Introduction
Myocardial infarction (MI) is the death of
cardiomyocytes caused by obstruction of one or more arteries
supplying the heart (1,2). Clinically, patients with MI often
show a number of different symptoms including pain, fever and
increased white blood cell count (3). The pivotal pathological
characteristic after MI is cardiac fibrosis, which leads to cardiac
remodeling and failure (4,5). In addition, there are various
complications of MI, such as arrhythmia, pericarditis, heart
rupture and papillary muscle rupture (6). MI pathology can also be complicated
by cardiogenic shock, which can be life threatening (7). The recurrence rate of MI is high, and
the time of recurrence is the highest within two years after
remission. Moreover, each relapse can aggravate the disease
(8).
The pathological mechanism of MI is complex.
Frangogiannis et al (9) and
Nian et al (10) found that
MI was related to innate immunity programming, which is crucial in
cardiac remodeling. Angiogenesis is also a key event for
re-establishing the blood supply to the surviving myocardium
following MI, which contributes to recovery of cardiac function
(11). In addition, age, sex,
smoking status, hypertension, atherosclerosis, dyslipidemia,
diabetes mellitus and hypercholesterolemia are reported to be the
primary risks of MI (12,13).
MicroRNAs (miRNAs/miRs) can negatively regulate
target mRNA expression. Previous reports demonstrated that miRNAs
could be crucial regulators in cardiovascular diseases, including
hypertrophy, heart failure and ischemic cardiomyopathy (14–17).
It is worth noting that some miRNAs, such as hsa-miR-1, hsa-miR-21,
hsa-miR-206 and hsa-miR-499-5p, are significantly dysregulated in
the hearts of patients with acute MI (18–22).
In view of the important roles of miRNAs in MI, the aim of the
present study was to find potential miRNAs and target mRNAs in MI,
which may be useful in understanding the molecular mechanism of MI
pathology. The present findings demonstrated that the epigenetic
modifications of hsa-miR-142-2p-ACSL1, hsa-miR-15a-3p-NAMPT,
hsa-miR-17-3p-JDP2, hsa-miR-33-5p-RGS2, hsa-miR- 24-1-5p-AQP9 and
hsa-miR-34a-5p-STAT1/AKT3 and four signaling pathways (‘chemokine
signaling pathway’, ‘Jak-STAT signaling pathway’, ‘MAPK signaling
pathway’ and ‘T cell receptor signaling pathway’) may be associated
with the development of MI. In addition, ACSL1, NAMPT, RGS2, JDP2,
AQP9, STAT1 and AKT3 could help monitor the recurrence risk of
patients with MI.
Materials and methods
Data retrieval
The miRNA and mRNA expression profiles were
downloaded from the Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo). The
key words of ‘Myocardial infarction’ and ‘Homo sapiens’ was used to
search for related datasets. Screening requirements for the
included datasets were as follows: i) The selected datasets must be
genome-wide miRNA/mRNA transcriptome expression data; and ii) both
standardized and original datasets were considered in this study.
According to the aforementioned screening criteria, one miRNA
dataset (GSE24548) and four mRNA datasets (GSE97320, GSE24519,
GSE66360 and GSE60993) were selected for integrated analysis, which
is listed in Table I.
 | Table I.One miRNA dataset and four mRNA
datasets used for integrated analysis. |
Table I.
One miRNA dataset and four mRNA
datasets used for integrated analysis.
| First author,
year | GEO ID | Platform | Samples | Type | Omics | (Refs.) |
|---|
| Bellin et
al, 2017 | GSE24548 | GPL 8227 | MI:Control,
4:3 | Blood | miRNA | Unpublished |
| Fan et al,
2017 | GSE97320 | GPL 570 | MI:Control,
3:3 | Blood | mRNA | (84) |
| Bellin et
al, 2017 | GSE24519 | GPL 2895 | MI:Control,
34:4 | Blood | mRNA | Unpublished |
| Muse et al,
2015 | GSE66360 | GPL 570 | MI:Control,
49:50 | Blood | mRNA | (85) |
| Park et al,
2015 | GSE60993 | GPL6884 | MI:Control,
17:7 | Blood | mRNA | (86) |
Identification of differentially expressed miRNAs
and mRNAs. Firstly, scale standardization was performed for all
datasets. The limma (version 3.36.5) (23) and metaMA (version 3.1.2) (24) packages in R were then used to
identify differentially expressed miRNAs and mRNAs. The inverse
normal method was used to combine the P-value in metaMA. The false
discovery rate (FDR) was performed for multiple testing corrections
of raw P-values through the Benjamin and Hochberg method (25,26).
