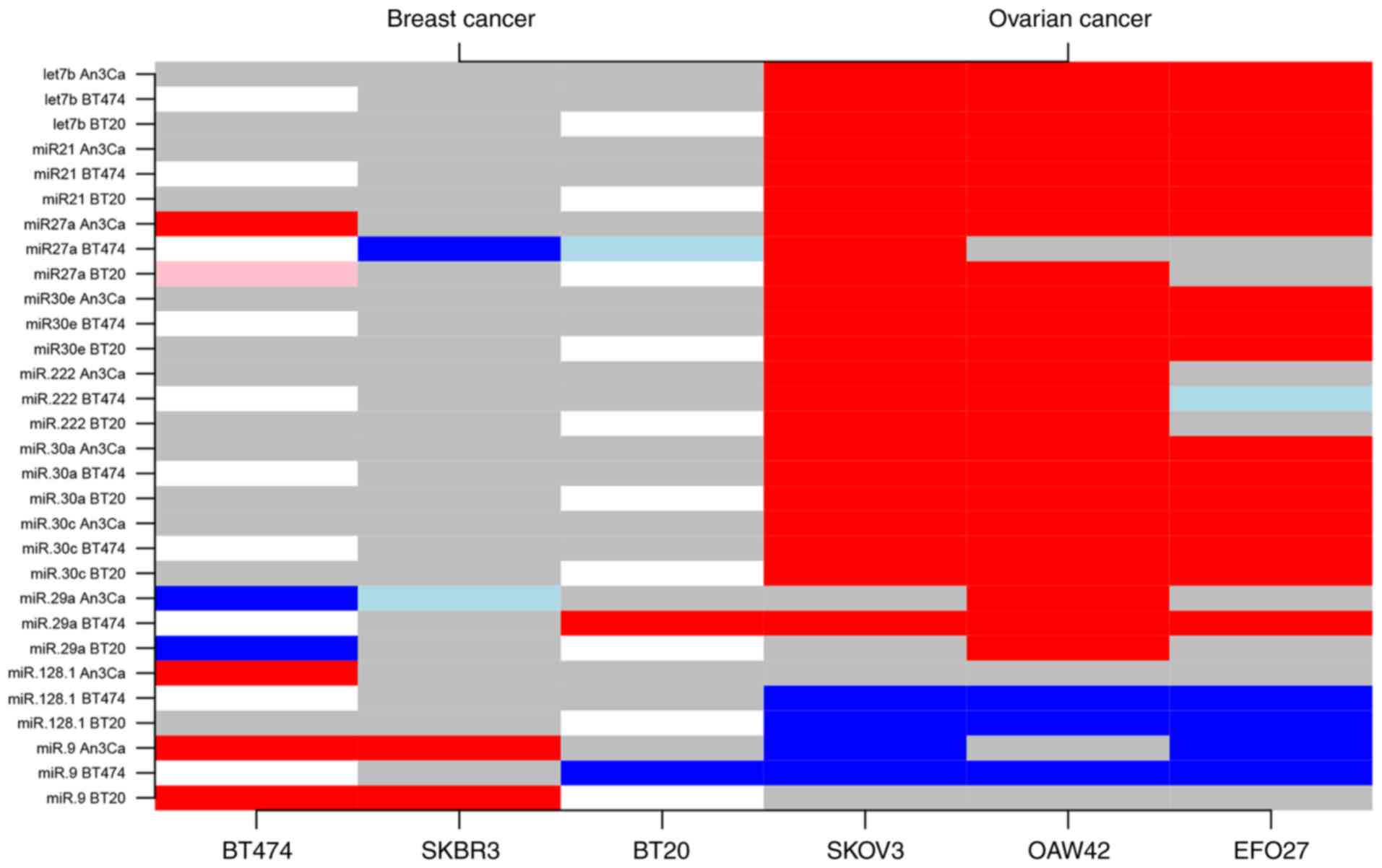
expression data clearly revealed a BC-associated miRNA subpanel
with significantly distinct expression levels compared with the
gynecological tumor types EC and OC (miRs: let-7b, −21, −27a, −30a,
−30c, −30e). Consecutively, miRNA clusters with statistical
relevance were defined to allow for discrimination between the
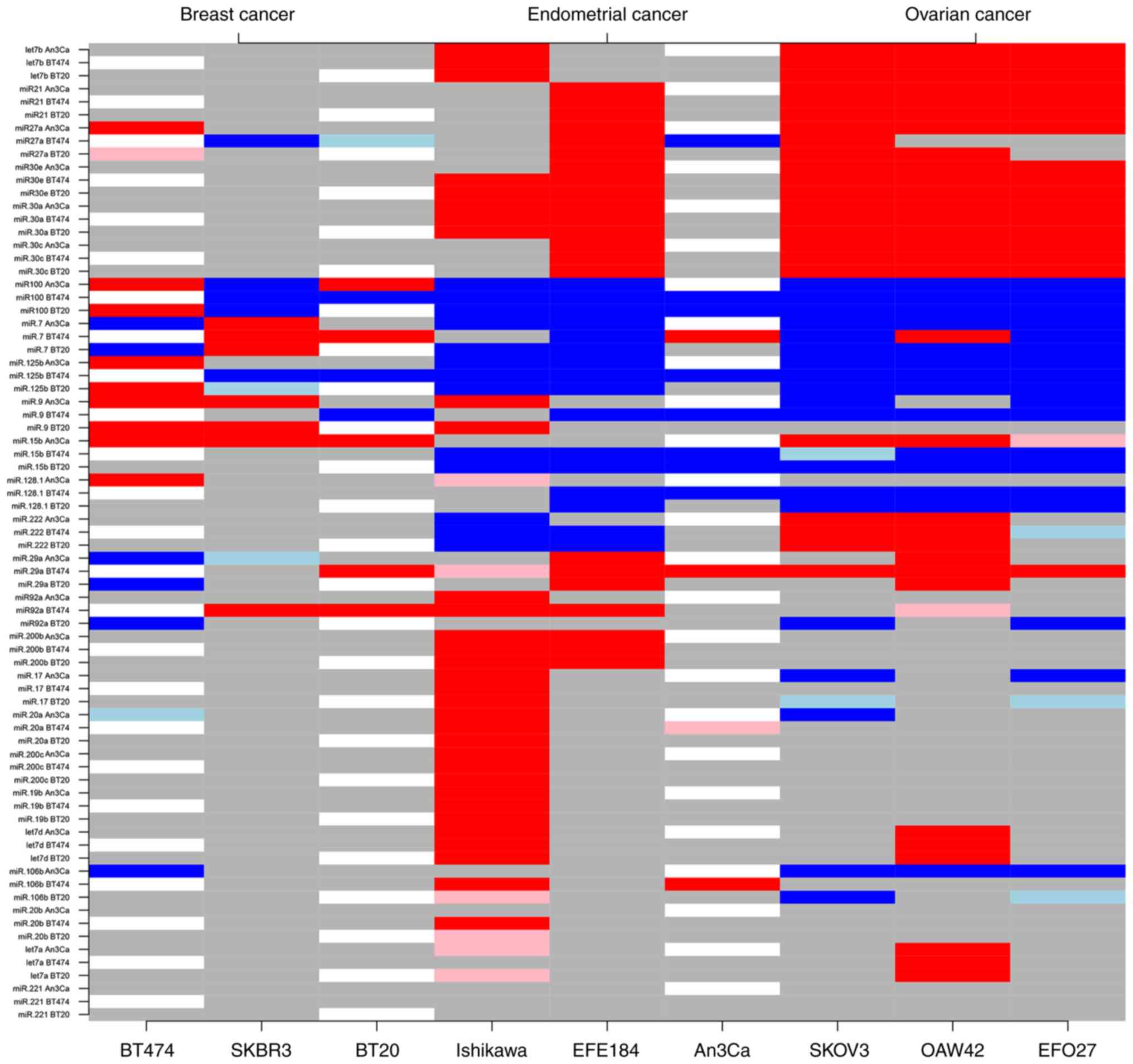
three tumor entities in a one-versus-one approach (Fig. 1 and Tables SI–SIII).
Expression analyses could determine a subgroup of
ten different miRNAs (miRs: let-7b, −21, 30a, −30c, −30e, −27a,
−222, −29a, −128.1, −9) that facilitated an expression level-based
discrimination between the BC and OC cell types. The notable types
included let-7b, miR-21 and the miR-30 family genes, which were
uniformly upregulated in OC cells compared with BC cells. For
example, compared with AN3CA cells, miR-let-7b was upregulated by a
mean value of 5.08 (95% confidence interval, 4.51, 5.65;
P<0.001) in SK-OV-3, 8.37 (7.80, 8.94; P<0.001) in OAW-42
cells, and 2.53 (1.96, 3.10; P<0.001) in EFO-27 OC cells
(Table SI). In contrast,
regression analyses demonstrated no significant difference of
