Introduction
Cataract is the most common eye disease that can
lead to blindness worldwide and is associated with several factors,
such as aging, genetic, malnutrition, immune and metabolic
abnormalities, trauma, radiation and poisoning (1). Separately or combined, these factors
induce high levels of lens epithelial cell apoptosis, which
ultimately results in cataract, which affects the eyesight of the
patients to some extent (2). With
regards to the risk factors of cataract, metabolic diseases,
especially diabetes, have become the second most important factor,
with age being the first, gradually increasing the incidence of
cataracts (3,4). Previous studies have shown that
reactive oxygen species (ROS) also play an important role in the
development of sugar cataract (5,6).
Thioredoxin (Trx), which combats against oxidative
damage, is widely distributed in the human body (7). Trx exists in almost all eukaryotic
cells and maintains the homeostasis of the organism against
oxidative damage caused by ROS (8). Trx contains disulfide/dithiol, which
acts as an active site for redox reactions, making it a scavenger
of ROS (9). Another protein,
thioredoxin interacting protein (Txnip), is closely linked to Trx
in human HL-60 cells and can give rise to 1,25-vitamin D3 gene
expression in high quantities, and thus, is referred to as vitamin
D3 upgrade protein (VDUP) (10).
Previous studies have also revealed the interaction between Trx and
Txnip using yeast two-hybrid method (11). Txnip serves a negative regulatory
role by changing the Trx disulfide bond conformation via binding
itself onto the redox active site of Trx, greatly weakening the Trx
redox function (12,13). Furthermore, a high-glucose
environment is a trigger of Txnip production (14).
The main role of ubiquitination is to degrade target
proteins. Under the environment of high glucose, ubiquitination, as
one of protein modification after translation, is very active
(15). Therefore, it was
speculated that the specificity of ubiquitin modification may
degrade Txnip to restore the function of Trx. The process of
ubiquitin degradation protein is a tertiary enzyme linked process:
Ubiquitin activating enzyme-E1 makes a thioester bond with the
carboxy-terminal glycine residue of ubiquitin. Then, the
ATP-activated ubiquitin is transferred to ubiquitin conjugating
enzyme-E2 via a covalent thioester linkage with the aid of E1
(16). Finally, ubiquitin
ligase-E3 completes the transfer of activated ubiquitin onto the
substrates and degrades them. There are only two types of E1 and a
few types of E2, and both of these are abundant in vivo,
which is similar to the ubiquitin molecules (17). However, the most important enzymes
in this pathway are E3s, as they recognize specific substrates. E3
ubiquitin ligase, as the key regulator of target protein
degradation in the ubiquitin-proteasome pathway, can specifically
identify and bind to the substrate protein (18).
Atrophin-1 interacting protein 4 (AIP4) or ITCH gene
was identified at the agouti locus in mouse coat color study and
encodes a 113 kDa HECT type Nedd4-like E3 ubiquitin ligase
(19). Moreover, ITCH plays an
important role in regulating apoptosis and other biological
processes (19). It has also been
shown that in myocardial cells, Txnip is a target protein of
ubiquitin molecules (20). By
interacting with E3 ligase-ITCH, Txnip protein expression is
degraded by ubiquitin, thus reversing the degree of myocardial cell
apoptosis caused by myocardial infarction (20). However, to the best of our
knowledge, whether this mechanism can be applied in the model of
sugar cataract has not been previously reported. Given that Txnip
is a key factor of diabetes (14)
and the effect of ITCH on Txnip degradation, this mechanism may be
suitable for application in sugar cataracts, which are caused by
oxidative stress.
In the present study, in order to explore the
pathogenesis of sugar cataract, it was proposed that ITCH may
regulate high glucose-induced oxidative stress through Txnip in
sugar cataracts. Following which, the changes and interactions of
Txnip, Trx and ITCH in human lens epithelia cells (HLECs) were
examined in a high glucose environment. It was determined that
oxidative stress was regulated through ITCH and altered the
apoptosis of cells. This may help to identify a novel methods to
guide clinical treatment and help the development of effective and
high safety anti-cataract drugs.
Materials and methods
Cell culture and grouping
The human lens epithelial cell (HLEC) line
(SRA01/04) was provided by the Ophthalmic Lens Key Laboratory of
The Fourth Affiliated Hospital of the Chinese Medical University.
Cells were incubated in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) with 25 mM glucose (Sigma-Aldrich; Merck KGaA), 10% FBS
(Serana Europe, GmbH), 100 U/ml penicillin and 100 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.), at 5%
CO2 saturation and 37°C in a cell incubator. When the
cell confluence reached ~70%, the glucose concentration for cells
was changed and cells were randomly divided into the following
groups: Low-glucose group (LG; 5.5 mM glucose concentration in
culture); high-glucose 1 group (HG1; 12.5 mM glucose concentration
in culture); high-glucose 2 group (HG2; 25 mM glucose concentration
in culture); high-glucose 3 group (HG3; 50 mM glucose concentration
in culture) and mannitol (Sangon Biotech Co., Ltd.) group (5.5 mM
glucose concentration and 50 mM mannitol in culture) The
aforementioned culture conditions were used and cells were cultured
for 24 h. The original cells were treated with 50 mM glucose for
different incubation time (0, 6, 12 and 24 h) (21).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA of each group of cells was extracted with
the TRIzol® reagent (Takara Bio, Inc.) via
centrifugation at 3,000 × g for 10 min at room temperature. After
total RNA was quantified, RT was performed to synthesize cDNA using
PrimeScript RT Master Mix (Takara Bio, Inc.). RT was conducted at
37°C for 15 min and 85°C for 5 sec. qPCR was conducted using cDNA,
forward and reverse primers, SYBR Premix Ex TaqII (Takara Bio,
Inc.) and ROX Reference Dye II (50X; Takara Bio, Inc.) under the
following conditions: Initial denaturation at 95°C for 30 sec,
followed by 40 cycles at 95°C for 5 sec and 60°C for 34 sec.
Relative expression levels of mRNA were calculated using the
2−ΔΔCq method (22).
The following primers were designed and synthesized
to detect Trx, Txnip, ITCH and β-actin cDNA: Trx forward,
5′-TGTGGGCCTTGCAAAATGA-3′; Trx reverse,
5′-GGAATATCACGTTGGAATACTTTTCA-3′; Txnip forward,
5′-ACCTGCCCCTGGTAATTGG-3′; Txnip reverse, 5′-TTCGGCTGGCCATGCT-3′;
ITCH forward, 5′-ACCGGCTGCCATCTTAGTCT-3′; ITCH reverse,
5′-GGAAAACCTGAAGTTCTCACAGT-3′; β-actin forward,
5′-GTGGGGCGCCCAGGCACCA-3′; and β-actin reverse,
5′-CTCCTTAATGTCACGCAGGATTTC-3′. Then, RT-qPCR amplification and
analysis were performed using an Applied Biosystems 7500 RT-qPCR
machine (Applied Biosystems; Thermo Fisher Scientific, Inc.).
β-actin was used as the internal reference.
Western blotting
When cell confluence reached ~90%, total protein was
extracted using RIPA protein lysis with PMSF (Beijing Solarbio
Science & Technology Co., Ltd.). After bicinchoninic acid (BCA;
Wanleibio Co., Ltd.) measurement, the proteins (40 µg/lane) were
added to 10% SDS-PAGE (Nanjing KeyGen Biotech Co., Ltd.) and
separated, and then transferred to PVDF membrane (EMD Millipore).
After blocking with 5% non-fat milk at room temperature for 2 h,
membranes were incubated overnight at 4°C with primary antibodies
and secondary antibodies at room temperature for 2 h. Antibody
incubations were performed at the following dilutions: Trx (cat.
no. ab26320; 1:5,000; Abcam), Txnip (cat. no. 14715s; 1:1,000; Cell
Signaling Technology, Inc.), ITCH (cat. no. 12117s; 1:1,000; Cell
Signaling Technology, Inc.), GAPDH (cat. no. 10494-1-AP; 1:5,000;
ProteinTech Group, Inc.), caspase-3 (cat. no. 19677-1-AP; 1:5,000;
ProteinTech Group, Inc.) and horseradish peroxidase-conjugated
secondary antibodies against rabbit (cat. no. 10285-1-AP; 1:10,000;
ProteinTech Group, Inc.). The blots were visualized using ECL
(Thermo Fisher Scientific, Inc.) and an ECL system (Fluor Chem FC2,
Alpha Innotech, Inc.) was used to visualize the bands.