The threshold of differentially expressed miRNAs and mRNAs was set
as FDR<0.01 and FDR<0.05, respectively. The pheatmap R
package (cran.r-project.org/web/packages/pheatmap/index.html)
was used for hierarchical clustering analysis of the differentially
expressed miRNAs and mRNAs (27).
Correlation analysis between
differentially expressed miRNAs and mRNAs
The miRWalk3.0 (http://mirwalk.umm.uni-heidelberg.de/) software was
used to predict the differentially expressed target mRNAs of
differentially expressed miRNAs. The establishment of a miRNA-mRNA
regulatory network was visualized using Cytoscape 3.7.1 software
(28).
Functional analysis of differentially
expressed mRNAs
Gene Ontology (GO) (29,30)
and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (31) pathway enrichment via GeneCodis 3
(http://genecodis.cnb.csic.es) software
were used to analyze the biological functions of the differentially
expressed target mRNAs of differentially expressed miRNAs. The
criterion for selecting significantly enriched functional terms of
target differentially expressed mRNAs was P<0.05.
Screening of differentially expressed
target mRNAs associated with MI prognosis
In the process of screening for datasets that
contained miRNA and mRNA expression profiles, the GSE59867,
GSE97320, GSE24519, GSE66360 and GSE60993 datasets were initially
retrieved. As the GSE59867 dataset recorded the mRNA expression
profile in the blood of patients at admission (111 cases), at
discharge (101 cases), one month after discharge (95 cases) and six
months after discharge (83 cases), this dataset was used for
screening mRNAs associated with prognosis from identified targeted
differentially expressed mRNAs, with the aim of finding reliable
biomarkers to monitor the risk of recurrence in patients with
MI.
Diagnostic analysis of differentially
expressed target mRNAs associated with prognosis
In order to study whether identified differentially
expressed target mRNAs associated with prognosis have diagnostic
value, receiver operating characteristic (ROC) analysis was
performed using pROC package in R language (32). The area under the curve (AUC) under
binomial exact confidence interval was calculated and the ROC curve
was generated.
In vitro validation of key miRNAs and
target mRNAs
A total of five patients with MI and five normal
controls were enrolled from October to November 2019 at Jinan
Jigang Hospital. The inclusion criteria of patients with MI was as
follows: i) Time of chest pain or distress was >30 min within 24
h, and the levels of creatine kinase-MB and troponin T were higher
than the normal range; ii) no history of MI; iii) no drug treatment
or surgical treatment before admission; iv) patients had to provide
blood samples before hospitalization, at discharge and 6 months
after MI; and v) patients had to provide complete clinical data,
including sex, age, height and weight. The exclusion criteria was
as follows: i) Patients with myocarditis and other diseases caused
by chest pain or distress; ii) patients had a history of renal
failure, advanced liver disease, malignant tumor and other
inflammatory diseases, such as psoriasis and rheumatoid arthritis;
iii) patients with recurring MI; iv) patients with incomplete
clinical data; and v) patients who did not provide blood samples
before hospitalization, at discharge and 6 months after MI. Ethical
approval was obtained from the Ethics Committee of Jinan Jigang
Hospital (approval no. JG-001) and informed written consent was
obtained from all individuals.