miR-let-7b in all investigated BC cell lines (Fig. 2 and Table SII). The expression levels of
miR-30a, miR-30c and miR-30e were also increased in all three
investigated OC cell lines. Specifically, compared with AN3CA
cells, the miR-30a was significantly increased by a mean value of
0.22 (0.21, 0.23; P<0.001) in SK-OV-3, 0.12 (0.10, 0.13;
P<0.001) in OAW-42 cells, and 0.43 (0.42, 0.44; P<0.001) in
EFO-27 OC cells. The expression levels of miR-30c and miR-30e were
also upregulated by a mean value of 0.21 (0.17, 0.25; P<0.001)
and 0.07 (0.06, 0.08; P<0.001) in SK-OV-3 cells, 0.12 (0.08,
0.16; P<0.001) and 0.03 (0.02, 0.04; P<0.001) in OAW-42
cells, and 0.50 (0.46, 0.55; P<0.001) and 0.16 (0.15, 0.17;
P<0.001) in EFO-27 OC cells (Table
SI). No significant differences were identified among all BC
cell lines (Table SII). miR-27
and miR-29a exhibited a moderate downregulation in BC cells, with
few inconsistent results depending on the cell line comparison
(AN3CA or BT-474). By contrast, miR-9 and miR-128.1 exhibited a
general moderate downregulation in OC cells compared with BC cells
(Fig. 2 and Tables SI and SII).
