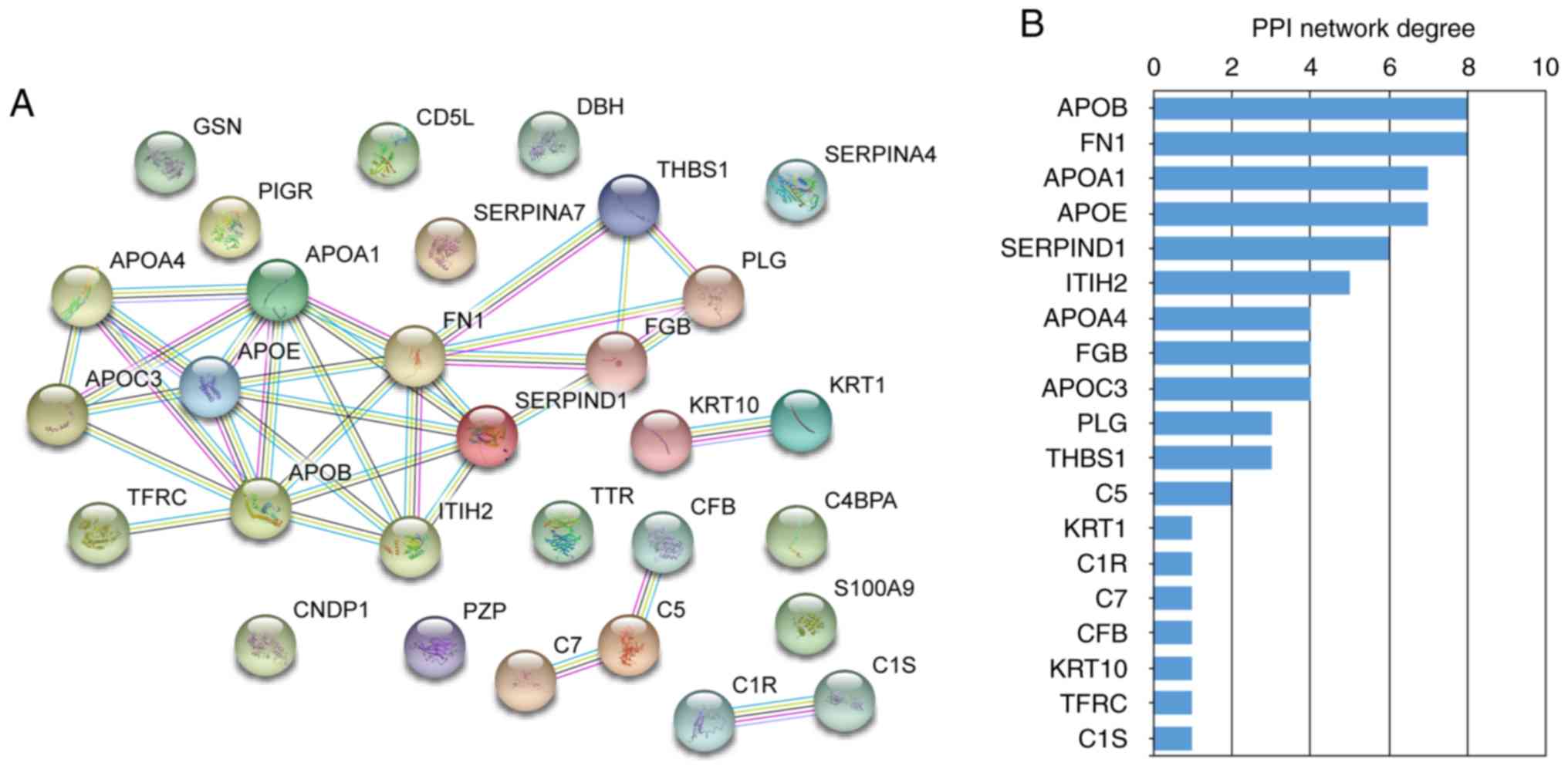
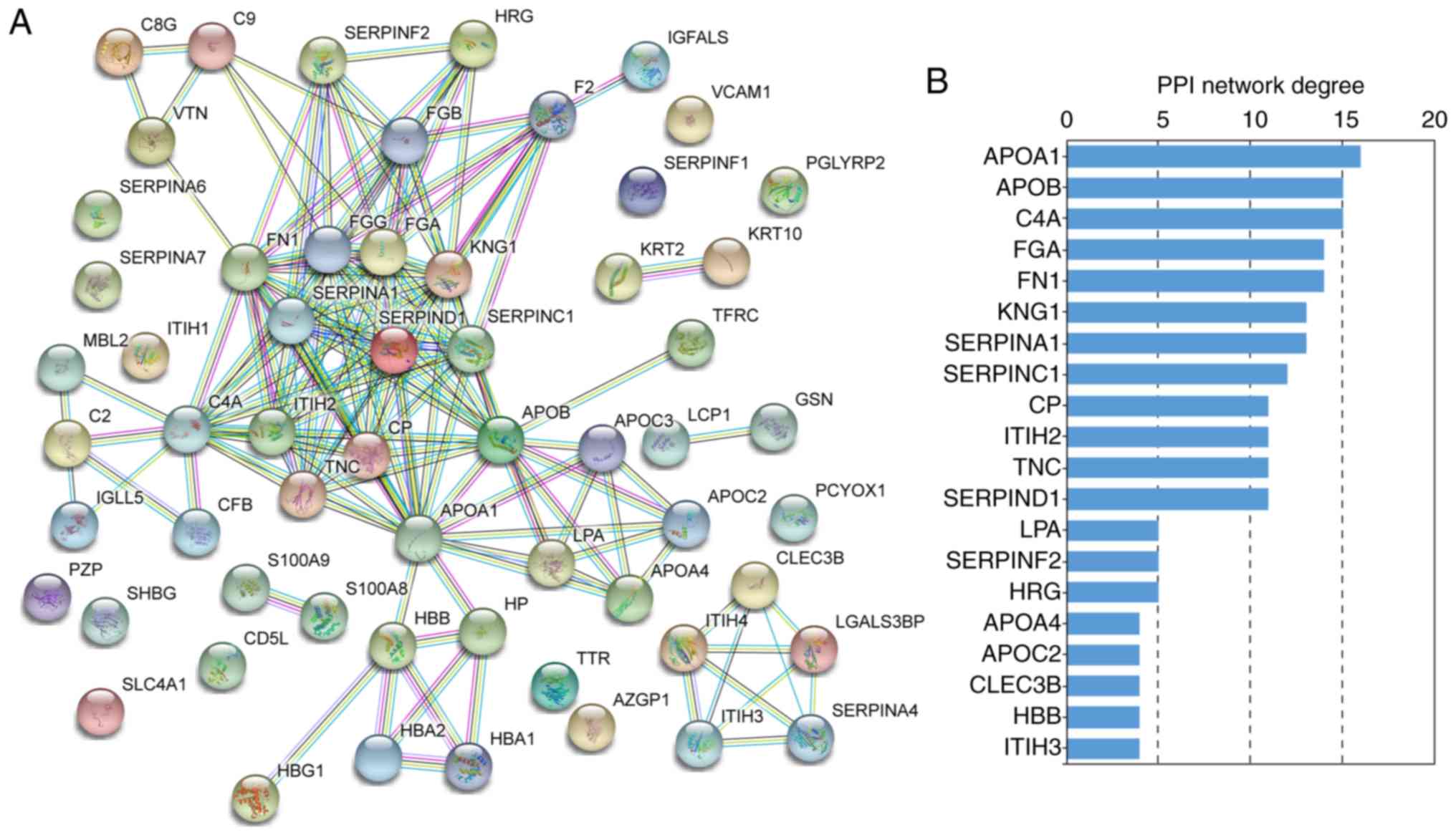
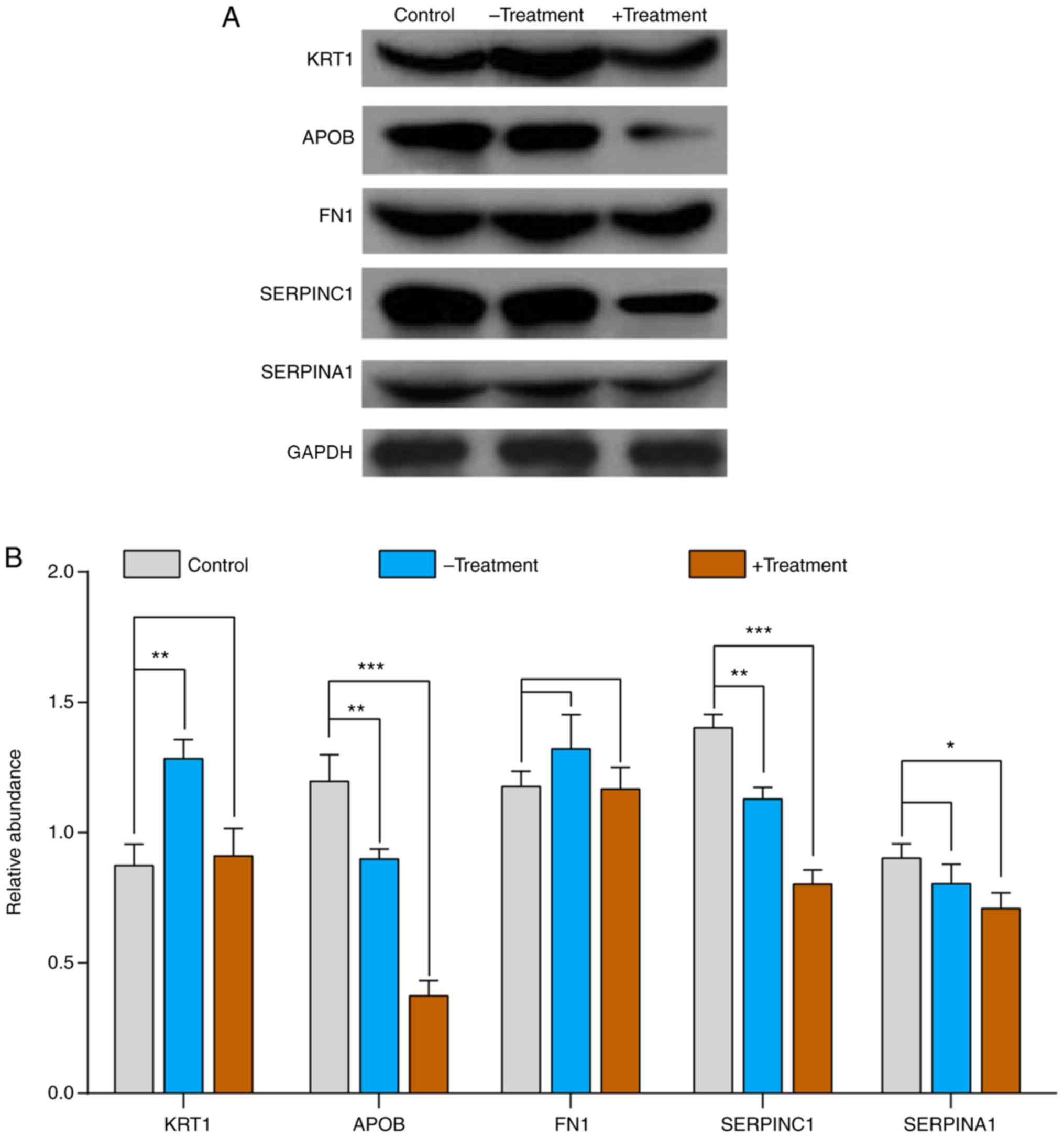
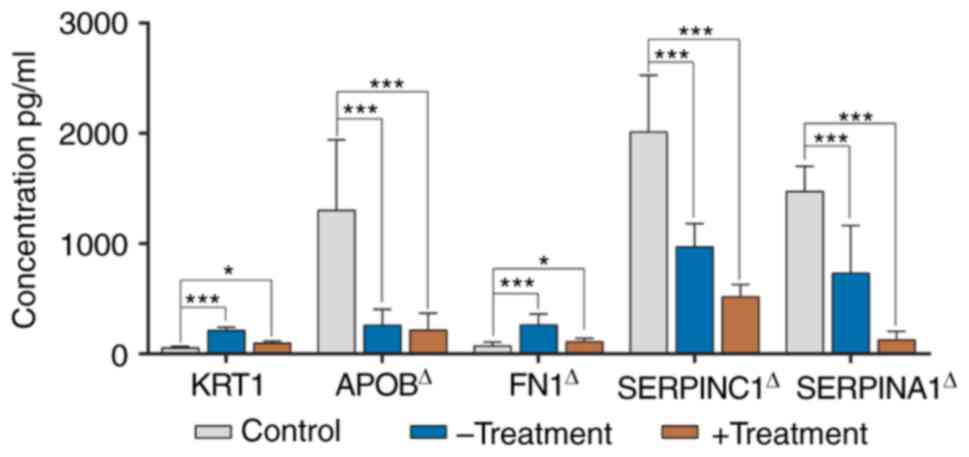
|
1
|
Global Initiative for Asthma, . Global
Strategy for Asthma Management and Prevention. 2018, http://www.ginasthma.org
|
|
2
|
Asthma Group, Society of Respiratory
Diseases, Chinese Medical Association, . Guidelines for the
prevention and treatment of bronchial asthma. Chin J Tuberculosis
Respir. 39:675–697. 2016.