Total RNA from the blood samples was extracted using
the RNAliquid reagent (Tiangen Biotech Co., Ltd.), according to the
manufacturer's protocols. RNA (1 µg) was applied to synthesize cDNA
for 3 min at 42°C, followed by 15 min at 42°C and 3 min at 95°C
using FastQuant RT kit (Tiangen Biotech Co., Ltd.). Then, reverse
transcription-quantitative PCR (RT-qPCR) was performed in an ABI
7300 Real-time PCR system with SYBR-Green PCR Master Mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions consisted of: i) Initial denaturation of 15 min at 95°C;
iii) 40 cycles of 10 sec at 95°C, 30 sec at 55°C, 32 sec at 72°C;
and iii) 15 sec at 95°C, 60 sec at 60°C, then 15 sec at 95°C. All
reactions were carried out in triplicate β-actin and GAPDH were
used as the internal reference genes. Relative gene expression was
analyzed by the fold-change method (33). The sequences of reverse and forward
primers for all of the genes analyzed were as follows: i)
ACSL1-forward, 5′-TGTGGGACCGGCTGATCTT-3′; ii) ACSL1-reverse,
5′-AGTGCACTCTGTCTGTCCGT-3′; iii) NAMPT-forward,
5′-TCTTCTGAGGCAGCGGTTG-3′; iv) NAMPT-reverse,
5′-GGCTGGAGTAGTGGGAAAGC-3′; v) RGS2-forward,