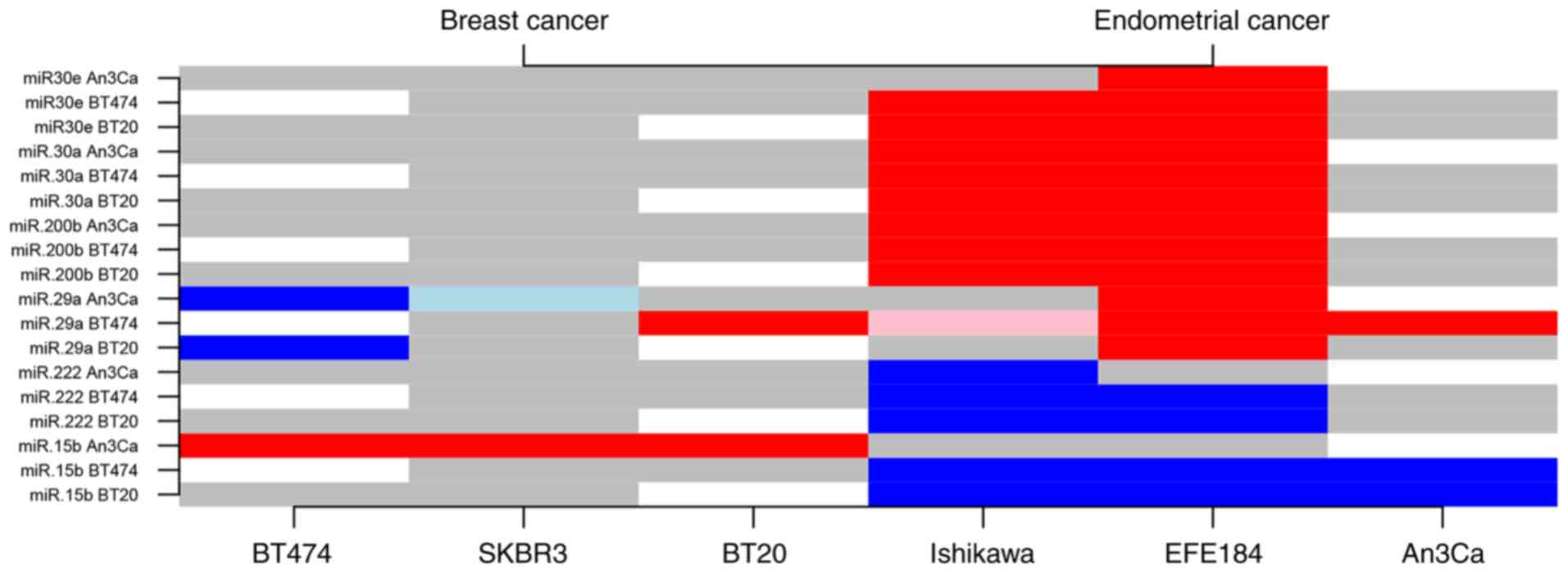
Among the 25 miRNAs evaluated in the present study,
six exhibited distinguishing characteristics in regard to BC
compared with EC cell expression profiles (miRs: −30a, −30e, −29a,
−15b, −200b, −222). While miR-29a, −30a, −30c and −200b were found
to be upregulated in EC cells, miR-15b and miR-222 demonstrated
downregulated expression levels in comparison to BC cells. For
example, miR-200b was upregulated by a mean value of 5.43 (5.27,
5.58; P<0.001) in Ishikawa cells and by 0.97 (0.81, 1.12;
P<0.001) in EFE-184 EM cells. Notably, AN3CA cells did not fully
comply to the EC-specific expression level trends, which may be
explained by cell-specific molecular characteristics (Fig. 3 and Tables SII and SIII).
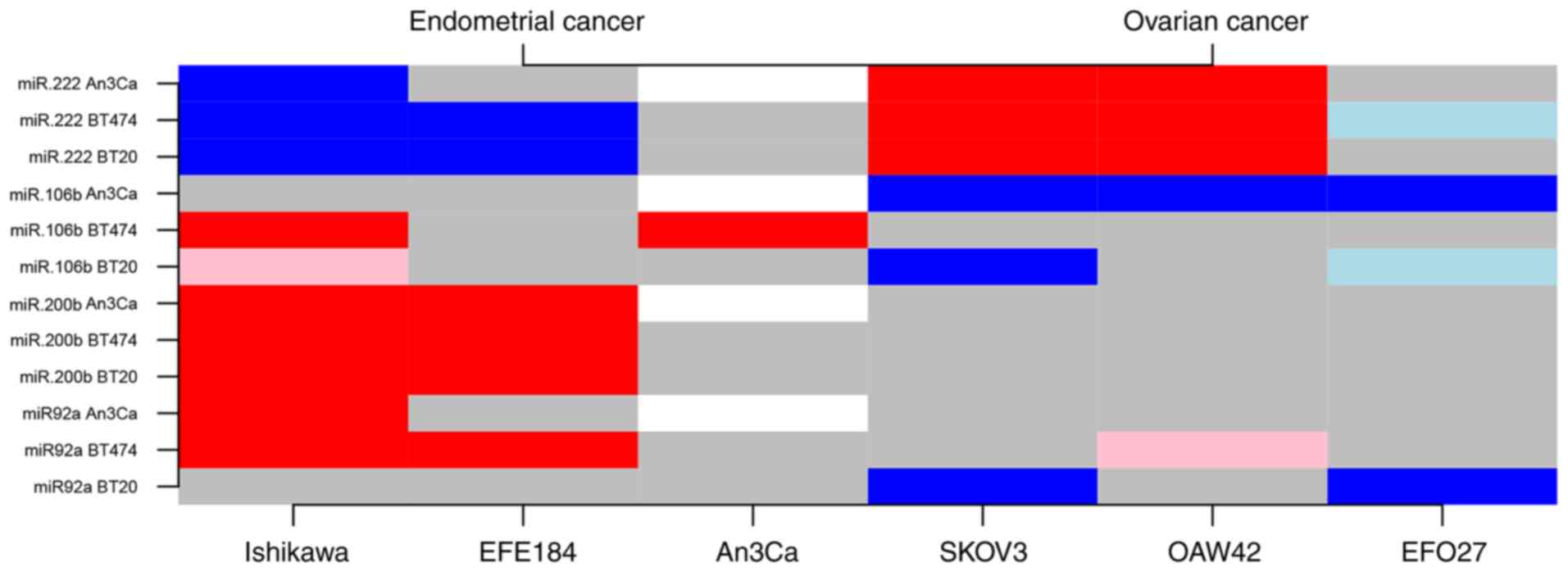
A total of four miRNAs (miR-92a, −106b, −200b, −222)
with altered expression levels that may serve a role in the
determination of endometrial compared with ovarian malignancies
were identified based on this in vitro approach. Upregulated
expression levels of miR-92a, −106b and −200b in EC cell types, as
well as an upregulation of miR-222 in OC cells may help to mutually
distinguish between these tumor types. Compared with AN3CA cells,
miR-222 expression was increased by a mean value of 0.66 (0.52,
0.80; P<0.001) in SK-OV-3 cells and by 0.85 (0.71, 0.99;
P<0.001) in OAW-42 OC cells. By contrast, a downregulation was
identified in two EC cell lines by a mean value of 0.48 (−0.62,
−0.34; P<0.001) in Ishikawa cells and by 0.18 (−0.32, −0.04;
P=0.018) in EFE-184 EM cells. However, individual cell
line-specific differences need to be taken into account in the
assessment of tumor type determination of a potential miRNA
subpanel with diagnostic power in this regard (Fig. 4 and Tables SI and SIII).
