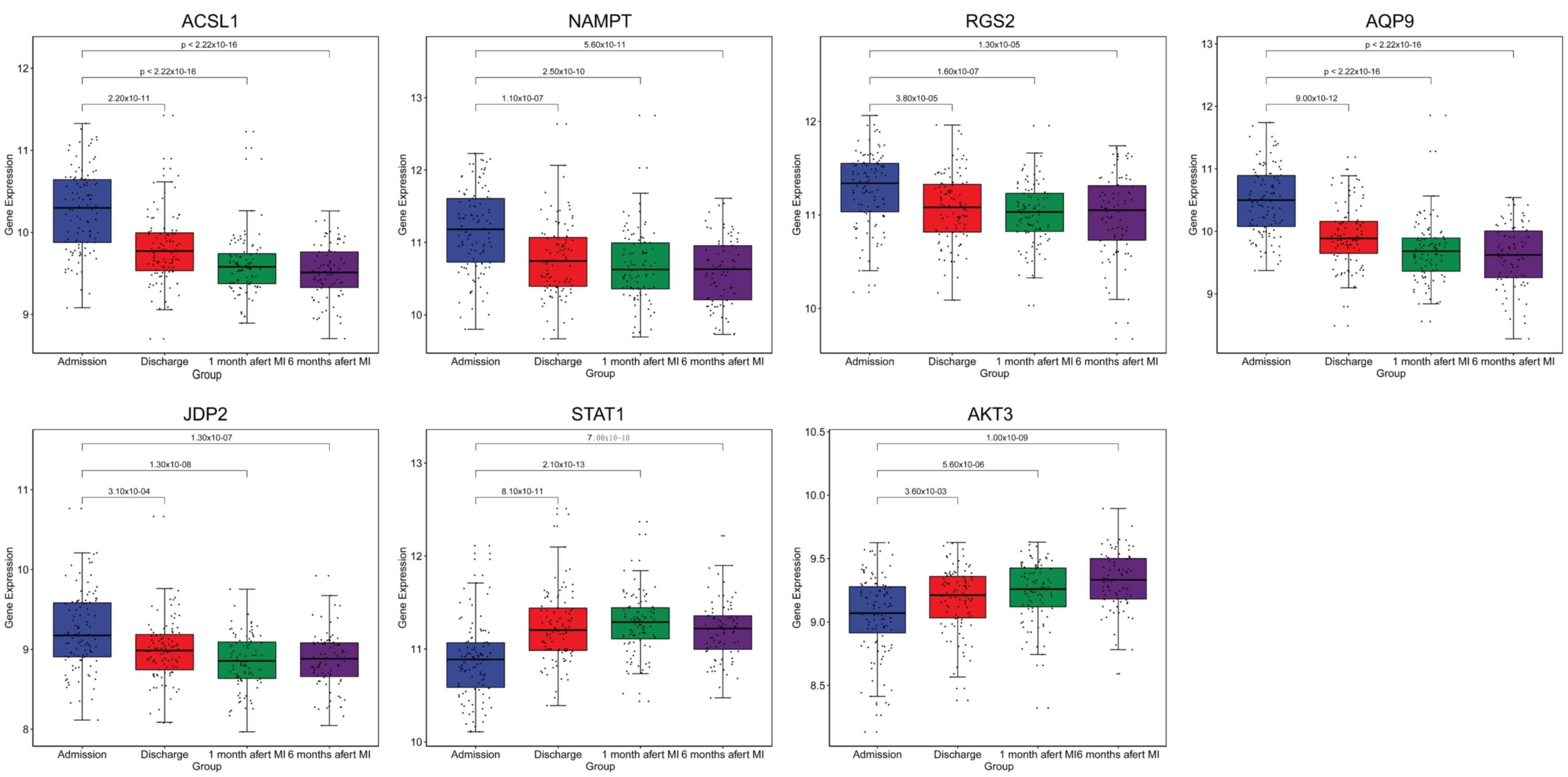
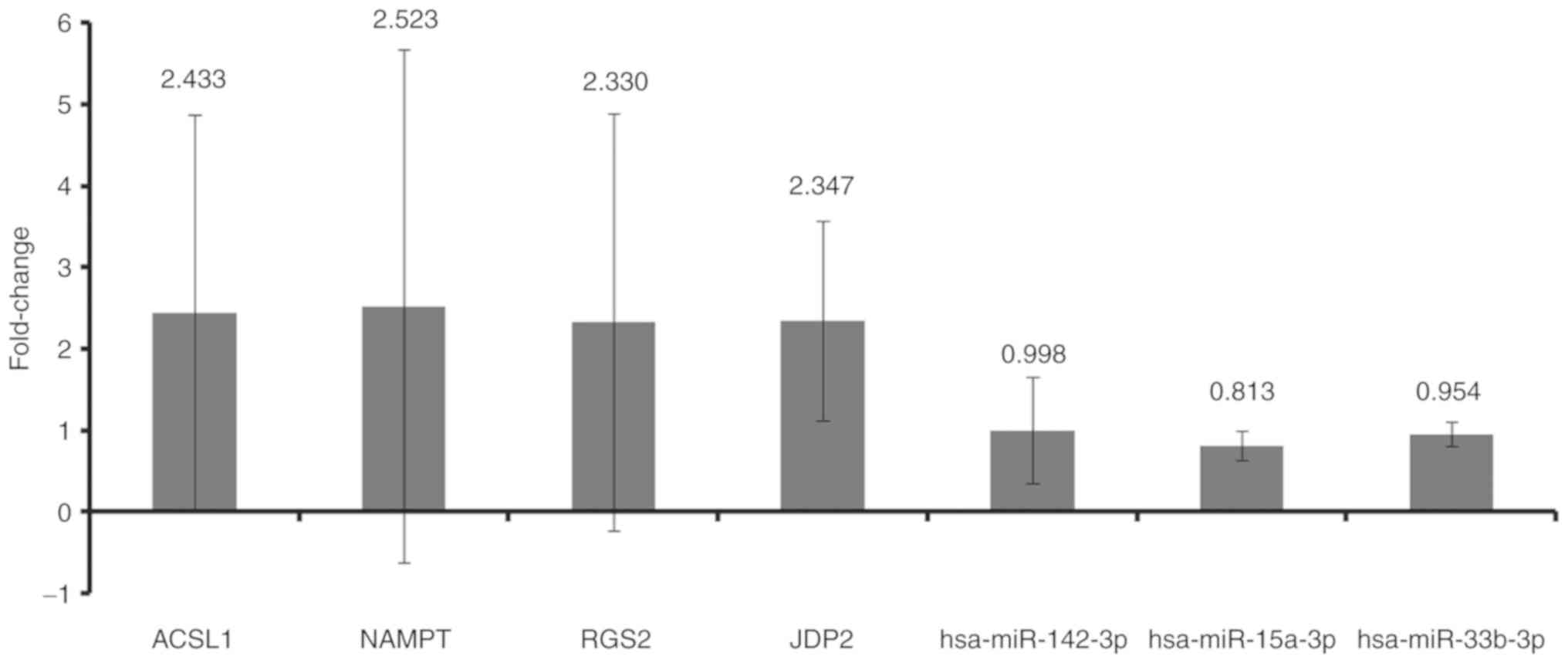
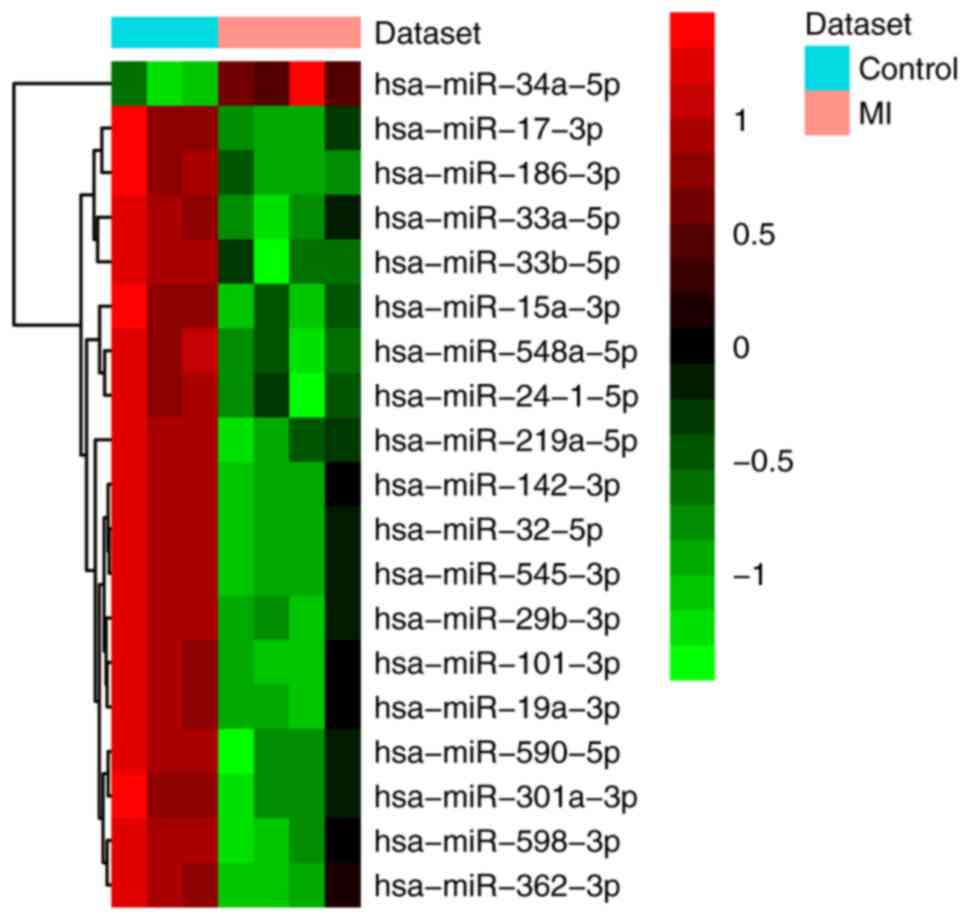
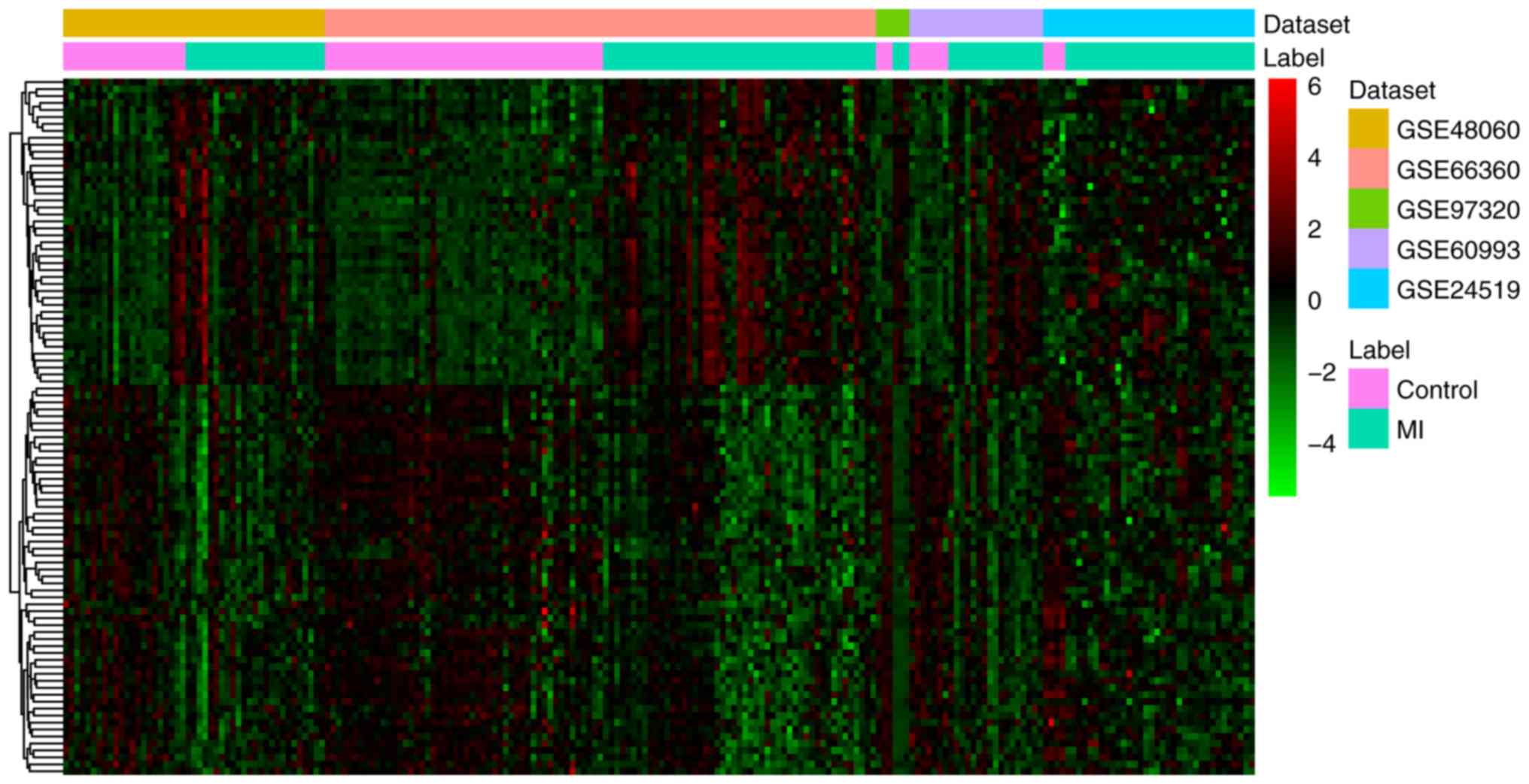
|
1
|
Bainey KR, Fresco C, Zheng Y, Halvorsen S,
Carvalho A, Ostojic M, Goldstein P, Gershlick AH, Westerhout CM,
Van de Werf F, et al: Implications of ischaemic area at risk and
mode of reperfusion in ST-elevation myocardial infarction. Heart.
102:527–533. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goto Y, Masaki I, Yamada A, Uno M,
Nakamori S, Nagata M, Ichikawa Y, Kitagawa K, Ito M and Sakuma H:
Native T1 mapping allows for the accurate detection of the segments
with chronic myocardial infarction in patients with known or