Densitometric analysis was performed by ImageJ software (version
1.8.0; National Institutes of Health). GAPDH was used to normalize
the expression data.
Immunofluorescence and
co-immunofluorescence
Cells were plated into 24-well plates at
5×104 cells per well and cultured for 24 h. When the
cell confluence reached ~50%, cells were fixed with 4%
paraformaldehyde (Sangon Biotech Co., Ltd.) at room temperature for
30 min and solubilized in 0.1% Triton X-100 (Sangon Biotech Co.,
Ltd.) at room temperature for 10 min. After blocking with 5%
non-fat milk at room temperature for 2 h, the cells were incubated
with primary antibodies at 37°C for 4 h and secondary antibodies at
37°C for 2 h in a dark environment. For immunofluorescence, the
primary antibody of Trx were added, and for co-immunofluorescence,
the primary antibodies of Txnip and Itch were added at the same
time. Antibody incubations were performed at the following
dilutions: Trx (cat. no. ab26320; 1:5,000; Abcam), Txnip (cat. no.
14715s; 1:100; Cell Signaling Technology, Inc.), ITCH (cat. no.
sc-28367; 1:100; Santa Cruz Biotechnology, Inc.), donkey anti-mouse
IgG highly cross-adsorbed secondary antibody-Alexa Fluor 488 (cat.
no. A-21202; 1:2,000; Invitrogen; Thermo Fisher Scientific, Inc.)
and goat anti-rabbit IgG highly cross-adsorbed secondary
antibody-Alexa Fluor 594 (cat. no. A-11012; 1:1,000; Invitrogen;
Thermo Fisher Scientific, Inc.). DAPI (Beyotime Institute of
Biotechnology) was used for staining at room temperature for 3 min.
Subsequently, a fluorescence microscope (magnification, ×400) was
used to observe and collect the images.
Co-immunoprecipitation (COIP)
Cells were plated at 5×106 cells per well
into three dishes with 10-cm diameter and cultured until cell
confluence reached ~100%. Subsequently, total protein was extracted
as aforementioned in the western blotting protocol and the protein
concentration was determined using the BCA method. Total protein
lysate, primary antibodies or IgG, incubation buffer (ProteinTech
Group, Inc.) and Protein A sepharose beads slurry (ProteinTech
Group, Inc.) were added into the spin columns and incubated at 4°C
overnight. Subsequently, protein A sepharose beads slurry were
added to the mixture for another incubation overnight at 4°C.
Antibody incubations were performed at the following dilutions:
Txnip (cat. no. 14715s; 1:50; Cell Signaling Technology, Inc.),
ITCH (cat. no. 12117s; 1:200; Cell Signaling Technology, Inc.), Trx
(cat. no. ab26320; 1:5,000; Abcam) and IgG (cat. no. sc-2025;
1:100; Santa Cruz Biotechnology, Inc.). The products were washed
with elution buffer (ProteinTech Group, Inc.) and mixed with Alkali
neutralization buffer (ProteinTech Group, Inc.) and protein loading
buffer (ProteinTech Group, Inc.) and boiled for 5 min. Finally, the
products were collected for western blotting.
Overexpression and knockdown of ITCH
in HLECs
ITCH full-length cDNA was amplified from HLECs and
ligated to a pEGFP-C1 vector (Thermo Fisher Scientific, Inc.) with
the restriction sites of SAC1 and BamH1. For overexpression of
ITCH, pEGFP-C1-ITCH or empty vector [negative control (NC)] was
mixed with Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) in serum-free DMEM for 5 min. The mixtures
were then added into each well in 6-well plates (1×106
cells) and incubated for 48 h at 37°C with 50 mM glucose, before
the cells were harvested for subsequent experiments. For knockdown
of ITCH, small interfering (si)RNA or siNC (Guangzhou RiboBio Co.,
Ltd.) was mixed with RiboFECT™ CP Reagent (Guangzhou RiboBio Co.,
Ltd.) and RiboFECT™ CP Buffer (Guangzhou RiboBio Co., Ltd.) in
serum-free DMEM medium for 15 min. The mixtures were added into
each well in 6-well plates (1×106 cells) and incubated
for 48 h at 37°C with 50 mM glucose, and then subsequent
experiments were conducted.
The following primers were designed and synthesized
to detect ITCH full-length cDNA. ITCH forward,
5′-CGAGCTCAAATGTCTGACAGTGGATCACAA-3′ and reverse,
5′-CGCGGATCCTTACTCTTGTCCAAATCCTTCTGTTTC-3′. PCR was performed under
the following conditions: Initial denaturation at 98°C for 5 min,
followed by 35 cycles at 98°C for 10 sec, 55°C for 15 sec, 68°C for
1 min and 68°C for 10 min.
siRNA sequences were as follows: si-ITCH_01:
GGAGCAACATCTGGATTAA; si-ITCH_02: CAGCAATGGCAGAGTATAT; and
si-ITCH_03: GAGAAGAAGGTTTAGATTA.
MG132 treatment for cells
A specific ubiquitination inhibitor MG132 (Sangon
Biotech Co., Ltd.) (23) 10 µM was
added to cells that had been transfected with pEGFP-C1-ITCH after
incubation for 44 h.
Cell proliferation assay by cell
counting Kit-8 (CCK-8)
Cell suspension was counted and inoculated in
96-well plates with 5×103 cells per well. Cells were
adherent to the wall and cultured for 48 h in groups according to
the following: LG, HG1, HG2, HG3, siNC, siRNA-ITCH, pEGFP-C1-ITCH,
pEGFP-C1 empty vector and MG132-pEGFP-C1-ITCH. The glucose
concentration of each transfected group was 50 mM. Each group was
divided into five multiple wells and a blank control group (only
the culture medium without cells) was set up. 48 h after
transfection, the new DMEM was replaced and 10 µl of CCK-8 reagent
(Beyotime Institute of Biotechnology) was added for a further
culture for 1 h. Then, 450 nm absorbance was detected by the
automatic microplate reader (ELX-800, BioTek Instruments,
Inc.).
Intracellular superoxide assay
Cells were plated in 96-well plates at
5×103 cells per well, cultured and grouped as mentioned
for the CCK-8 assay, and were incubated with 200 µl of the
superoxide-detecting reagents, including WST-1 and catalase
(Beyotime Institute of Biotechnology) at 37°C for 3 min. An
automatic microplate reader (ELX-800; BioTek Instruments, Inc.) was
used to detect absorbance in 450 nm.
Insulin disulfide reduction experiment
for Trx activity assay
Trx activity was evaluated with insulin disulfide
reduction assay, as previously reported (24,25).
NP40 protein lysis with PMSF (Beyotime Institute of Biotechnology)
was added to the cells to extract the total protein. Lysate was
incubated with 2 µl DTT active buffer composed of 50 mM HEPES (pH
7.6), 1 mM EDTA, 1 mg/ml BSA and 2 mM DTT solution (Beyotime
Institute of Biotechnology) at 37°C for 20 min. Then, the reaction
buffer containing 200 µl 1 M HEPES (pH 7.6), 40 µl 0.2 M EDTA, 40
µl 40 mg/ml NADPH (Beyotime Institute of Biotechnology) and 500 µl
10 mg/ml insulin (Beyotime Institute of Biotechnology) was added.
The reaction was started by the addition of 10 µl recombinational
Trx reductase (Abcam) and incubated for 20 min at 37°C. The
reaction was terminated by the addition 0.5 ml 6 mol/l
guanidine-HCl and 1 mmol DTNB (Beyotime Institute of Biotechnology)
and absorbance at 412 nm was measured using an automatic microplate
reader (ELX-800; BioTek Instruments, Inc.).
Flow cytometry for cell apoptosis
assay
Cells were cultured and treated for 48 h in the same
condition as mentioned for the CCK-8 assay, and then the procedure
was performed according manufacturer's instructions of the Annexin
V-APC/7-AAD apoptosis kit (Wanleibio Co., Ltd.). Then, flow
cytometry (Accuri C6; BD Biosciences) was used to determine the
apoptotic rate of each group.