5′-AAGATTGGAAGACCCGTTTGAG-3′; vi) RGS2-reverse,
5′-GCAAGACCATATTTGCTGGCT-3′; vii) JDP2-forward,
5′-AGACCCAGATTGAGGAGCTG-3′; JDP2-reverse,
5′-AGTGGGTTGCCTTCTGACCTC-3′; viii) hsa-miR-142-3p-forward,
5′-TGTAGTGTTTCCTACTTTATGGA-3; ix) hsa-miR-15a-3p-forward,
5′-CAGGCCATATTGTGCTGCCTCA-3; x) hsa-miR-33b-3p-forward,
5′-GTGCATTGCTGTTGCATTGC-3; xi) GAPDH-forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′; xii) GAPDH-reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′; xiii) ACTB-forward,
5′-CATGTACGTTGCTATCCAGGC-3′; and vix) ACTB-reverse,
5′-CTCCTTAATGTCACGCACGAT-3′. The sequences of the reverse primers
for hsa-miR-142-3p, hsa-miR-15a-3p and hsa-miR-33b-3p were not
disclosed by the manufacturer.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism (version 2.0; GraphPad Software, Inc.). For the
result described in Figs. 7 and
9, one-way ANOVA, followed by
Bonferroni correction, were used to assess statistical significance
among different groups. Results are presented as the mean ± SD. All
experiments were repeated independently at least three times.
P<0.05 was considered to indicate a statistically significant