suspected coronary artery disease. J Cardiovasc Magn Reson.
18:P702016. View Article : Google Scholar
|
|
3
|
Orlic D, Ostojic M, Beleslin B,
Milasinovic D, Tesic M, Borovic M, Vukcevic V, Stojkovic S,
Nedeljkovic M and Stankovic G: The randomized physiologic
assessment of thrombus aspiration in patients with acute ST-segment
elevation myocardial infarction trial (PATA STEMI): Study rationale
and design. J Interv Cardiol. 27:341–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Furtado MB, Costa MW and Rosenthal NA: The
cardiac fibroblast: Origin, identity and role in homeostasis and
disease. Differentiation. 92:93–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kurose H and Mangmool S: Myofibroblasts
and inflammatory cells as players of cardiac fibrosis. Arch Pharm
Res. 39:1100–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao M, Yin D, Chen J and Qu X: Activating
the interleukin-6-Gp130-STAT3 pathway ameliorates ventricular
electrical stability in myocardial infarction rats by modulating
neurotransmitters in the paraventricular nucleus. BMC Cardiovasc
Disord. 20:602020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ouweneel DM, Eriksen E, Sjauw KD, van
Dongen IM, Hirsch A, Packer EJ, Vis MM, Wykrzykowska JJ, Koch KT,
Baan J, et al: Percutaneous mechanical circulatory support versus
intra-aortic balloon pump in cardiogenic shock after acute
myocardial infarction. J Am Coll Cardiol. 69:278–287. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rodríguez-Jiménez AE, Cruz-Inerarity H,
Negrín-Valdés T, Fardales-Rodríguez R and Chávez-González E:
Corrected QT-Interval Dispersion: An Electrocardiographic Tool to
Predict Recurrence of Myocardial Infarction. MEDICC Rev. 21:22–25.
2019.PubMed/NCBI
|
|
9
|
Frangogiannis NG, Smith CW and Entman ML:
The inflammatory response in myocardial infarction. Cardiovasc Res.
53:31–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nian M, Lee P, Khaper N and Liu P:
Inflammatory cytokines and postmyocardial infarction remodeling.