Statistical analysis
SPSS 24.0 statistical software (IBM Corp.) and
GraphPad Prism 8.0 (GraphPad Software, Inc.) were used for data
statistics and analysis. One-way ANOVA followed by Tukey-Kramer
multiple comparison post hoc tests was used for multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression levels of Txnip, Trx and
ITCH were affected by high glucose in HLECs
It was found that the mRNA and protein expression
levels of Txnip in HLECs were increased in a time-dependent and
concentration-dependent manner after glucose stimulation
(P<0.05; Figs. 1A and B and
2A and B). However, the trends of
Trx and ITCH showed an opposite effect to Txnip (P<0.05;
Figs. 1C-F and 2C-F). The mRNA expression levels of
Txnip, Trx and ITCH showed no significant difference between LG and
mannitol groups (Fig. S1A-C). The
insulin disulfide reduction assay results indicated that the
activity of Trx protein in the LG group was the highest, which
gradually decreased with the increase in glucose concentration
(P<0.05; Fig. 2G). Using
immunofluorescence localization, it was demonstrated that the
expression localization of Trx protein in LG group was in nucleus
and cytoplasm, and the main expression location was in the nucleus
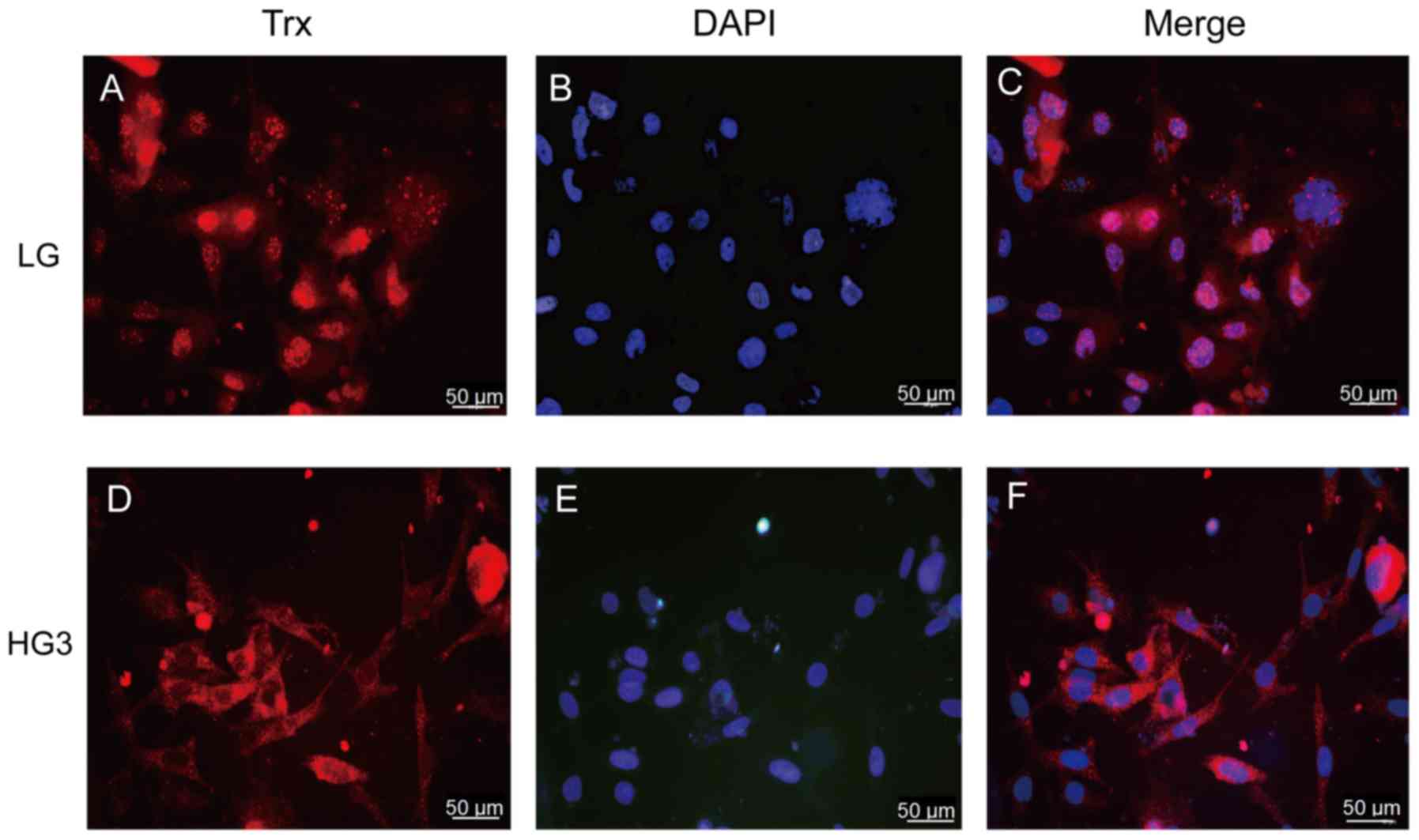
(Fig. 3A-C). In the HG3 group,
with the increase of glucose concentration, the expression of Trx
was transferred from the nucleus to the cytoplasm, and the main
expression was located in the cytoplasm (Fig. 3D-F).
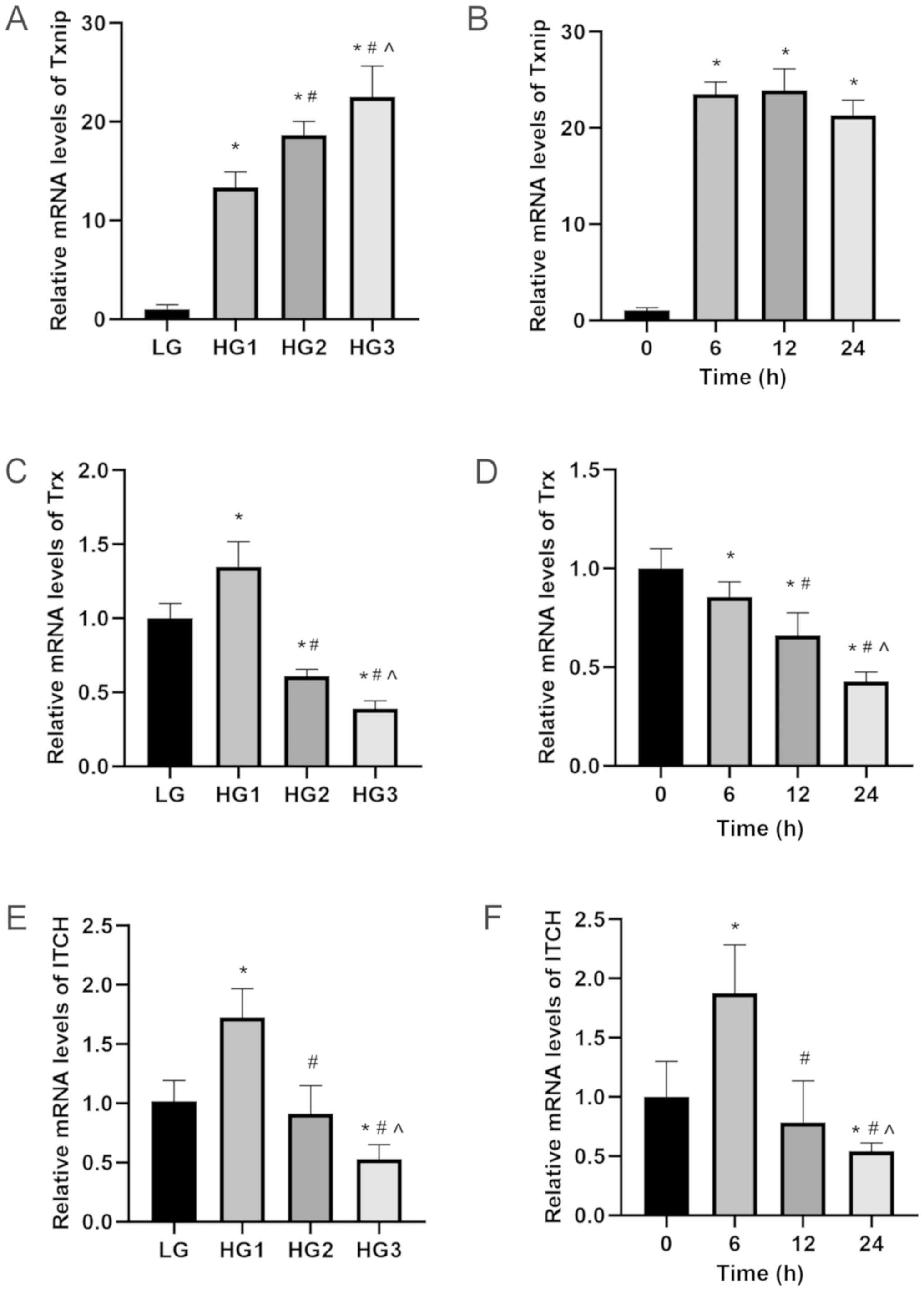 | Figure 1.mRNA expression levels of Txnip, Trx
and ITCH in HLECs. HLECs were treated with different concentration
of glucose (LG, 5.5 mM; HG1, 12.5 mM; HG2, 25 mM; HG3, 50 mM) for
24 h or treated with 50 mM glucose for different incubation time
(0, 6, 12 and 24 h). (A and B) Txnip. (C and D) Trx. (E and F)
ITCH. Data are presented as the mean ± standard deviation and
analyzed by ANOVA followed by Tukey-Kramer multiple comparison post
hoc tests. *P<0.05 vs. LG or 0 h; #P<0.05 vs. HG1
or 6 h; ^P<0.05 vs. HG2 or 12 h. Txnip, thioredoxin