The search for clinically applicable biomarkers
necessitates a stringent multi-step selection process to
singularize, evaluate and validate the usability of a potential
biomolecule, or even grouped biomolecule expression profiles or
signatures for clear diagnostic purposes. The present study focused
on one possible initial step in the determination of potential
novel biomarkers that help to detect and distinguish healthy women
from patients with malignant disease of the breast, endometrium or
ovaries. Based on an in vitro model approach, the proof of
principle was accomplished to corroborate the initial hypothesis of
discriminating diagnostic features of miRNA signatures in the
diagnosis of breast and gynecological malignancies. In general, the
detected intracellular miRNA expression levels can be transferred
to the extracellular setting of secreted miRNAs, as shown in a
previous study (20). Therefore,
the experimental design of the present study was targeted on the
identification of miRNA signatures, based on tumor-specific
expression differences, that enable the mutual discrimination of
the three common female cancer types BC, OC and EC. Since miRNAs
are robust and easily accessible biomolecules that can be
quantified in a wide range of biomaterials, including tissue and
liquid biopsies, they meet important requirements for modern and
applicable diagnostic biomarkers (44). Thousands of different human miRNAs
have been described, of which a clinically relevant subset of 25
different miRNAs with potential impacts in BC, EC and/or OC was
pre-selected for the present analytical in vitro approach.
Although certain differences in cell line-based and in vivo
settings need to be kept in consideration, the current study is
intended to provide initial findings that guide further
investigations in a promising direction.
Global expression profile analyses in the present
study resulted in the identification of cancer type-specific miRNA
subgroups. These clusters of distinct miRNAs were characterized by
differences in expression levels that can significantly
discriminate between the tumor types BC, OC and EC. However, no
significant subtype-specific miRNA expression signature differences
could be detected among the respective cancer types analyzed.
A parallel comparison of entity-specific clustering
habits highlighted a BC-specific miRNA subpanel of six miRNAs that
exhibited significantly different expression levels compared with
those observed in EC and OC in vitro models. In
particularly, members of the miR-30 family were identified in this
respect.
Comparisons of miRNA expression signatures in either
BC/OC, BC/EC or EC/OC clearly revealed the most miRNA expression
profile differences in the comparison of BC vs. OC, with ten of the
25 miRNAs exhibiting significantly different expression levels in
these tumor types. Members of the miR-30 family were identified to
be significantly differentially expressed, in addition to few more
types, including miR-9, which has previously been described as a
prognostic marker in OC (45), as
well as miR-222 and miR-29a, which are known triggers in breast
cancer therapy resistance mechanisms (46). In previous studies, the let-7
family has been reported to exhibit decreased expression levels in
OC tissues as well as in OC cell lines, and has been identified to
serve a role in OC progression (47,48).
In contrast to the literature, in the present study, let-7b was
found to be upregulated in OC cells compared with BC cells.
A direct comparison of EC and BC miRNA expression
signatures revealed six miRNAs with significantly different
expression levels. Again, members of the miR-30 family were
prominent, but also miR-15b and mi-200b were identified in this
comparison. miR-15b has been described as an aberrantly regulated
tumor suppressor (49), whereas
miR-200b has a role in epithelial-mesenchymal transition processes
(50).
The miRNAs miR-92a, miR-106b, miR-200b and miR-222
compose the smaller subgroup of four miRNAs that exhibited
significant expression differences in EC compared with OC in
vitro models. Consistent with a previous study by Záveský et
al (51), the present data
confirmed the differential expression of the miRNAs miR-92a,
miR-106b and miR-200b in EC compared with OC. Upregulated
expression levels of miR-222 in OC cells were found to associated
with epithelial OC in a previous investigation (52).
In conclusion, the diagnostic power and validity of
entity-specific miRNA clusters is partially limited due to
individual cell type characteristics, such as receptor status or
tumor origin (primary tumor or metastasis). For instance, in the
present study the estrogen receptor (ER) EC cell line exhibited a
different miRNA expression compared with two ER+ EC cell
lines. In addition, EFO-27 deviated from the other OC in
vitro models to a certain extent, thus an intra-entity
variation in miRNA expression signatures has to be taken into
account. Furthermore, the present analyses revealed a notable
difference in the molecular relationship of EC and OC compared with
BC. Therefore, the number of miRNAs with distinguishing expression
levels was markedly increased in EC and OC vs. BC than in the
comparison between EC vs. OC.
To pursue the identification of a clinically
valuable highly entity-specific signature panel with diagnostic
power for implementation in routine screening, the obtained data of
the present discovery phase approach require further verification
and validation. Thus, additional and extended in vitro
analyses, followed by translational studies using patients' tissues
and liquid biopsy materials should be performed in further analyses
to provide substantial evidence for miRNA-based biomarker
expression signatures that enable tumor detection, characterization
and potential therapy monitoring.
Not applicable.
No funding was received.
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
The project idea and experimental design was
conceived by MH, MJ, DW, SM, BK and TE. In vitro experiments
were performed by MJ, CN and DW. Data and statistical analyses were
performed by GR, supported by DW, TE and MH. MH, IG, JW, TE, GR,
MV, KB and JA interpreted the results and wrote the manuscript. GR,
MV, BK, KB and SM critically revised the final version of the
manuscript. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Heywang-Koebrunner S, Bock K, Heindel W,
Hecht G, Regitz-Jedermann L, Hacker A and Kaeaeb-Sanyal V:
Mammography Screening-as of 2013. Geburtshilfe Frauenheilkd.
73:1007–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clarke MA, Long BJ, Del Mar Morillo A,
Arbyn M, Bakkum-Gamez JN and Wentzensen N: Association of
endometrial cancer risk with postmenopausal bleeding in Women: A
systematic review and meta-analysis. JAMA Intern Med.