Circ Res. 94:1543–1553. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Landmesser U, Wollert KC and Drexler H:
Potential novel pharmacological therapies for myocardial
remodelling. Cardiovasc Res. 81:519–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamada Y, Ichihara S and Nishida T:
Molecular genetics of myocardial infarction. Genomic Med. 2:7–22.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Swirski FK and Nahrendorf M: Leukocyte
behavior in atherosclerosis, myocardial infarction, and heart
failure. Science. 339:161–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng Y and Zhang C: MicroRNA-21 in
cardiovascular disease. J Cardiovasc Transl Res. 3:251–255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marques FZ, Vizi D, Khammy O, Mariani JA
and Kaye DM: The transcardiac gradient of cardio-microRNAs in the
failing heart. Eur J Heart Fail. 18:1000–1008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oliveira-Carvalho V, da Silva MM,
Guimarães GV, Bacal F and Bocchi EA: MicroRNAs: new players in
heart failure. Mol Biol Rep. 40:2663–2670. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang JB, Mei J, Jiang LY, Jiang ZL, Liu
H, Zhang JW and Ding FB: miR-196a2 rs11614913 T>C polymorphism
is associated with an increased risk of tetralogy of fallot in a
Chinese population. Acta Cardiol Sin. 31:18–23. 2015.PubMed/NCBI
|
|
18
|
Meder B, Keller A, Vogel B, Haas J,
Sedaghat-Hamedani F, Kayvanpour E, Just S, Borries A, Rudloff J,
Leidinger P, et al: MicroRNA signatures in total peripheral blood
as novel biomarkers for acute myocardial infarction. Basic Res
Cardiol. 106:13–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bostjancic E, Zidar N and Glavac D:
MicroRNA microarray expression profiling in human myocardial
infarction. Dis Markers. 27:255–268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang H, Zhang C, Ban T, Liu Y, Mei L,
Piao X, Zhao D, Lu Y, Chu W and Yang B: A novel reciprocal loop
between microRNA-21 and TGFβRIII is involved in cardiac fibrosis.
Int J Biochem Cell Biol. 44:2152–2160. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL,
Zhu JN, Liu XY, Zhang YY, Li Y, Lin SG and Yu XY: Upregulated
expression of miR-1/miR-206 in a rat model of myocardial
infarction. Biochem Biophys Res Commun. 381:597–601. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi B, Guo Y, Wang J and Gao W: Altered
expression of microRNAs in the myocardium of rats with acute
myocardial infarction. BMC Cardiovasc Disord. 10:112010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reiner-Benaim A: FDR control by the BH
procedure for two-sided correlated tests with implications to gene
expression data analysis. Biom J. 49:107–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
1995.
|
|
27
|
Wu Y, Zhang L, Zhang Y, Zhen Y and Liu S:
Bioinformatics analysis to screen for critical genes between
survived and non-survived patients with sepsis. Mol Med Rep.
18:3737–3743. 2018.PubMed/NCBI
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
The Gene Ontology Consortium, . The Gene
Ontology Resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ellis KL, Cameron VA, Troughton RW,
Frampton CM, Ellmers LJ and Richards AM: Circulating microRNAs as
candidate markers to distinguish heart failure in breathless
patients. Eur J Heart Fail. 15:1138–1147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Su Q, Liu Y, Lv XW, Ye ZL, Sun YH, Kong BH
and Qin ZB: Inhibition of lncRNA TUG1 upregulates miR-142-3p to
ameliorate myocardial injury during ischemia and reperfusion via
targeting HMGB1- and Rac1-induced autophagy. Mol Cell Cardiol.
133:12–25. 2019. View Article : Google Scholar
|
|
36
|
Zhu Y, Lin Y, Yan W, Sun Z, Jiang Z, Shen
B, Jiang X and Shi J: Novel biomarker MicroRNAs for subtyping of
acute coronary syndrome: A Bioinformatics Approach. Biomed Res Int.
2016:46183232016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Durgan DJ, Smith JK, Hotze MA, Egbejimi O,
Cuthbert KD, Zaha VG, Dyck JR, Abel ED and Young ME: Distinct
transcriptional regulation of long-chain acyl-CoA synthetase
isoforms and cytosolic thioesterase 1 in the rodent heart by fatty
acids and insulin. Am J Physiol Heart Circ Physiol.