interaction protein; Trx, thioredoxin; HLECs, human lens epithelial
cells; LG, low glucose; HG, high glucose. |
 | Figure 2.Protein expression levels of Txnip,
Trx and ITCH in HLECs. HLECs were treated with different glucose
concentration (LG, 5.5 mM; HG1, 12.5 mM; HG2, 25 mM; HG3, 50 mM)
for 24 h or treated with 50 mM glucose for different incubation
time (0, 6, 12 and 24 h). (A and B) Txnip. (C and D) Trx. (E and F)
ITCH. (G) Activity levels of Trx protein via insulin disulfide
reduction experiment in different groups. Data are presented as the
mean ± standard deviation and analyzed by ANOVA followed by
Tukey-Kramer multiple comparison post hoc tests. *P<0.05 vs. LG
or 0 h; #P<0.05 vs. HG1 or 6 h; ^P<0.05
vs. HG2 or 12 h. Txnip, thioredoxin interaction protein; Trx,
thioredoxin; HLECs, human lens epithelial cells; LG, low glucose;
HG, high glucose. |
Cell proliferation, intracellular
superoxide accumulation and apoptosis were affected by high glucose
in HLECs
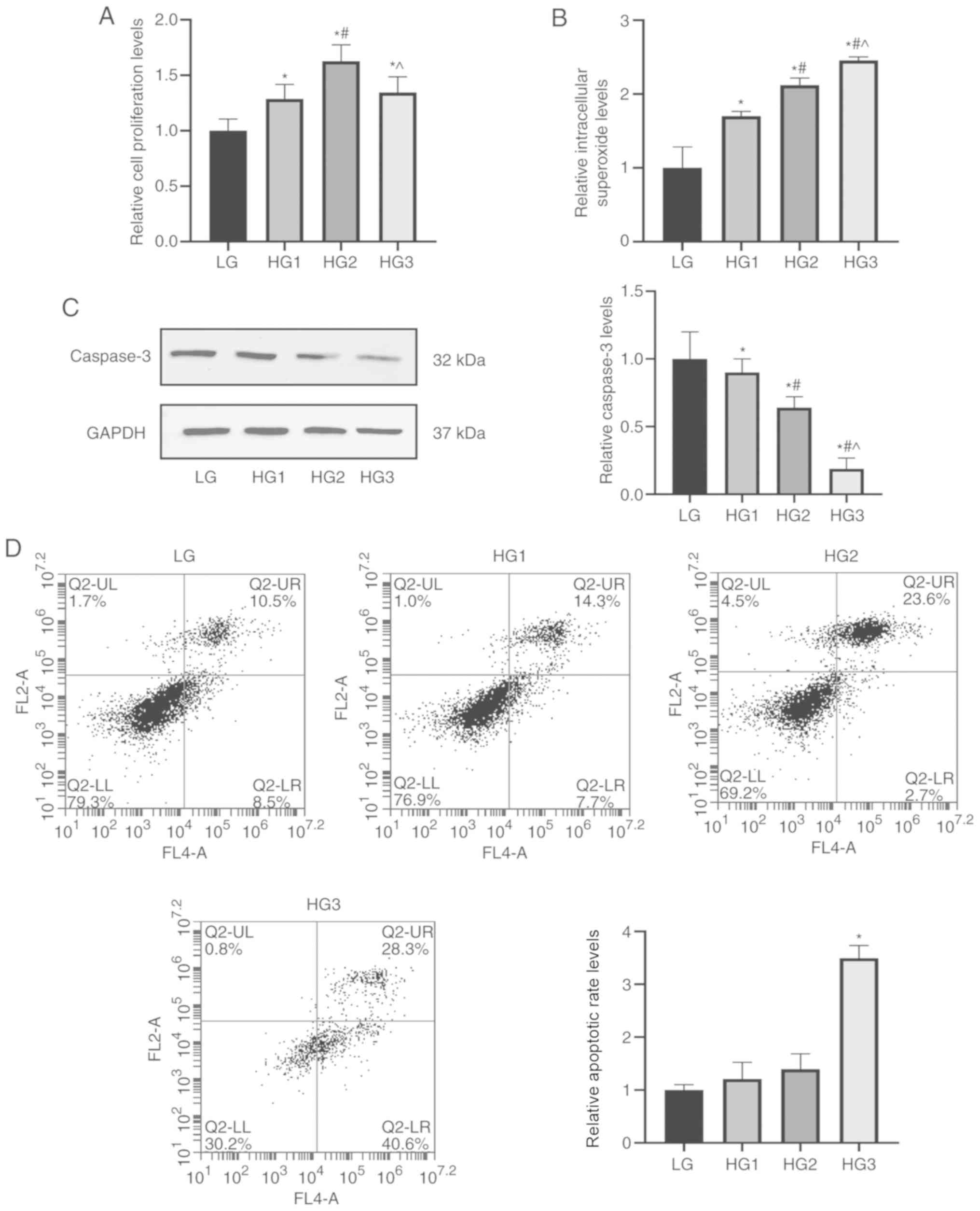
The CCK-8 results suggested that the proliferation
rate of cells showed a significant increase with rising glucose
concentration (P<0.05), while the 25-mM concentration (HG2
group) was an inflection point. Subsequently, with the continuing
increase in concentration of glucose, proliferation was decreased
(HG3 group; P<0.05; Fig. 4A).
The intracellular superoxide detection assay results identified an
increase of superoxide in the HG1 group compared with the LG group,
and this continued to increase with the elevated glucose
concentration (P<0.05; Fig.
4B).
For cell apoptosis, the content of the classical
apoptotic protein caspase-3 in each group was detected by western
blotting, and it was found that compared with the LG group,
caspase-3 expression gradually decreased in HG1, HG2 and HG3 groups
(P<0.05; Fig. 4C). Furthermore,
flow cytometry was used to quantitatively determine the apoptotic
rate of each group of cells, which demonstrated the same trend as
western blotting (P<0.05; Fig.
4D).