|
|
3
|
Bateman ED, Hurd SS, Barnes PJ, Bousquet
J, Drazen JM, FitzGerald JM, Gibson P, Ohta K, O'Byrne P, Pedersen
SE, et al: Global strategy for asthma management and prevention:
GINA executive summary. Eur Respir J. 31:143–178. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang FL, Liao EC and Yu SJ: House dust
mite allergy: Its innate immune response and immunotherapy.
Immunobiology. 223:300–302. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eyerich S, Metz M, Bossios A and Eyerich
K: New biological treatments for asthma and skin allergies.
Allergy. 75:546–560. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pfaar O, Lou H, Zhang Y, Klimek L and
Zhang L: Recent developments and highlights in allergen
immunotherapy. Allergy. 73:2274–2289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shamji M, Kappen J, Akdis M,
Jensen-Jarolim E, Knol EF, Kleine-Tebbe J, Bohle B, Chaker AM, Till
SJ, Valenta R, et al: Biomarkers for monitoring clinical efficacy
of allergen immunotherapy for allergic rhinoconjunctivitis and
allergic asthma: An EAACI position paper. Allergy. 72:1156–1173.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Keles S, Karakoc-Aydiner E, Ozen A, Izgi
AG, Tevetoglu A, Akkoc T, Bahceciler NN and Barlan I: A novel
approach in allergen-specific immunotherapy: Combination of
sublingual and subcutaneous routes. J Allergy Clin Immunol.
128:808–815 e7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Celis JE, Gromov P, Cabezon T, Moreira
JMA, Ambartsumian N, Sandelin K, Rank F and Gromova I: Proteomic
characterization of the interstitial fluid perfusing the breast
tumor microenvironment: A novel resource for biomarker and
therapeutic target discovery. Mol Cell Proteomics. 3:327–344. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ray S, Reddy PJ, Jain R, Gollapalli K,
Moiyadi A and Srivastava S: Proteomic technologies for the
identification of disease biomarkers in serum: Advances and
challenges ahead. Proteomics. 11:2139–2161. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thomas PD, Campbell MJ, Kejariwal A, Mi H,
Karlak B, Daverman R, Diemer K, Muruganujan A and Narechania A:
PANTHER: A library of protein families and subfamilies indexed by
function. Genome Res. 13:2129–2141. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mi H, Muruganujan A and Thomas PD: PANTHER
in 2013: Modeling the evolution of gene function, and other gene
attributes, in the context of phylogenetic trees. Nucleic Acids
Res. 41:D377–D386. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mi H, Muruganujan A, Ebert D, Huang X and
Thomas PD: PANTHER version 14: More genomes, a new PANTHER GO-slim
and improvements in enrichment analysis tools. Nucleic Acids Res.
47:D419–D426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tripathi S, Pohl M, Zhou Y,
Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che
J, Mulder LCF, et al: Meta- and orthogonal integration of influenza
‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host
Microbe. 18:723–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aggarwal K, Choe LH and Lee KH: Shotgun
proteomics using the iTRAQ isobaric tags. Brief Funct Genomics
Proteomic. 5:112–120. 2006. View Article : Google Scholar
|
|
21
|
Roth W, Kumar V, Beer HD, Richter M,
Wohlenberg C, Reuter U, Thiering S, Staratschek-Jox A, Hofmann A,
Kreusch F, et al: Keratin 1 maintains skin integrity and
participates in an inflammatory network in skin through
interleukin-18. J Cell Sci. 125:5269–5279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choate KA, Lu Y, Zhou J, Elias PM, Zaidi
S, Paller AS, Farhi A, Nelson-Williams C, Crumrine D, Milstone LM
and Lifton RP: Frequent somatic reversion of KRT1 mutations in
ichthyosis with confetti. J Clin Invest. 125:1703–1707. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Depianto D, Kerns M, Dlugosz A and
Coulombe PA: Keratin 17 promotes epithelial proliferation and tumor
growth by polarizing the immune response in skin. Nat Genet.