290:H2480–H2497. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chiu HC, Kovacs A, Ford DA, Hsu FF, Garcia
R, Herrero P, Saffitz JE and Schaffer JE: A novel mouse model of
lipotoxic cardiomyopathy. J Clin Invest. 107:813–822. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang L, Yang Y, Si D, Shi K, Liu D, Meng H
and Meng F: High expression of long chain acyl-coenzyme A
synthetase 1 in peripheral blood may be a molecular marker for
assessing the risk of acute myocardial infarction. Exp Ther Med.
14:4065–4072. 2017.PubMed/NCBI
|
|
40
|
Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai
LS, Zhang L and Hu Y: miR-15a and miR-16 affect the angiogenesis of
multiple myeloma by targeting VEGF. Carcinogenesis. 34:426–435.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jackstadt R and Hermeking H: MicroRNAs as
regulators and mediators of c-MYC function. Biochim Biophys Acta.
1849:544–553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chamorro-Jorganes A, Araldi E, Penalva LO,
Sandhu D, Fernández-Hernando C and Suárez Y: MicroRNA-16 and
microRNA-424 regulate cell-autonomous angiogenic functions in
endothelial cells via targeting vascular endothelial growth factor
receptor-2 and fibroblast growth factor receptor-1. Arterioscler
Thromb Vasc Biol. 31:2595–2606. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chan LS, Yue PY, Wong YY and Wong RN:
MicroRNA-15b contributes to ginsenoside-Rg1-induced angiogenesis
through increased expression of VEGFR-2. Biochem Pharmacol.
86:392–400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Abu-Halima M, Poryo M, Ludwig N, Mark J,
Marsollek I, Giebels C, Petersen J, Schäfers HJ, Grundmann U,
Pickardt T, et al: Differential expression of microRNAs following
cardiopulmonary bypass in children with congenital heart diseases.
J Transl Med. 15:1172017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dahl TB, Holm S, Aukrust P and Halvorsen
B: Visfatin/NAMPT: A multifaceted molecule with diverse roles in
physiology and pathophysiology. Annu Rev Nutr. 32:229–243. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qiu L and Liu X: Identification of key
genes involved in myocardial infarction. Eur J Med Res. 24:222019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Montecucco F, Bauer I, Braunersreuther V,
Bruzzone S, Akhmedov A, Lüscher TF, Speer T, Poggi A, Mannino E,
Pelli G, et al: Inhibition of nicotinamide
phosphoribosyltransferase reduces neutrophil-mediated injury in
myocardial infarction. Antioxid Redox Signal. 18:630–641. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang X, Chen J and Huang X: Rosuvastatin
attenuates myocardial ischemia-reperfusion injury via upregulating
miR-17-3p-mediated autophagy. Cell Reprogram. 21:323–330. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tao H, Chen ZW, Yang JJ and Shi KH:
MicroRNA-29a suppresses cardiac fibroblasts proliferation via
targeting VEGF-A/MAPK signal pathway. Int J Biol Macromol.
88:414–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Romaine SP, Tomaszewski M, Condorelli G
and Samani NJ: MicroRNAs in cardiovascular disease: An introduction
for clinicians. Heart. 101:921–928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dews M, Homayouni A, Yu D, Murphy D,
Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT and
Thomas-Tikhonenko A: Augmentation of tumor angiogenesis by a
Myc-activated microRNA cluster. Nat Genet. 38:1060–1065. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Landskroner-Eiger S, Qiu C, Perrotta P,
Siragusa M, Lee MY, Ulrich V, Luciano AK, Zhuang ZW, Corti F,
Simons M, et al: Endothelial miR-17~92 cluster negatively regulates
arteriogenesis via miRNA-19 repression of WNT signaling. Proc Natl
Acad Sci USA. 112:12812–12817. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Maciejak A, Kiliszek M, Michalak M, Tulacz
D, Opolski G, Matlak K, Dobrzycki S, Segiet A, Gora M and Burzynska
B: Gene expression profiling reveals potential prognostic
biomarkers associated with the progression of heart failure. Genome
Med. 7:262015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Heger J, Bornbaum J, Würfel A, Hill C,
Brockmann N, Gáspár R, Pálóczi J, Varga ZV, Sárközy M, Bencsik P,
et al: JDP2 overexpression provokes cardiac dysfunction in mice.