178:1210–1222. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Steiner E, Eicher O, Sagemüller J, Schmidt
M, Pilch H, Tanner B, Hengstler JG, Hofmann M and Knapstein PG:
Multivariate independent prognostic factors in endometrial
carcinoma: A clinicopathologic study in 181 patients: 10 years
experience at the department of obstetrics and gynecology of the
Mainz University. Int J Gynecol Cancer. 13:197–203. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tejerizo-Garcia A, Jiménez-López JS,
Muñoz-González JL, Bartolomé-Sotillos S, Marqueta-Marqués L,
López-González G and Gómez JF: Overall survival and disease-free
survival in endometrial cancer: Prognostic factors in 276 patients.
Onco Targets Ther. 9:1305–1313. 2013.PubMed/NCBI
|
|
5
|
Das PM and Bast RC Jr: Early detection of
ovarian cancer. Biomark Med. 2:291–303. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Howlader N, Noone AM, Krapcho M, Miller D,
Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS,
Feuer EJ and Cronin KA: SEER Cancer Statistics Review. 1975-2017,
National Cancer Institute; Bethesda, MD, USA: https://seer.cancer.gov/csr/1975_2017/
|
|
7
|
Frangogiannis NG: Biomarkers: Hopes and
challenges in the path from discovery to clinical practice. Transl
Res. 159:197–204. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Makarova JA, Shkurnikov MU, Wicklein D,
Lange T, Samatov TR, Turchinovich AA and Tonevitsky AG:
Intracellular and extracellular microRNA: An update on localization
and biological role. Prog Histochem Cytochem. 51:33–49. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends in
Molecular Medicine. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanekura K, Nishi H, Isaka K and Kuroda M:
MicroRNA and gynecologic cancers. J Obstet Gynaecol Res.
42:612–617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira
M, Matsumoto H and Horiguchi J: Recent trends in microRNA research
into breast cancer with particular focus on the associations
between microRNAs and intrinsic subtypes. J Hum Genet. 62:15–24.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nakamura K, Sawada K, Yoshimura A, Kinose
Y, Nakatsuka E and Kimura T: Clinical relevance of circulating
cell-free microRNAs in ovarian cancer. Mol Cancer. 15:482016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rapisuwon S, Vietsch EE and Wellstein A:
Circulating biomarkers to monitor cancer progression and treatment.
Comput Struct Biotechnol J. 14:211–222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Torres A, Torres K, Pesci A, Ceccaroni M,
Paszkowski T, Cassandrini P, Zamboni G and Maciejewski R:
Diagnostic and prognostic significance of miRNA signatures in
tissues and plasma of endometrioid endometrial carcinoma patients.
Int J Cancer. 132:1633–1645. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Widodo, Djati MS and Rifa'i M: Role of
microRNAs in carcinogenesis that potential for biomarker of
endometrial cancer. Ann Med Surg (Lond. 7:9–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yanokura M, Banno K, Iida M, Irie H, Umene
K, Masuda K, Kobayashi Y, Tominaga E and Aoki D: MicroRNAS in
endometrial cancer: Recent advances and potential clinical
applications. Excli J. 14:190–198. 2015.PubMed/NCBI
|
|
18
|
Hirschfeld M, Rücker G, Weiß D, Berner K,
Ritter A, Jäger M and Erbes T: Urinary exosomal MicroRNAs as
potential non-invasive biomarkers in breast cancer detection. Mol
Diagn Ther. 24:215–232. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ritter A, Hirschfeld M, Berner K, Jaeger
M, Grundner-Culemann F, Schlosser P, Asberger J, Weiss D, Noethling
C, Mayer S and Erbes T: Discovery of potential serum and
urine-based microRNA as minimally-invasive biomarkers for breast
and gynecological cancer. Cancer Biomark. 27:225–242. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritter A, Hirschfeld M, Berner K, Rücker
G, Jäger M, Weiss D, Medl M, Nöthling C, Gassner S, Asberger J and
Erbes T: Circulating noncoding RNA-biomarker potential in
neoadjuvant chemotherapy of triple negative breast cancer? Int J
Oncol. 56:47–68. 2020.PubMed/NCBI
|
|
21
|
Lou Y, Yang X, Wang F, Cui Z and Huang Y:
MicroRNA-21 promotes the cell proliferation, invasion and migration
abilities in ovarian epithelial carcinomas through inhibiting the
expression of PTEN protein. Int J Mol Med. 26:819–827. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin X, Yan L, Zhao X, Li C and Fu Y:
MicroRNA-21 overexpression contributes to cell proliferation by
targeting PTEN in endometrioid endometrial cancer. Oncol Lett.
4:1290–1296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Au Yeung CL, Co NN, Tsuruga T, Yeung TL,
Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, et al: Exosomal
transfer of stroma-derived miR21 confers paclitaxel resistance in
ovarian cancer cells through targeting APAF1. Nat Commun.
7:11502016. View Article : Google Scholar
|
|
26
|
Greene SB, Herschkowitz JI and Rosen JM:
Small players with big roles: microRNAs as targets to inhibit
breast cancer progression. Curr Drug Targets. 11:1059–1073. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Encarnacion J, Ortiz C, Vergne R, Vargas
W, Coppola D and Matta JL: High DRC Levels are associated with
Let-7b overexpression in women with breast cancer. Int J Mol Sci.
17:8652016. View Article : Google Scholar
|
|
28
|
Chung YW, Bae HS, Song JY, Lee JK, Lee NW,
Kim T and Lee KW: Detection of microRNA as novel biomarkers of
epithelial ovarian cancer from the serum of ovarian cancer
patients. Int J Gynecol Cancer. 23:673–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang Z, Ow GS, Thiery JP, Ivshina AV and
Kuznetsov VA: Meta-analysis of transcriptome reveals let-7b as an
unfavorable prognostic biomarker and predicts molecular and
clinical subclasses in high-grade serous ovarian carcinoma. Int J
Cancer. 134:306–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong
C, Huang Y, Hu X, Su F, Lieberman J and Song E: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng CW, Wang HW, Chang CW, Chu HW, Chen
CY, Yu JC, Chao JI, Liu HF, Ding SL and Shen CY: MicroRNA-30a
inhibits cell migration and invasion by downregulating vimentin
expression and is a potential prognostic marker in breast cancer.