Interaction between ITCH and Txnip,
Txnip and Trx
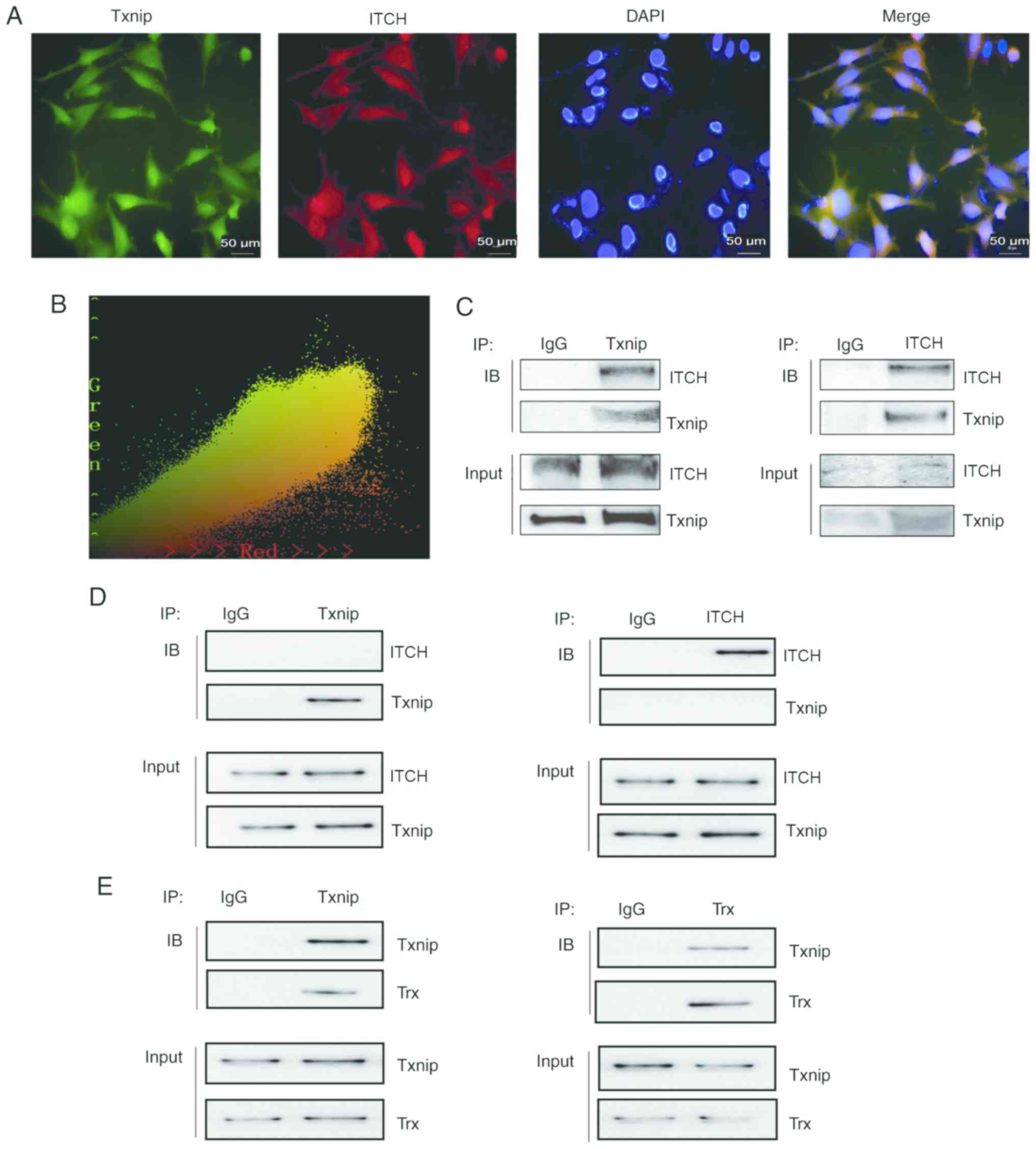
Immunofluorescence co-localization reflected that
Txnip and ITCH proteins were expressed in both the nucleus and
cytoplasm of HLECs cultured with 50 mM glucose (Fig. 5A). Subsequently, Image Pro Plus 6.0
was used for co-location analysis of these two proteins and the
results were as follow: Pearson's correlation, r=0.953276,
suggested a strong correlation between the two proteins in HLECs
(Fig. 5B). For the HG3 group, COIP
analysis indicated a prominent band expression of ITCH at 105 kDa
after IP Txnip (rabbit monoclonal antibody) and adding ITCH
antibody (rabbit monoclonal antibody) (Fig. 5C). Furthermore, Txnip antibody
(mouse monoclonal antibody) was added and a significant band of
Txnip expression was observed at 55 kDa. After IP with ITCH (rabbit
monoclonal antibody), Txnip antibody (mouse monoclonal antibody)
was added and the expression of Txnip protein band was identified
at 55 kDa (Fig. 5C). In addition,
ITCH antibody of rabbit origin was also added, and there was a
significant ITCH protein band expression at 105 kDa (Fig. 5C). For the LG group, COIP analysis
indicated that after IP Txnip, there was no prominent ITCH band at
105 kDa and after IP ITCH, the Txnip band was barely visible at 55
kDa (Fig. 5D). For the HG3 group,
COIP analysis also demonstrated that after IP Txnip (rabbit
monoclonal antibody), and Trx antibody (rabbit monoclonal antibody)
was added, a prominent band of Trx at 12 kDa was shown (Fig. 5E). After IP Trx (rabbit monoclonal
antibody), and Txnip antibody (mouse monoclonal antibody) was
added, the expression of Txnip protein band at 55 kDa was observed
(Fig. 5E).
Transfection of pEGFP-C1-ITCH plasmid
and siRNA-ITCH causes changes in the expression of associated
proteins in high-glucose environment
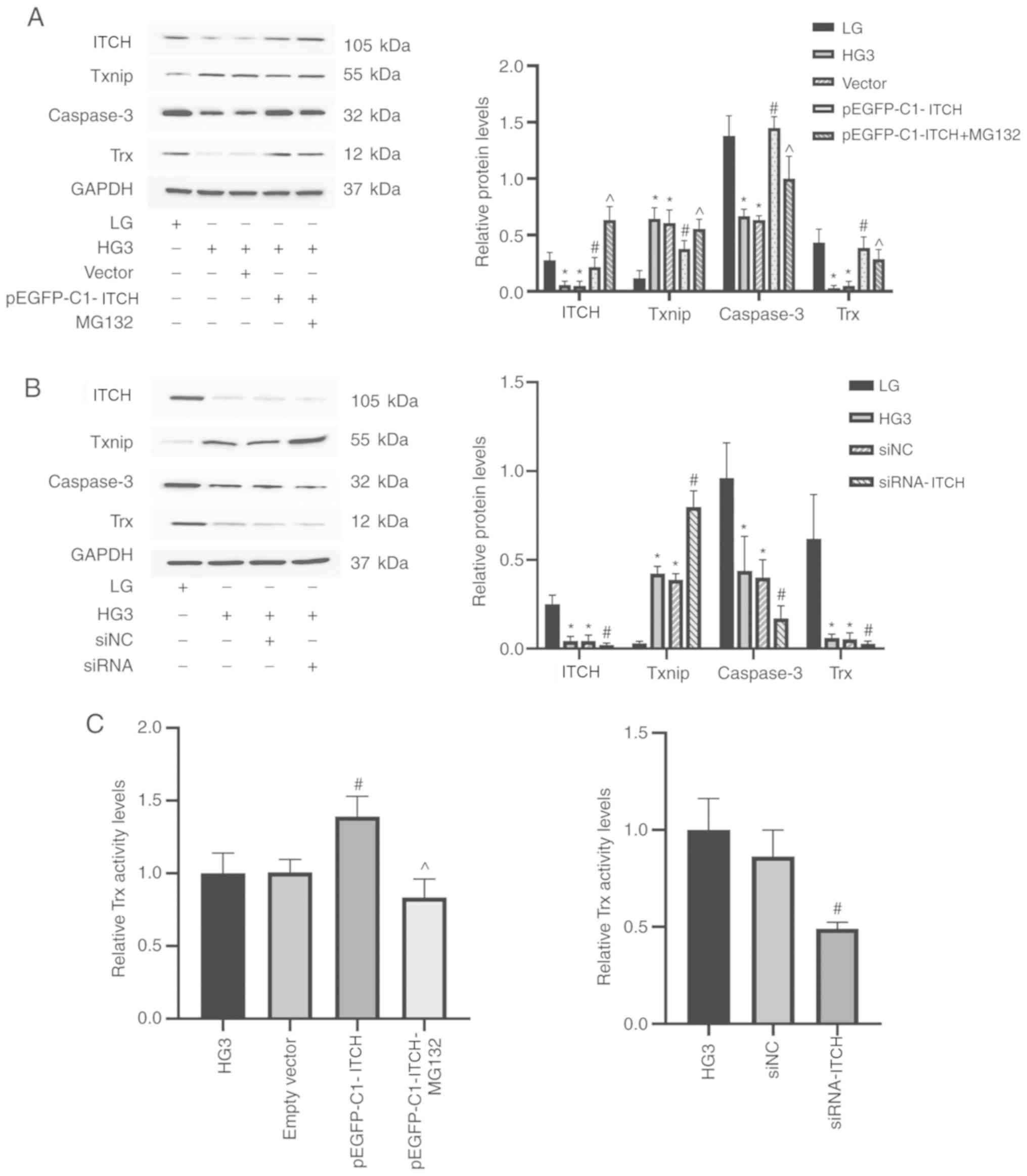
Compared with the HG3 group, ITCH protein
overexpression after transfection of pEGFP-C1-ITCH led to a series
of changes in the downstream proteins: After the increase in ITCH
expression, Txnip was significantly degraded, while Trx expression
was increased (P<0.05; Fig.