Sci Rep. 8:76472018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pfeiffer L, Wahl S, Pilling LC, Reischl E,
Sandling JK, Kunze S, Holdt LM, Kretschmer A, Schramm K, Adamski J,
et al: DNA methylation of lipid-related genes affects blood lipid
levels. Circ Cardiovasc Genet. 8:334–342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Heximer SP, Knutsen RH, Sun X, Kaltenbronn
KM, Rhee MH, Peng N, Oliveira-dos-Santos A, Penninger JM, Muslin
AJ, Steinberg TH, et al: Hypertension and prolonged vasoconstrictor
signaling in RGS2-deficient mice. J Clin Invest. 111:445–452. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang KM, Wang GR, Lu P, Karas RH,
Aronovitz M, Heximer SP, Kaltenbronn KM, Blumer KJ, Siderovski DP,
Zhu Y and Mendelsohn ME: Regulator of G-protein signaling-2
mediates vascular smooth muscle relaxation and blood pressure. Nat
Med. 9:1506–1512. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wieland T, Lutz S and Chidiac P:
Regulators of G protein signalling: A spotlight on emerging
functions in the cardiovascular system. Curr Opin Pharmacol.
7:201–207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang J, Huang W, Xu R, Nie Y, Cao X, Meng
J, Xu X, Hu S and Zheng Z: MicroRNA-24 regulates cardiac fibrosis
after myocardial infarction. J Cell Mol Med. 16:2150–2160. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Inouye M, Ripatti S, Kettunen J,
Lyytikäinen LP, Oksala N, Laurila PP, Kangas AJ, Soininen P,
Savolainen MJ, Viikari J, et al: Novel Loci for metabolic networks
and multi-tissue expression studies reveal genes for
atherosclerosis. PLoS Genet. 8:e10029072012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Thuny F, Textoris J, Amara AB, Filali AE,
Capo C, Habib G, Raoult D and Mege JL: The gene expression analysis
of blood reveals S100A11 and AQP9 as potential biomarkers of
infective endocarditis. PLoS One. 7:e314902012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Huang X, Yu X, Li H, Han L and Yang X:
Regulation mechanism of aquaporin 9 gene on inflammatory response
and cardiac function in rats with myocardial infarction through
extracellular signal-regulated kinase1/2 pathway. Heart Vessels.
34:2041–2051. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan
M, Lin YD, Fisch S, Unno K, Sereti KI and Liao R: MicroRNA-34a
plays a key role in cardiac repair and regeneration following
myocardial infarction. Circ Res. 117:450–459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huang Y, Qi Y, Du JQ and Zhang DF:
MicroRNA-34a regulates cardiac fibrosis after myocardial infarction
by targeting Smad4. Expert Opin Ther Targets. 18:1355–1365.
2014.PubMed/NCBI
|
|
65
|
Matsumoto S, Sakata Y, Suna S, Nakatani D,
Usami M, Hara M, Kitamura T, Hamasaki T, Nanto S, Kawahara Y and
Komuro I: Circulating p53-responsive microRNAs are predictive
indicators of heart failure after acute myocardial infarction. Circ
Res. 113:322–326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bernardo BC, Gao XM, Winbanks CE, Boey EJ,
Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, et
al: Therapeutic inhibition of the miR-34 family attenuates
pathological cardiac remodeling and improves heart function. Proc
Natl Acad Sci USA. 109:17615–17620. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bernardo BC, Gao XM, Tham YK, Kiriazis H,
Winbanks CE, Ooi JY, Boey EJ, Obad S, Kauppinen S, Gregorevic P, et
al: Silencing of miR-34a attenuates cardiac dysfunction in a
setting of moderate, but not severe, hypertrophic cardiomyopathy.
PLoS One. 9:e903372014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Stephanou A, Brar BK, Scarabelli TM,
Jonassen AK, Yellon DM, Marber MS, Knight RA and Latchman DS:
Ischemia-induced STAT-1 expression and activation play a critical
role in cardiomyocyte apoptosis. J Biol Chem. 275:10002–10008.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Stephanou A: Role of STAT-1 and STAT-3 in
ischaemia/reperfusion injury. J Cell Mol Med. 8:519–525. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang S, Liu W, Liu X, Qi J and Deng C:
Biomarkers identification for acute myocardial infarction detection
via weighted gene co-expression network analysis. Medicine
(Baltimore). 96:e83752017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chang PC, Lin SF, Chu Y, Wo HT, Lee HL,
Huang YC, Wen MS and Chou CC: LCZ696 therapy reduces ventricular
tachyarrhythmia inducibility in a myocardial infarction-induced
heart failure rat model. Cardiovasc Ther. 2019:60326312019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Li SY, Li ZX, He ZG, Wang Q, Li YJ, Yang
Q, Wu DZ, Zeng HL and Xiang HB: Quantitative proteomics reveal the
alterations in the spinal cord after myocardial
ischemia-reperfusion injury in rats. Int J Mol Med. 44:1877–1887.