difference.
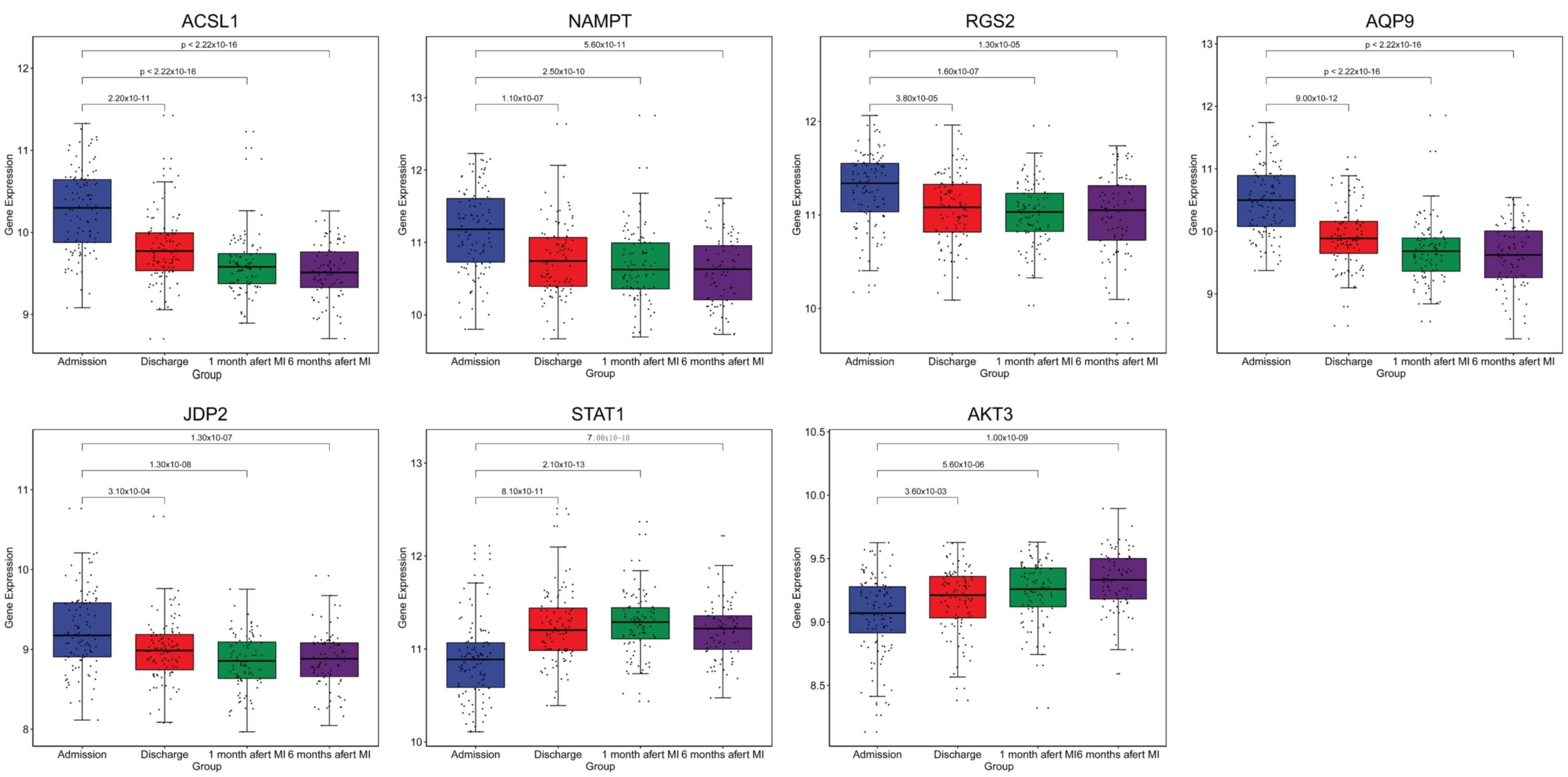 | Figure 7.Box plots of ACSL1, NAMPT, RGS2,
AQP9, JDP2, STAT1 and AKT3 expression. The expression levels of
ACSL1, NAMPT, RGS2, AQP9, JDP2, STAT1 and AKT3 were determined in
the blood of patients with MI at admission (n=111), at discharge
(n=101), 1 month after discharge (n=95) and 6 months after
discharge (n=83). ACSL1, long-chain-fatty-acid-CoA ligase 1; NAMPT,
nicotinamide phosphoribosyltransferase; RGS2, regulator of
G-protein signaling 2; JDP2, Jun dimerization protein 2; AQP9,
aquaporin-9; MI, myocardial infarction. |
Results
Expression pattern of miRNA and
mRNA
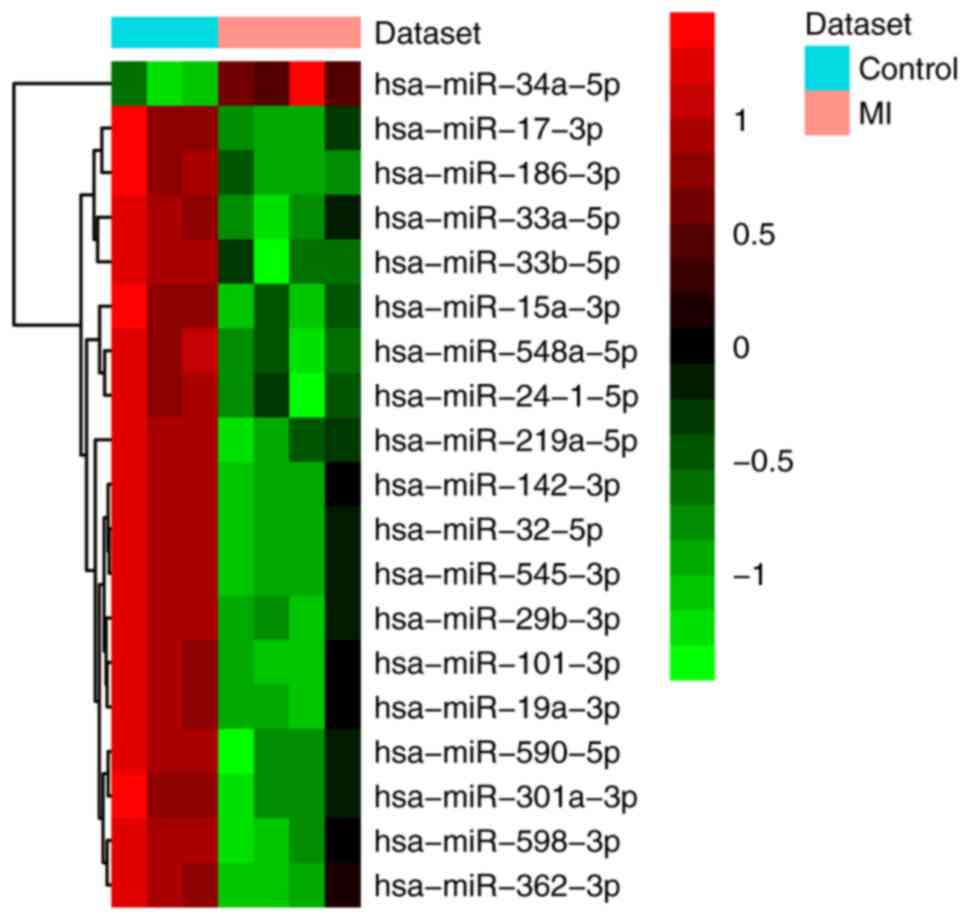
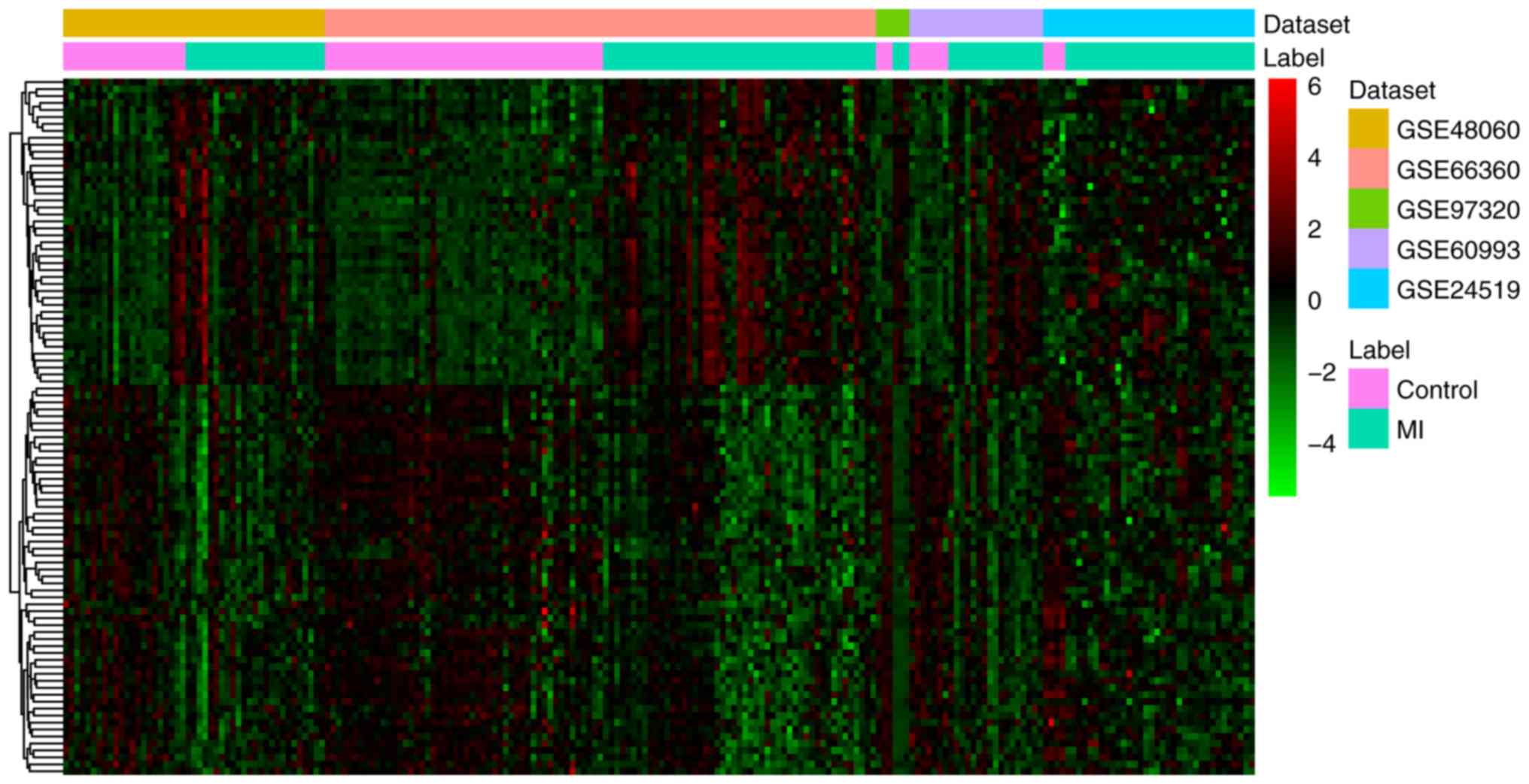
A total of 19 differentially expressed (one
upregulated and 18 downregulated) miRNAs and 1,007 differentially
expressed (287 upregulated and 720 downregulated) mRNAs were
identified in MI. All differentially expressed miRNAs and the top
20 differentially expressed mRNAs are shown in Tables II and III, respectively. The heat map of all
differentially expressed miRNAs and the top 100 differentially
expressed mRNAs are shown in Figs.