Breast Cancer Res Treat. 134:1081–1093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rodriguez-Gonzalez FG, Sieuwerts AM, Smid
M, Look MP, Meijer-van Gelder ME, de Weerd V, Sleijfer S, Martens
JW and Foekens JA: MicroRNA-30c expression level is an independent
predictor of clinical benefit of endocrine therapy in advanced
estrogen receptor positive breast cancer. Breast Cancer Res Treat.
127:43–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bockhorn J, Dalton R, Nwachukwu C, Huang
S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, et al:
MicroRNA-30c inhibits human breast tumour chemotherapy resistance
by regulating TWF1 and IL-11. Nat Commun. 4:13932013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee H, Park CS, Deftereos G, Morihara J,
Stern JE, Hawes SE, Swisher E, Kiviat NB and Feng Q: MicroRNA
expression in ovarian carcinoma and its correlation with
clinicopathological features. World J Surg Oncol. 10:1742012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cabarcas SM, Thomas S, Zhang X, Cherry JM,
Sebastian T, Yerramilli S, Lader E, Farrar WL and Hurt EM: The role
of upregulated miRNAs and the identification of novel mRNA targets
in prostatospheres. Genomics. 99:108–117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Li L, Qu Z, Li R, Bi T, Jiang J
and Zhao H: The expression of miR-30a* and miR-30e* is associated
with a dualistic model for grading ovarian papillary serious
carcinoma. Int J Oncol. 44:1904–1914. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shang C, Lu YM and Meng LR: MicroRNA-125b
down-regulation mediates endometrial cancer invasion by targeting
ERBB2. Med Sci Moni. 18:BR149–BR155. 2012.
|
|
38
|
Li C, Gao Y, Zhang K, Chen J, Han S, Feng
B, Wang R and Chen L: Multiple Roles of MicroRNA-100 in human
cancer and its therapeutic potential. Cellular Physiology and
Biochemistry. 37:2143–2159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer research. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mattie MD, Benz CC, Bowers J, Sensinger K,
Wong L, Scott GK, Fedele V, Ginzinger D, Getts R and Haqq C:
Optimized high-throughput microRNA expression profiling provides
novel biomarker assessment of clinical prostate and breast cancer
biopsies. Mol Cancer. 5:242006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guan Y, Yao H, Zheng Z, Qiu G and Sun K:
miR-125b targets BCL3 and suppresses ovarian cancer proliferation.
Int J Cancer. 128:2274–2283. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper-Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang H, Peng R, Wang J, Qin Z and Xue L:
Circulating microRNAs as potential cancer biomarkers: The advantage
and disadvantage. Clin Epigenetics. 10:592018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun H, Shao Y, Huang J, Sun S, Liu Y, Zhou
P and Yang H: Prognostic value of microRNA-9 in cancers: A
systematic review and meta-analysis. Oncotarget. 7:67020–67032.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhong S, Li W, Chen Z, Xu J and Zhao J:
miR-222 and miR-29a contribute to the drug-resistance of breast
cancer cells. Gene. 531:8–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dahiya N and Morin PJ: MicroRNAs in
ovarian carcinomas. Endocr Relat Cancer. 17:F77–F89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dahiya N, Sherman-Baust CA, Wang TL,
Davidson B, Shih IeM, Zhang Y, Wood W III, Becker KG and Morin PJ:
MicroRNA expression and identification of putative miRNA targets in
ovarian cancer. PLoS One. 3:e24362008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao C, Wang G, Zhu Y, Li X, Yan F, Zhang
C, Huang X and Zhang Y: Aberrant regulation of miR-15b in human
malignant tumors and its effects on the hallmarks of cancer. Tumour
Biol. 37:177–183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Madhavan D, Peng C, Wallwiener M, Zucknick
M, Nees J, Schott S, Rudolph A, Riethdorf S, Trumpp A, Pantel K, et
al: Circulating miRNAs with prognostic value in metastatic breast
cancer and for early detection of metastasis. Carcinogenesis.
37:461–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Záveský L, Jandáková E, Turyna R,
Langmeierová L, Weinberger V, Záveská Drábková L, Hůlková M,
Hořínek A, Dušková D, Feyereisl J, et al: Evaluation of Cell-Free
Urine microRNAs expression for the use in diagnosis of ovarian and
endometrial cancers. A Pilot Study. Pathol Oncol Res. 21:1027–1035.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sun C, Li N, Zhou B, Yang Z, Ding D, Weng
D, Meng L, Wang S, Zhou J, Ma D and Chen G: miR-222 is upregulated
in epithelial ovarian cancer and promotes cell proliferation by
downregulating P27kip1. Oncol Lett. 6:507–512. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lin Z, Li JW, Wang Y, Chen T, Ren N, Yang
L, Xu W, He H, Jiang Y, Chen X, et al: Abnormal miRNA-30e
expression is associated with breast cancer progression. Clin Lab.
62:121–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW,
Nam SJ and Chun KH: MicroRNA let-7a suppresses breast cancer cell
migration and invasion through downregulation of C-C chemokine
receptor type 7. Breast Cancer Res. 14:1–12. 2012. View Article : Google Scholar
|
|
55
|
Liu K, Zhang C, Li T, Ding Y, Tu T, Zhou
F, Qi W, Chen H and Sun X: Let-7a inhibits growth and migration of
breast cancer cells by targeting HM. Int J Oncol. 46:2526–2534.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang L, Zheng W, Zhang S, Chen X and
Hornung D: Expression of monocyte chemotactic protein-1 in human
endometrial cancer cells and the effect of treatment with tamoxifen
or buserelin. J Int Med Res. 34:284–290. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chun SM, Park HJ, Kim CH and Kim I: The
Significance of MicroRNA Let-7b, miR-30c, and miR-200c Expression
in Breast Cancers. J Pathol Transl Med. 45:354–360. 2011.
|
|
58
|
Liu P, Qi M, Ma C, Lao G and Liu Y and Liu
Y and Liu Y: Let7a inhibits the growth of endometrial carcinoma
cells by targeting Aurora-B. FEBS Lett. 587:2523–2529. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bayani J, Kuzmanov U, Saraon P, Fung WA,
Soosaipillai A, Squire JA and Diamandis EP: Copy number and
expression alterations of miRNAs in the ovarian cancer cell line
OVCAR-3: Impact on kallikrein 6 protein expression. Clin Chem.