6A). In addition, caspase-3 expression was increased
(P<0.05; Fig. 6A). However, the
relative expression of each protein in the cells transfected with
empty plasmid was not statistically different compared with the HG3
group (P<0.05; Fig. 6A). When
MG132 was added to the transfection group, Txnip expression was
increased, which reversed the trend of the overexpression of ITCH
(P<0.05; Fig. 6A). Compared
with the HG3 group, ITCH expression was significantly decreased
following the knockdown of ITCH mRNA, while the expression of
downstream proteins Txnip, Trx and caspase-3 proteins demonstrated
an opposite trend compared with the pEGFP-C1-ITCH group (P<0.05;
Fig. 6B). The activity of Trx in
different groups were detected under high-glucose concentration (50
mM) by the insulin disulfide reduction experiment. Trx activity was
highest in pEGFP-C1-ITCH group, while MG132 reversed the increase
and caused a decrease in Trx activity (P<0.05); whilst Trx
activity was significantly decreased in the siRNA-ITCH group
(P<0.05; Fig. 6C).
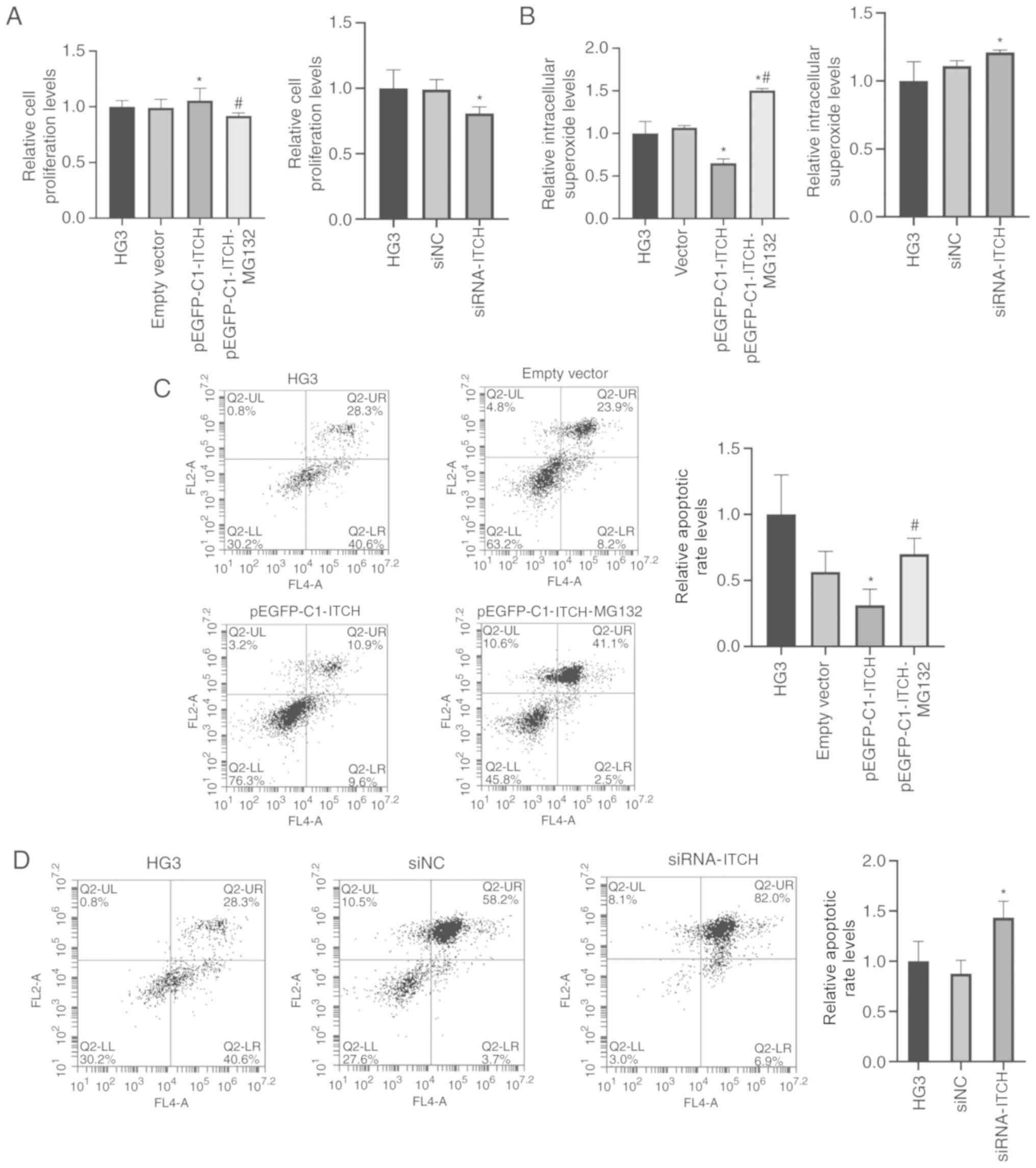
Effects of overexpression and
knockdown of ITCH on cell proliferation, superoxide content and
apoptosis in high-glucose environment
The CCK-8 results indicated that the cell
proliferation in the pEGFP-C1-ITCH group was higher compared with
the HG3 group, but this trend was reversed by MG132 and the
proliferation rate decreased (P<0.05). Furthermore, the
proliferation rate of the siRNA-ITCH group was lower compared with
the HG3 group (P<0.05; Fig.
7A).
With regards to the detection of intracellular
superoxide, it was found that the content of superoxide in the
pEGFP-C1-ITCH group was lower compared with the HG3 group, but
increased significantly after MG132 treatment (P<0.05); the
superoxide content of the siRNA-ITCH group was also higher compared
with the HG3 group (P<0.05; Fig.
7B). The flow cytometry assay demonstrated that the apoptotic
rate of the cells were consistent with the results of the
superoxide content (P<0.05; Fig. 7C
and D).
Discussion
To the best of our knowledge, the present study was
the first to demonstrate the association between oxidative stress
and Trx and Txnip in HLECs cultured in a high-glucose-induced
oxidative stress status. Cataract is the most common eye disease
causing blindness worldwide (1),
and its occurrence is associated with numerous factors, among which
oxidative stress is an important pathway (26). In addition, as a disease associated
with oxidative stress, diabetes accelerates and aggravates the
development and progression of cataract (27).
Trx is an important protein that widely exists in
eukaryotic cells to protect against oxidative stress. Trx has a
stable and unique catalytic group-cys-gly-pro-cys and through this
group, Trx in a reduced state can provide H+ for other
proteins containing disulfide bonds, which reduces the oxidation of
the target protein (28). Txnip
was originally identified as a protein that induces high levels of
vitamin D3 expression in human HL-60 cells, and hence was named
VDUP1 (29). Txnip is synthesized
in large quantities in a high-glucose environment, and strongly
binds cysteine residues to the reduce Trx via a disulfide exchange
reaction, resulting in Trx losing its ability to resist oxidative
stress (11). The present study
quantitatively analyzed the changes in the expression levels of
Txnip and Trx in culture environments with different glucose
concentrations at different durations of action. It was
demonstrated that with the increase in glucose concentration in the
medium and the extension of the time of action, the expression of
Txnip was increased, while Trx decreased. Although the mRNA
expression of Trx in the HG1 group increased, it was speculated
that it was an adaption to the rise in high glucose concentration
in the early stage of oxidative stress. In line with previous
studies, the present results suggested that Txnip and Trx were
closely associated with glucose concentration in HLECs cultured
in vitro, and showed an opposite trend, thus indicating that
they may have antagonistic effects. The mRNA expression levels of
Txnip, Trx and ITCH showed no significant difference between LG and
mannitol groups, which suggested that the changes should not be
caused by osmotic pressure. Spielberger's study (30) showed that the expression of Trx1
was also correlated with the cell density: The amount of Trx1 was
greater in sparse (low density; 20% confluent) cultures compared
with confluent (high density; 95% confluent) cultures in HeLa
cells, A549 cells and 293 cells. In human lens epithelial cells, it
was unknown whether cell density affected the expression of Trx1.
Therefore, the same initial cell density (70% confluence) was used
in order to measured Trx expression between cultures with similar
cell density, to avoid the deviation by the effect of cell
proliferation. The present results suggested that the changes in
Txnip expression induced by glucose affected Trx expression.
Immunofluorescence experiments demonstrated that Trx
was located both in nucleus and cytoplasm in the LG group, mainly
with nuclear expression. However, in the HG3 group, its expression
was transferred from the nucleus to the cytoplasm, and thus it was
speculated that Trx was transferred to mitochondria, the main site
of oxidative stress response, to play its role. The insulin
disulfide reduction experiment also identified that the activity of
Trx was decreased in the high-glucose environment.