42:910–914. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao Z, Ji X, Gu J, Wang XY, Ding L and
Zhang H: microRNA-107 protects against inflammation and endoplasmic
reticulum stress of vascular endothelial cells via KRT1-dependent
Notch signaling pathway in a mouse model of coronary
atherosclerosis. J Cell Physiol. 234:12029–12041. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang HC, Wu BQ, Hao YL, Luo Y, Zhao HL,
Zhang WY, Zhang ZL, Liang JJ, Liu W and Chen XH: KRT1 gene
silencing ameliorates myocardial ischemia-reperfusion injury via
the activation of the Notch signaling pathway in mouse models. J
Cell Physiol. 234:3634–3646. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tao H, Cox D and Frazer K: Allele-specific
KRT1 expression is a complex trait. PLoS Genet. 2:e932006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao P, Grigoryev D, Breslin L, Cheadle C,
Mathias RA, Beaty TH, Togias A and Barnes K: Keratins: Important
candidate genes for asthma and immune responsiveness to cockroach.
J Allergy Clin Immunol. 119 (Suppl):S1762007. View Article : Google Scholar
|
|
28
|
Whitfield AJ, Barrett PH, Van Bockxmeer FM
and Burnett JR: Lipid disorders and mutations in the APOB gene.
Clin Chem. 50:1725–1732. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duvillard L, Pont F, Florentin E,
Galland-Jos C, Gambert P and Vergès B: Metabolic abnormalities of
apolipoprotein B-containing lipoproteins in non-insulin-dependent
diabetes: A stable isotope kinetic study. Eur J Clin Invest.
30:685–694. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Walldius G and Jungner I: The apoB/apoA-I
ratio: A strong, new risk factor for cardiovascular disease and a
target for lipid-lowering therapy-a review of the evidence. J
Intern Med. 259:493–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tort O, Escribà T, Egaña-Gorroño L, de
Lazzari E, Cofan M, Fernandez E, Gatell JM, Martinez E, Garcia F
and Arnedo M: Cholesterol efflux responds to viral load and CD4
counts in HIV+ patients and is dampened in HIV exposed. J Lipid
Res. 59:2108–2115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee J, Kang M, Choi J, Park JS, Park JK,
Lee EY, Lee EB, Pap T, Yi EC and Song YW: Apolipoprotein B binds to
enolase-1 and aggravates inflammation in rheumatoid arthritis. Ann
Rheum Dis. 77:1480–1489. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nagel G, Weiland S, Rapp K, Link B,
Zoellner I and Koenig W: Association of apolipoproteins with
symptoms of asthma and atopy among schoolchildren. Int Arch Allergy
Immunol. 149:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pettersson ME, Koppelman GH, Flokstra-de
Blok BM, van Ginkel CD, Roozendaal C, Muller-Kobold AC, Kollen BJ
and Dubois AEJ: Apolipoprotein B: A possible new biomarker for
anaphylaxis. Ann Allergy Asthma Immunol. 118:515–516. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Perelman B, Adil A and Vadas P:
Relationship between platelet activating factor acetylhydrolase
activity and apolipoprotein B levels in patients with peanut
allergy. Allergy Asthma Clin Immunol. 10:202014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang BJ, Wang GL, Chen DH, Wang WX, Huang
J, Rong JY, Liu XT and Yang S: Association of ORMDL3 single
nucleotide polymorphisms with lysophosphatidylcholine and
apolipoprotein B levels in children with asthma. Zhongguo Dang Dai
Er Ke Za Zhi. 17:241–244. 2015.(In Chinese). PubMed/NCBI
|
|
37
|
Barochia AV, Kaler M, Cuento RA, Gordon
EM, Weir NA, Sampson M, Fontana JR, MacDonald S, Moss J,
Manganiello V, et al: Serum apolipoprotein A-I and large
high-density lipoprotein particles are positively correlated with
FEV1 in atopic asthma. Am J Respir Crit Care Med. 191:990–1000.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bochkov Y, Hanson K, Keles S,
Brockman-Schneider RA, Jarjour NN and Gern JE: Rhinovirus-induced
modulation of gene expression in bronchial epithelial cells from
subjects with asthma. Mucosal Immunol. 3:69–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang M, Zhao X, Liu Y, Tian Y, Ran X and
Jiang Y: A role for WNT1-inducible signaling protein-1 in airway
remodeling in a rat asthma model. Int Immunopharmacol. 17:350–357.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo Z, Wu J, Zhao J, Liu F, Chen Y, Bi L
and Dong L: IL-33/ST2 promotes airway remodeling in asthma by
activating the expression of fibronectin 1 and type 1 collagen in
human lung fibroblasts. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi.