59:296–305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dong P, Ihira K, Xiong Y, Watari H, Hanley
SJ, Yamada T, Hosaka M, Kudo M, Yue J and Sakuragi N: Reactivation
of epigenetically silenced miR-124 reverses the
epithelial-to-mesenchymal transition and inhibits invasion in
endometrial cancer cells via the direct repression of IQGAP1
expression. Oncotarget. 7:20260–20270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Boren T, Xiong Y, Hakam A, Wenham R, Apte
S, Wei Z, Kamath S, Chen DT, Dressman H and Lancaster JM: MicroRNAs
and their target messenger RNAs associated with endometrial
carcinogenesis. Gynecol Oncol. 110:206–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kowalewska M, Bakula-Zalewska E,
Chechlinska M, Goryca K, Nasierowska-Guttmejer A, Danska-Bidzinska
A and Bidzinski M: microRNAs in uterine sarcomas and mixed
epithelial-mesenchymal uterine tumors: A preliminary report. Tumour
Biol. 34:2153–2160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Markou A, Zavridou M, Sourvinou I, Yousef
G, Kounelis S, Malamos N, Georgoulias V and Lianidou E: Direct
comparison of metastasis-related miRNAs expression levels in
circulating tumor cells, corresponding plasma, and primary tumors
of breast cancer patients. Clin Chem. 62:1002–1011. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Echevarria-Vargas IM, Valiyeva F and
Vivas-Mejia PE: Upregulation of miR-21 in cisplatin resistant
ovarian cancer via JNK-1/c-Jun pathway. PLoS One. 9:e970942014.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Myatt SS, Wang J, Monteiro LJ, Christian
M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S and Lam EW:
Definition of microRNAs that repress expression of the tumor
suppressor gene FOXO1 in endometrial cancer. 70:367–377.
2010.PubMed/NCBI
|
|
68
|
Ouzounova M, Vuong T, Ancey PB, Ferrand M,
Durand G, Le-Calvez Kelm F, Croce C, Matar C, Herceg Z and
Hernandez-Vargas H: MicroRNA miR-30 family regulates non-attachment
growth of breast cancer cells. BMC Genomics. 14:1392013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chang CW, Yu JC, Hsieh YH, Yao CC, Chao
JI, Chen PM, Hsieh HY, Hsiung CN, Chu HW, Shen CY and Cheng CW:
MicroRNA-30a increases tight junction protein expression to
suppress the epithelial-mesenchymal transition and metastasis by
targeting Slug in breast cancer. Oncotarget. 7:16462–16478. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Berber U, Yilmaz I, Narli G, Haholu A,
Kucukodaci Z and Demirel D: miR-205 and miR-200c: Predictive Micro
RNAs for lymph node metastasis in triple negative breast cancer. J
Breast Cancer. 17:143–148. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tsukamoto O, Miura K, Mishima H, Abe S,
Kaneuchi M, Higashijima A, Miura S, Kinoshita A, Yoshiura K and
Masuzaki H: Identification of endometrioid endometrial
carcinoma-associated microRNAs in tissue and plasma. Gynecol Oncol.
132:715–721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Nagaraja AK, Creighton CJ, Yu Z, Zhu H,
Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM,
et al: A link between mir-100 and FRAP1/mTOR in clear cell ovarian
cancer. Mol Endocrinol. 24:447–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Calura E, Fruscio R, Paracchini L,
Bignotti E, Ravaggi A, Martini P, Sales G, Beltrame L, Clivio L,
Ceppi L, et al: MiRNA landscape in stage I epithelial ovarian
cancer defines the histotype specificities. Clin Cancer Res.
19:4114–4123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen
Y, Wang J, Liu Y, Chen P, Wu X and Wen J: Urinary microRNA-30a-5p
is a potential biomarker for ovarian serous adenocarcinoma. Oncol
Rep. 33:2915–2923. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ayaz L, Çayan F, Balci Ş, Görür A, Akbayir
S, Yıldırım Yaroğlu H, Doğruer Unal N and Tamer L: Circulating
microRNA expression profiles in ovarian cancer. J Obstet Gynaecol.
34:620–624. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hausler SF, Keller A, Chandran PA, Ziegler
K, Zipp K, Heuer S, Krockenberger M, Engel JB, Hönig A, Scheffler
M, et al: Whole blood-derived miRNA profiles as potential new tools
for ovarian cancer screening. Br J Cancer. 103:693–700. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Shukla K, Sharma AK, Ward A, Will R,
Hielscher T, Balwierz A, Breunig C, Münstermann E, König R,
Keklikoglou I and Wiemann S: MicroRNA-30c-2-3p negatively regulates
NF-κB signaling and cell cycle progression through downregulation
of TRADD and CCNE1 in breast cancer. Mol Oncol. 9:1106–1119. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tanic M, Yanowsky K, Rodriguez-Antona C,
Andrés R, Márquez-Rodas I, Osorio A, Benitez J and Martinez-Delgado
B: Deregulated miRNAs in hereditary breast cancer revealed a role
for miR-30c in regulating KRAS oncogene. PLoS One. 7:e388472012.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gong Y, He T, Yang L, Yang G, Chen Y and
Zhang X: The role of miR-100 in regulating apoptosis of breast
cancer cells. Sci Rep. 5:116502015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zaman MS, Maher DM, Khan S, Jaggi M and
Chauhan SC: Current status and implications of microRNAs in ovarian
cancer diagnosis and therapy. J Ovarian Res. 5:442012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Okuda H, Xing F, Pandey PR, Sharma S,
Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y, et al:
miR-7 suppresses brain metastasis of breast cancer stem-like cells
by modulating KLF4. Cancer Res. 73:1434–1444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Banno K, Yanokura M, Iida M, Adachi M,
Nakamura K, Nogami Y, Umene K, Masuda K, Kisu I, Nomura H, et al:
Application of microRNA in diagnosis and treatment of ovarian
cancer. Biomed Res Int. 2014:2328172014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ramon LA, Braza-Boïls A, Gilabert J,
Chirivella M, España F, Estellés A and Gilabert-Estellés J:
microRNAs related to angiogenesis are dysregulated in endometrioid
endometrial cancer. Hum Reprod. 27:3036–3045. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kinose Y, Sawada K, Nakamura K and Kimura
T: The Role of MicroRNAs in Ovarian Cancer. Biomed Res Int.