The CCK-8 results indicated that with the increased
concentration of glucose, cell proliferation rate at first
increased, but then began to decrease at the inflection point at 25
mM glucose. Therefore, it was suggested that this may be a response
caused by overnutrition before 25 mM glucose. Moreover, the results
demonstrated that superoxide content was closely associated with
glucose. It has been reported that Txnip protein induces apoptosis
via several pathways, including the classical caspase pathway, the
MAPK pathway and the NLR family pyrin domain containing 3 pathway
(31,32). Therefore, the present study
examined the caspase-3 protein expression in the caspase pathway
and found that with the increase in glucose concentration, the
expression of caspase-3 was gradually decreased. Subsequently, flow
cytometry experiments further indicated that with the gradual
increase in glucose, the apoptotic rate of cells showed an
increasing trend.
Ubiquitination is a reversible covalent modification
and a multistage enzyme-linked reaction process, which is completed
via the continuous action of ubiquitin-activated enzyme E1,
ubiquitin-conjugating enzyme E2 and ubiquitin ligase E3 (18). ITCH, as one of the E3 ligases in
the HECT family, has been reported to be closely associated with
several diseases, such as autoimmune disease, chronic pulmonary
disease, rheumatoid arthritis and allergic dermatitis, as well as
abnormalities in the morphology of cranial and facial muscles
(33). A previous study has shown
that ITCH consumption can cause intracellular oxidative stress and
induce apoptosis (34). However,
its presence, mechanism and effect in ocular lens epithelial cells
have not been previously reported. In the present study, ITCH mRNA
and protein expression levels in HLECs were detected, and the
changes in expression, which was similar to Trx, was confirmed in
the different groups. In line with previous studies, it was
speculated that ITCH was associated with oxidative stress in HLECs.
It has been reported that Txnip is the specific target protein of
ITCH in 293T, H1299 and U2OS cells, which mediates the degradation
of Txnip and regulates apoptosis (34). Therefore, in the present study, the
co-location analysis at 50 mM glucose concentration suggested a
strong correlation between Txnip and ITCH. Moreover, the
interaction between Txnip and ITCH in HLECs was assessed by
co-localization of immunofluorescence and COIP in the HG3 group; it
was demonstrated that the interaction between ITCH and Txnip was
very weak at 5.5 mM glucose concentrations compared with 50 mM. In
addition, the interaction between Txnip and Trx was observed in
HLECs in 50 mM glucose concentration.
In the present study, ITCH was used to regulate the
oxidative stress process caused by Txnip. ITCH was overexpressed by
transfection, which downregulated Txnip and caused the release of
active Trx, leading to the increase in proliferation rate, the
decline in superoxide content and apoptotic rate in cells. To
investigate whether the changes were caused by ubiquitination,
MG132, a specific ubiquitination inhibitor, was added and it was
found that MG132 completely reversed the changes in protein
expression and cell function caused by the transfection. It was
also demonstrated that MG132 mainly inhibited the binding of
ubiquitin molecules to target proteins instead of degrading them.
In the MG132 treatment group, it was identified that with the
increase in ITCH expression, there was no reduction in Txnip.
Moreover, the degradation of ITCH itself is dependent on the
ubiquitin-proteasome proteolytic digestion system, and the specific
protein identification of ITCH by ubiquitin E3 ligase is via ITCH
itself (35). Therefore, MG132
inhibited ubiquitination and ITCH degradation at the same time,
which lead to an increase in ITCH expression, while no decrease in
Txnip expression, and this process also identified that the
regulation of oxidative stress was accomplished via ubiquitination.
Collectively, it was speculated that the ubiquitination of ITCH by
transfection with ITCH overexpression plasmids reversed the
oxidative stress of Txnip. Furthermore, the knockout of ITCH found
that the downstream associated protein showed an opposite change in
trend compared with the overexpression of ITCH.
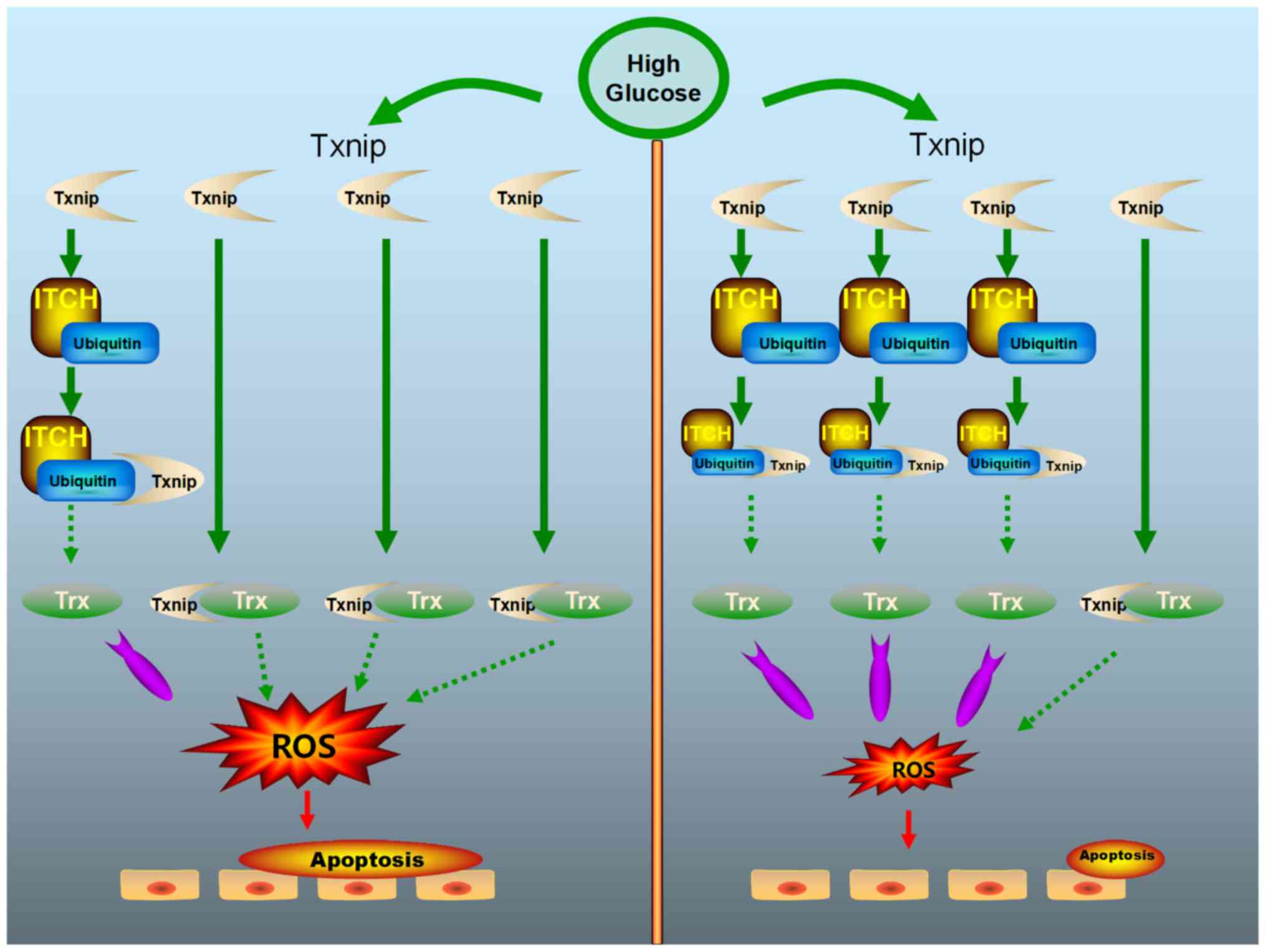
In conclusion, the present results indicated a
potential anti-oxidative stress function of the protein ITCH in
sugar cataract. The overexpression of ITCH inhibited the apoptosis
of HLECs (Fig. 8). Therefore, the
upregulation of ITCH may be a potential treatment approach for
patients with sugar cataract.
In the present study, although it was verified that
osmotic pressure had no significant effect on the results at the
RNA level, further experiments were not conducted and thus, this
was only a cursory conclusion. Further studies will be conducted in
the future to investigate the role of osmotic pressure in ITCH,
Txnip and Trx in HLECs. In addition, the effect of cell density on
the expression of Trx in HLECs was also not studied, which was
another limitation of this study, and will be investigated in
future studies.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81170836) and
Clinical Genetics (Ophthalmology), Subject Construction Project of
China Medical University (grant no. 3110118049).
Availability of data and materials
The datasets used and or/analyzed during the present
study are available from the corresponding author on reasonable
requests.