2019.PubMed/NCBI
|
|
73
|
Getz GS and Reardon CA: Do the
Apoe−/- and Ldlr−/−Mice yield the same
insight on atherogenesis? Arterioscler Thromb Vasc Biol.
36:1734–1741. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Frangogiannis NG: Chemokines in the
ischemic myocardium: From inflammation to fibrosis. Inflamm Res.
53:585–595. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Frangogiannis NG and Entman ML: Chemokines
in myocardial ischemia. Trends Cardiovasc Med. 15:163–169. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Boengler K, Hilfiker-Kleiner D, Drexler H,
Heusch G and Schulz R: The myocardial JAK/STAT pathway: From
protection to failure. Pharmacol Ther. 120:172–185. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Fuglesteg BN, Suleman N, Tiron C, Kanhema
T, Lacerda L, Andreasen TV, Sack MN, Jonassen AK, Mjøs OD, Opie LH
and Lecour S: Signal transducer and activator of transcription 3 is
involved in the cardioprotective signalling pathway activated by
insulin therapy at reperfusion. Basic Res Cardiol. 103:444–453.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liu YH, Wang D, Rhaleb NE, Yang XP, Xu J,
Sankey SS, Rudolph AE and Carretero OA: Inhibition of p38
mitogen-activated protein kinase protects the heart against cardiac
remodeling in mice with heart failure resulting from myocardial
infarction. J Card Fail. 11:74–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ge ZW, Zhu XL, Wang BC, Hu JL, Sun JJ,
Wang S, Chen XJ, Meng SP, Liu L and Cheng ZY: MicroRNA-26b relieves
inflammatory response and myocardial remodeling of mice with
myocardial infarction by suppression of MAPK pathway through
binding to PTGS2. Int J Cardiol. 280:152–159. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu X, Chen K, Zhuang Y, Huang Y, Sui Y,
Zhang Y, Lv L and Zhang G: Paeoniflorin improves pressure
overload-induced cardiac remodeling by modulating the MAPK
signaling pathway in spontaneously hypertensive rats. Biomed
Pharmacother. 111:695–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li C, Li J, Xue K, Zhang J, Wang C, Zhang
Q, Chen X, Gao C, Yu X and Sun L: MicroRNA-143-3p promotes human
cardiac fibrosis via targeting sprouty3 after myocardial
infarction. J Mol Cell Cardiol. 129:281–292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Jia L, Zhu L, Wang JZ, Wang XJ, Chen JZ,
Song L, Wu YJ, Sun K, Yuan ZY and Hui R: Methylation of FOXP3 in
regulatory T cells is related to the severity of coronary artery
disease. Atherosclerosis. 228:346–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Abbate A, Bonanno E, Mauriello A, Bussani
R, Biondi-Zoccai GG, Liuzzo G, Leone AM, Silvestri F, Dobrina A,
Baldi F, et al: Widespread myocardial inflammation and
infarct-related artery patency. Circulation. 110:46–50. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Fan L, Meng H, Guo X, Li X and Meng F:
Differential gene expression profiles in peripheral blood in
northeast chinese han people with acute myocardial infarction.
Genet Mol Biol. 41:59–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Muse ED, Kramer ER, Wang H, Barrett P,
Parviz F, Novotny MA, Lasken RS, Jatkoe TA, Oliveira G, Peng H, et
al: A whole blood molecular signature for acute myocardial
infarction. Sci Rep. 7:122682017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Park HJ, Noh JH, Eun JW, Koh YS, Seo SM,
Park WS, Lee JY, Chang K, Seung KB, Kim PJ and Nam SW: Assessment
and diagnostic relevance of novel serum biomarkers for early
decision of ST-elevation myocardial infarction. Oncotarget.
6:12970–12983. 2015. View Article : Google Scholar : PubMed/NCBI
|