2014:112014. View Article : Google Scholar
|
|
86
|
Lu J, Zhang X, Zhang R and Ge Q: MicroRNA
heterogeneity in endometrial cancer cell lines revealed by deep
sequencing. Oncol Lett. 10:3457–3465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER- breast
cancer cell lines. J Cell Mol Med. 19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chong GO, Jeon HS, Han HS, Son JW, Lee YH,
Hong DG, Lee YS and Cho YL: Differential MicroRNA Expression
Profiles in Primary and Recurrent Epithelial Ovarian Cancer.
Anticancer Res. 35:2611–2617. 2015.PubMed/NCBI
|
|
89
|
Singh SR and Rameshwar P: MicroRNA in
development and in the progression of cancer. Springer-Verlag; New
York: 2014, View Article : Google Scholar
|
|
90
|
Brockhoff G, Heckel B, Schmidt-Bruecken E,
Plander M, Hofstaedter F, Vollmann A and Diermeier S: Differential
impact of Cetuximab, Pertuzumab and Trastuzumab on BT474 and
SK-BR-3 breast cancer cell proliferation. Cell Prolif. 40:488–507.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lasfargues EY, Coutinho WG and Redfield
ES: Isolation of two human tumor epithelial cell lines from solid
breast carcinomas. J Natl Cancer Inst. 61:967–978. 1978.PubMed/NCBI
|
|
92
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG and
Tang P: The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67
and AR by immunohistochemical analysis in breast cancer cell lines.
Breast Cancer. 4:35–41. 2010.PubMed/NCBI
|
|
93
|
Lee S, Yang W, Lan KH, Sellappan S, Klos
K, Hortobagyi G, Hung MC and Yu D: Enhanced sensitization to
taxol-induced apoptosis by herceptin pretreatment in
ErbB2-overexpressing breast cancer cells. Cancer Res. 62:5703–5710.
2002.PubMed/NCBI
|
|
94
|
Kim DJ, Lee WY, Park NW, Kim GS, Lee KM,
Kim J, Choi MK, Lee GH, Han W and Lee SK: Drug response of captured
BT20 cells and evaluation of circulating tumor cells on a silicon
nanowire platform. Biosens Bioelectron. 67:370–378. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kloten V, Schlensog M, Eschenbruch J,
Gasthaus J, Tiedemann J, Mijnes J, Heide T, Braunschweig T, Knüchel
R and Dahl E: Abundant NDRG2 expression is associated with
aggressiveness and unfavorable patients' outcome in basal-like
breast cancer. PLoS One. 11:e01590732016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Croxtall JD, Elder MG and White JO:
Hormonal control of proliferation in the Ishikawa endometrial
adenocarcinoma cell line. J Steroid Biochem. 35:665–669. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Nishida M, Kasahara K, Kaneko M, Iwasaki H
and Hayashi K: Establishment of a new human endometrial
adenocarcinoma cell line, Ishikawa cells, containing estrogen and
progesterone receptors. Nihon Sanka Fujinka Gakkai zasshi.
37:1103–1111. 1985.(In Japanese). PubMed/NCBI
|
|
98
|
Dawe CJ, Banfield WG, Morgan WD, Slatick
MS and Curth HO: Growth in continuous culture, and in hamsters, of
cells from a neoplasm associated with acanthosis nigricans. J Natl
Cancer Inst. 33:441–456. 1964.PubMed/NCBI
|
|
99
|
Korch C, Spillman MA, Jackson TA, Jacobsen
BM, Murphy SK, Lessey BA, Jordan VC and Bradford AP: DNA profiling
analysis of endometrial and ovarian cell lines reveals
misidentification, redundancy and contamination. Gynecol Oncol.
127:241–248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Basu M, Mukhopadhyay S, Chatterjee U and
Roy SS: FGF16 promotes invasive behavior of SKOV-3 ovarian cancer
cells through activation of mitogen-activated protein kinase (MAPK)
signaling pathway. J Biol Chem. 289:1415–1428. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Irmer G, Bürger C, Müller R, Ortmann O,
Peter U, Kakar SS, Neill JD, Schulz KD and Emons G: Expression of
the messenger RNAs for luteinizing hormone-releasing hormone (LHRH)
and its receptor in human ovarian epithelial carcinoma. Cancer Res.
55:817–822. 1995.PubMed/NCBI
|
|
102
|
Yoon J, Kim ES, Lee SJ, Park CW, Cha HJ,
Hong BH and Choi KY: Apoptosis-related mRNA expression profiles of
ovarian cancer cell lines following cisplatin treatment. J Gynecol
Oncol. 21:255–261. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Langdon SP and Lawrie SS: Establishment of
ovarian cancer cell lines. Methods Mol Med. 39:155–159.
2001.PubMed/NCBI
|
|
104
|
Shukla J, Sharma U, Kar R, Varma IK, Juyal
S, Jagannathan NR and Bandopadhyaya GP:
Tamoxifen-2-hydroxylpropyl-beta-cyclodextrin-aggregated
nanoassembly for nonbreast estrogen-receptor-positive cancer
therapy. Nanomedicine (Lond). 4:895–902. 2009. View Article : Google Scholar : PubMed/NCBI
|