1 and 2, respectively.
 | Table II.Differentially expressed miRNAs in
MI. |
Table II.
Differentially expressed miRNAs in
MI.
| Symbol | LogFC | Average
expression | P-value | FDR |
|---|
| hsa-miR-32-5p | −1.72 | 4.66 |
3.17×10−05 |
4.93×10−03 |
| hsa-miR-545-3p | −1.50 | 4.37 |
3.76×10−05 |
4.93×10−03 |
|
hsa-miR-219a-5p | −1.34 | 3.30 |
5.23×10−05 |
4.93×10−03 |
| hsa-miR-29b-3p | −1.46 | 7.45 |
1.04×10−04 |
6.01×10−03 |
|
hsa-miR-548a-5p | −1.05 | 2.59 |
1.11×10−04 |
6.04×10−03 |
| hsa-miR-17-3p | −1.04 | 6.45 |
2.03×10−04 |
6.04×10−03 |
| hsa-miR-33a-5p | −1.34 | 6.14 |
2.05×10−04 |
6.04×10−03 |
| hsa-miR-33b-5p | −1.17 | 2.70 |
2.14×10−04 |
6.04×10−03 |
| hsa-miR-186-3p | −0.90 | 2.14 |
2.32×10−04 |
6.04×10−03 |
| hsa-miR-101-3p | −1.19 | 7.41 |
2.46×10−04 |
6.04×10−03 |
| hsa-miR-142-3p | −1.08 | 1.05 |
2.57×10−04 |
6.04×10−03 |
| hsa-miR-598-3p | −1.14 | 4.69 |
2.84×10−04 |
6.04×10−03 |
| hsa-miR-362-3p | −1.51 | 3.30 |
2.91×10−04 |
6.04×10−03 |
| hsa-miR-590-5p | −1.20 | 7.49 |
3.10×10−04 |
6.04×10−03 |
|
hsa-miR-24-1-5p | −1.07 | 5.30 |