Authors' contributions
LFJ designed and performed the experiments,
conducted the statistical analysis of the data and drafted the
manuscript. WKZ and BL contributed towards the study design. QCY
contributed towards the study design and data analysis, revised the
manuscript critically for important intellectual content and gave
final approval of the version to be published. All authors read and
approved the final manuscript and agree to be accountable for all
aspects of the research in ensuing that the accuracy or integrity
of and part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fukuoka H and Afshari NA: The impact of
age-related cataract on measures of frailty in an aging global
population. Curr Opin Ophthalmol. 28:93–97. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Francis PJ and Moore AT: Genetics of
childhood cataract. Curr Opin Ophthalmol. 15:10–15. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Andley UP, Tycksen E, McGlasson-Naumann BN
and Hamilton PD: Probing the changes in gene expression due to
α-crystallin mutations in mouse models of hereditary human
cataract. PLoS One. 13:e01908172018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bahia S, Blais E, Murugkar S, Chauhan V
and Kumarathasan P: Oxidative and nitrative stress-related changes
in human lens epithelial cells following exposure to X-rays. Int J
Radiat Biol. 94:366–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhagat AK, Bhardwaj H, Bhardwaj BL, Goyal
S, Jaura S and Jain P: Acute bilateral cataract in patient with
type 1 diabetes mellitus. J Assoc Physicians India.
67:832019.PubMed/NCBI
|
|
6
|
Mirsky N, Cohen R, Eliaz A and Dovrat A:
Featured article: Inhibition of diabetic cataract by glucose
tolerance factor extracted from yeast. Exp Biol Med (Maywood).
241:817–829. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tinkov AA, Bjørklund G, Skalny AV,
Holmgren A, Skalnaya MG, Chirumbolo S and Aaseth J: The role of the
thioredoxin/thioredoxin reductase system in the metabolic syndrome:
Towards a possible prognostic marker? Cell Mol Life Sci.
75:1567–1586. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Montano SJ, Lu J, Gustafsson TN and
Holmgren A: Activity assays of mammalian thioredoxin and
thioredoxin reductase: Fluorescent disulfide substrates,
mechanisms, and use with tissue samples. Anal Biochem. 449:139–146.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J and Holmgren A: The thioredoxin
antioxidant system. Free Radic Biol Med. 66:75–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen KS and DeLuca HF: Isolation and
characterization of a novel cDNA from HL-60 cells treated with
1,25-dihydroxyvitamin D-3. Biochim Biophys Acta. 1219:26–32. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Patwari P, Higgins LJ, Chutkow WA,
Yoshioka J and Lee RT: The interaction of thioredoxin with Txnip.
Evidence for formation of a mixed disulfide by disulfide exchange.
J Biol Chem. 281:21884–21891. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abderrazak A, Syrovets T, Couchie D, El
Hadri K, Friguet B, Simmet T and Rouis M: NLRP3 inflammasome: From
a danger signal sensor to a regulatory node of oxidative stress and
inflammatory diseases. Redox Biol. 4:296–307. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nishiyama A, Matsui M, Iwata S, Hirota K,
Masutani H, Nakamura H, Takagi Y, Sono H, Gon Y and Yodoi J:
Identification of thioredoxin-binding protein-2/vitamin D(3)
up-regulated protein 1 as a negative regulator of thioredoxin
function and expression. J Biol Chem. 274:21645–21650. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pasternak Y, Ohana M, Biron-Shental T,
Cohen-Hagai K, Benchetrit S and Zitman-Gal T: Thioredoxin,
thioredoxin interacting protein and transducer and activator of
transcription 3 in gestational diabetes. Mol Biol Rep.
47:1199–1206. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao C, Huang W, Kanasaki K and Xu Y: The
role of ubiquitination and sumoylation in diabetic nephropathy.
Biomed Res Int. 2014:1606922014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lorenz S: Structural mechanisms of
HECT-type ubiquitin ligases. Biol Chem. 399:127–145. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Michelle C, Vourc'h P, Mignon L and Andres
CR: What was the set of ubiquitin and ubiquitin-like conjugating
enzymes in the eukaryote common ancestor? J Mol Evol. 68:616–628.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li W, Bengtson MH, Ulbrich A, Matsuda A,
Reddy VA, Orth A, Chanda SK, Batalov S and Joazeiro CA: Genome-wide
and functional annotation of human E3 ubiquitin ligases identifies
MULAN, a mitochondrial E3 that regulates the organelle's dynamics
and signaling. PLoS One. 3:e14872008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aki D, Zhang W and Liu YC: The E3 ligase
Itch in immune regulation and beyond. Immunol Rev. 266:6–26. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Otaki Y, Takahashi H, Watanabe T, Funayama
A, Netsu S, Honda Y, Narumi T, Kadowaki S, Hasegawa H, Honda S, et
al: HECT-type ubiquitin E3 ligase ITCH interacts with
thioredoxin-interacting protein and ameliorates reactive oxygen
species-induced cardiotoxicity. J Am Heart Assoc. 5:e0024852016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han X, Dong XX, Shi MY, Feng L, Wang XL,
Zhang JS and Yan QC: SUMOylation and deacetylation affect NF-κB p65
activity induced by high glucose in human lens epithelial cells.
Int J Ophthalmol. 12:1371–1379. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park YJ, Oanh NTK, Heo J, Kim SG, Lee HS,
Lee H, Lee JH, Kang HC, Lim W, Yoo YS and Cho H: Dual targeting of
RIG-I and MAVS by MARCH5 mitochondria ubiquitin ligase in innate
immunity. Cell Signal. 67:1095202020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Junn E, Han SH, Im JY, Yang Y, Cho EW, Um
HD, Kim DK, Lee KW, Han PL, Rhee SG and Choi I: Vitamin D3
up-regulated protein 1 mediates oxidative stress via suppressing
the thioredoxin function. J Immunol. 164:6287–6295. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, De Keulenaer GW and Lee RT:
Vitamin D(3)-up- regulated protein-1 is a stress-responsive gene
that regulates cardiomyocyte viability through interaction with
thioredoxin. J Biol Chem. 277:26496–26500. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Osnes-Ringen Ø, Berg KH, Moe MC,
Zetterström C, Røger M and Nicolaissen B: Cell death pattern in
lens epithelium of cataract patients. Acta Ophthalmol. 94:514–520.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bron AJ, Brown NA, Harding JJ and Ganea E:
The lens and cataract in diabetes. Int Ophthalmol Clin. 38:37–67.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Collet JF and Messens J: Structure,
function, and mechanism of thioredoxin proteins. Antioxid Redox
Signal. 13:1205–1216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen KS and DeLuca HF: Cloning of the
human 1 alpha, 25-dihydroxyvitamin D-3 24-hydroxylase gene promoter
and identification of two vitamin D-responsive elements. Biochim
Biophys Acta. 1263:1–9. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jeanine CS, Amie DM and Walter HW:
Oxidation and nuclear localization of thioredoxin-1 in sparse cell
cultures. J Cell Biochem. 104:1879–1889. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Zhang JH, Chen XY, Hu QH, Wang
MX, Jin R, Zhang QY, Wang W, Wang R, Kang LL, et al: Reactive
oxygen species-induced TXNIP drives fructose-mediated hepatic
inflammation and lipid accumulation through NLRP3 inflammasome
activation. Antioxid Redox Signal. 22:848–870. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Oslowski CM, Hara T, O'Sullivan-Murphy B,
Kanekura K, Lu S, Hara M, Ishigaki S, Zhu LJ, Hayashi E, Hui ST, et
al: Thioredoxin-interacting protein mediates ER stress-induced β
cell death through initiation of the inflammasome. Cell Metab.
16:265–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bernardini S, Gravina P, Croce N,
Perricone R, Knight RA, Valentini A, Melino G and Federici G: Itch
gene polymorphisms in healthy population and in patients affected
by rheumatoid arthritis and atopic dermatitis. Cell Cycle.
7:3607–3609. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang P, Wang C, Gao K, Wang D, Mao J, An
J, Xu C, Wu D, Yu H, Liu JO and Yu L: The ubiquitin ligase itch
regulates apoptosis by targeting thioredoxin-interacting protein
for ubiquitin-dependent degradation. J Biol Chem. 285:8869–8879.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mouchantaf R, Azakir BA, McPherson PS,
Millard SM, Wood SA and Angers A: The ubiquitin ligase itch is
auto-ubiquitylated in vivo and in vitro but is protected from
degradation by interacting with the deubiquitylating enzyme
FAM/USP9X. J Biol Chem. 281:38738–38747. 2006. View Article : Google Scholar : PubMed/NCBI
|