30:975–979. 2014.(In Chinese). PubMed/NCBI
|
|
41
|
Bezemer ID, Bare LA, Doggen CJ, Arellano
AR, Tong C, Rowland CM, Catanese J, Young BA, Reitsma PH, Devlin JJ
and Rosendaal FR: Gene variants associated with deep vein
thrombosis. JAMA. 299:1306–1314. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
de Haan H, Bezemer I, Doggen C, Cessie SL,
Reitsma PH, Arellano AR, Tong CH, Devlin JJ, Bare LA, Rosendaal FR
and Vossen CY: Multiple SNP testing improves risk prediction of
first venous thrombosis. Blood. 120:656–663. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luxembourg B, Pavlova A, Geisen C,
Spannagl M, Bergmann F, Krause M, Alesci S, Seifried E and
Lindhoff-Last E: Impact of the type of SERPINC1 mutation and
subtype of antithrombin deficiency on the thrombotic phenotype in
hereditary antithrombin deficiency. Thromb Haemost. 111:249–257.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rosendaal F: Causes of venous thrombosis.
Thromb J. 14 (Suppl 1):S242016. View Article : Google Scholar
|
|
45
|
Majoor CJ, Kamphuisen PW, Zwinderman AH,
Ten Brinke A, Amelink M, Rijssenbeek-Nouwens L, Sterk PJ, Büller HR
and Bel EH: Risk of deep vein thrombosis and pulmonary embolism in
asthma. Eur Respir J. 42:655–661. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sneeboer MMS, Majoor CJ, de Kievit A,
Meijers JCM, van der Poll T, Kamphuisen PW and Bel EH:
Prothrombotic state in patients with severe and
prednisolone-dependent asthma. J Allergy Clin Immunol.
137:1727–1732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu CJ, Söderhäll C, Bustamante M, Baïz N,
Gruzieva O, Gehring U, Mason D, Chatzi L, Basterrechea M, Llop S,
et al: DNA methylation in childhood asthma: An epigenome-wide
meta-analysis. Lancet Respir Med. 6:379–388. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cosio MG, Bazzan E, Rigobello C, Tinè M,
Turato G, Baraldo S and Saetta M: Alpha-1 antitrypsin deficiency:
Beyond the protease/antiprotease paradigm. Ann Am Thorac Soc. 4
(Suppl 13):S305–S310. 2016. View Article : Google Scholar
|
|
49
|
McCarthy C, Reeves EP and McElvaney NG:
The role of neutrophils in alpha-1 antitrypsin deficiency. Ann Am
Thorac Soc. 4 (Suppl 13):S297–S304. 2016. View Article : Google Scholar
|
|
50
|
De Serres F and Blanco I: Role of alpha-1
antitrypsin in human health and disease. J Intern Med. 276:311–335.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Janciauskiene S and Welte T: Well-known
and less well-known functions of alpha-1 antitrypsin. Its role in
chronic obstructive pulmonary disease and other disease
developments. Ann Am Thorac Soc. 4 (Suppl 13):S280–S288. 2016.
View Article : Google Scholar
|
|
52
|
Bragina EY, Goncharova IA, Garaeva AF,
Nemerov EV, Babovskaya AA, Karpov AB, Semenova YV, Zhalsanova IZ,
Gomboeva DE, Saik OV, et al: Molecular relationships between
bronchial asthma and hypertension as comorbid diseases. J Integr
Bioinform. 15:201800522018. View Article : Google Scholar
|
|
53
|
Suárez-Lorenzo I, de Castro FR,
Cruz-Niesvaara D, Herrera-Ramos E, Rodríguez-Gallego C and
Carrillo-Diaz T: Alpha 1 antitrypsin distribution in an allergic
asthmatic population sensitized to house dust mites. Clin Transl
Allergy. 8:442018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hernández-Pérez JM, Ramos-Díaz R and Pérez
JA: Identification of a new defective SERPINA1 allele (PI*Z la
palma) encoding an alpha-1-antitrypsin with altered glycosylation
pattern. Respir Med. 131:114–117. 2017. View Article : Google Scholar : PubMed/NCBI
|