3.20×10−04 |
6.04×10−03 |
| hsa-miR-34a-5p | 1.10 | 4.99 |
5.55×10−04 |
9.75×10−03 |
| hsa-miR-15a-3p | −0.84 | 2.89 |
5.86×10−04 |
9.75×10−03 |
|
hsa-miR-301a-3p | −0.91 | 7.82 |
6.47×10−04 |
9.90×10−03 |
| hsa-miR-19a-3p | −1.06 | 8.77 |
6.65×10−04 |
9.90×10−03 |
 | Table III.Top 20 differentially expressed mRNAs
in MI. |
Table III.
Top 20 differentially expressed mRNAs
in MI.
| Symbol | Combined ES | P-value | FDR | Up/down |
|---|
| ACSL1 | 1.59 |
<4.44×10−15 |
<6.76×10−12 | Up |
| S100A12 | 1.44 |
<4.44×10−15 |
<6.76×10−12 | Up |
| IL1R2 | 1.27 |
4.44×10−15 |
6.76×10−12 | Up |
| NAMPT | 1.24 |
1.22×10−14 |
1.69×10−11 | Up |
| RGS2 | 1.60 |
2.51×10−14 |
3.18×10−11 | Up |
| AQP9 | 1.19 |
3.22×10−14 |
3.77×10−11 | Up |
| JDP2 | 1.20 |
4.13×10−14 |
4.49×10−11 | Up |
| DUSP1 | 1.24 |
2.57×10−13 |
2.17×10−10 | Up |
| HAL | 1.12 |
5.64×10−13 |
4.30×10−10 | Up |
| S100A8 | 1.88 |
6.36×10−13 |
4.61×10−10 | Up |
| PLA2G12A | −1.34 |
2.09×10−13 |
1.87×10−10 | Down |
| GIMAP6 | −1.08 |
1.55×10−12 |
9.43×10−10 | Down |
| PAQR8 | −1.32 |
1.85×10−12 |
1.08×10−09 | Down |
| CHST12 | −1.17 |
1.67×10−11 |
7.83×10−09 | Down |
| GZMA | −1.03 |
3.45×10−11 |
1.31×10−08 | Down |
| RNASEH2B | −1.20 |
4.46×10−11 |
1.61×10−08 | Down |
| DENND2D | −1.03 |
4.95×10−11 |
1.71×10−08 | Down |
| RPAP2 | −1.12 |
8.16×10−11 |
2.53×10−08 | Down |
| GIMAP7 | −1.00 |
1.33×10−10 |
3.76×10−08 | Down |
| TRIM68 | −1.04 |
2.13×10−10 |
5.68×10−08 | Down |
Associations between miRNAs and
mRNAs
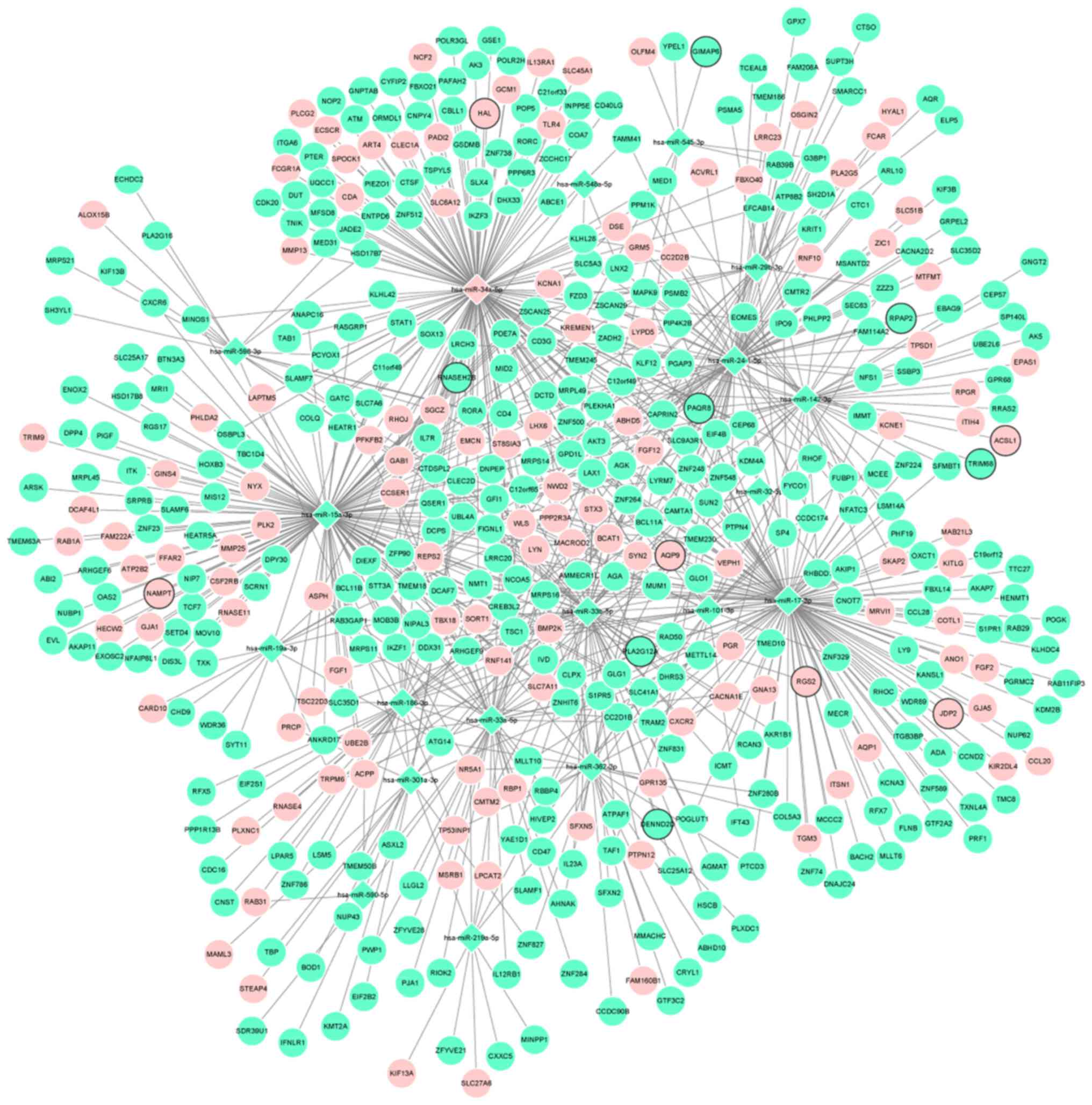
Depending on the target analysis, 2,137 miRNA-mRNA
pairs, including 19 miRNAs and 481 mRNAs, were identified. The
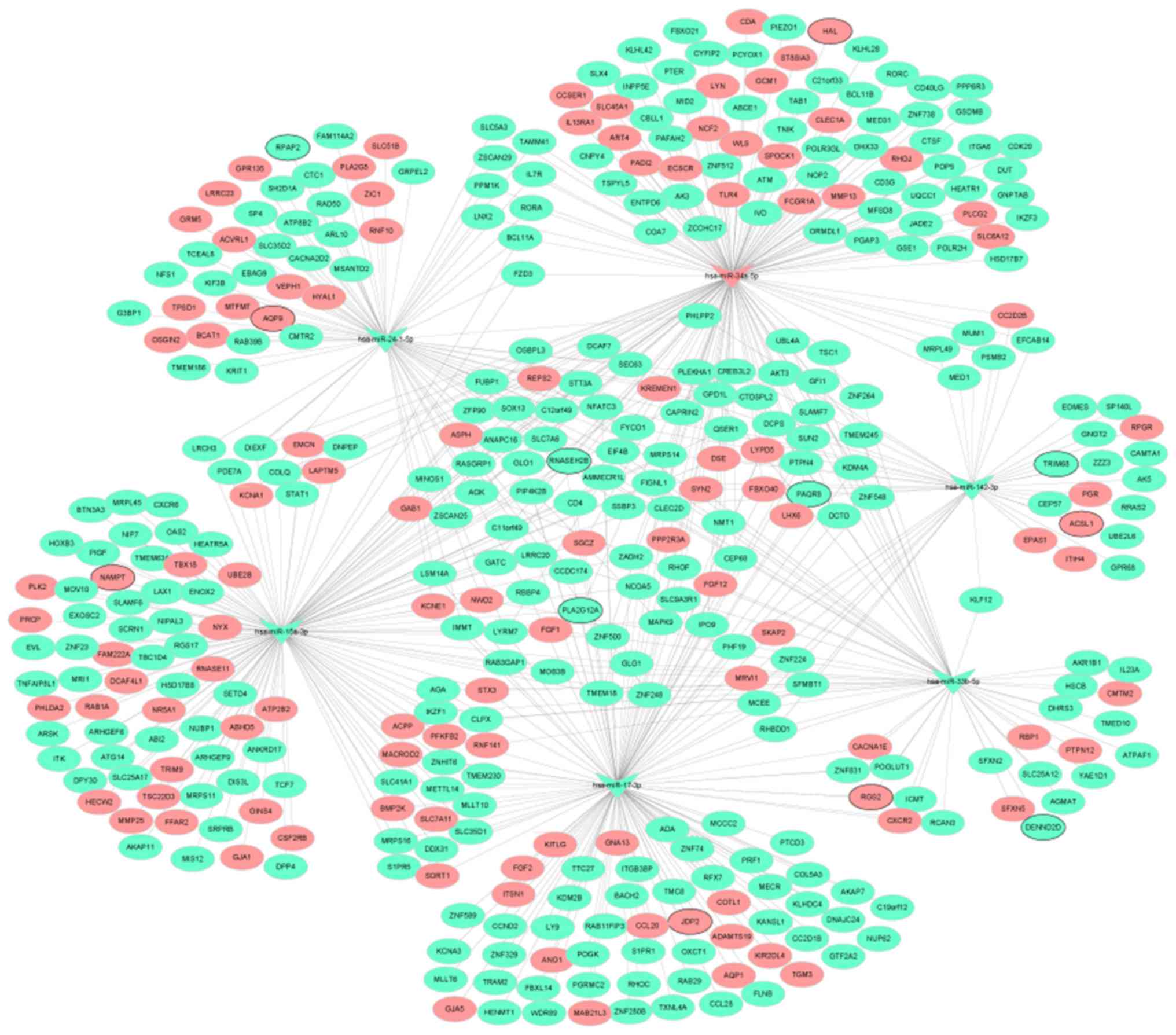
regulatory network of miRNA-mRNA is shown in Fig. 3. In addition, the regulatory
subnetworks of miRNAs-mRNAs between hsa-miR-142-3p, hsa-miR-15a-3p,
hsa-miR-17-3p, hsa-miR-33b-5p, hsa-miR-24-1-5p, hsa-miR-34a-5p and
their target mRNAs are shown in Fig.
4.
Functional analysis of differentially
expressed target mRNAs
In order to understand the biological function of
481 differentially expressed target mRNAs of 19 differentially
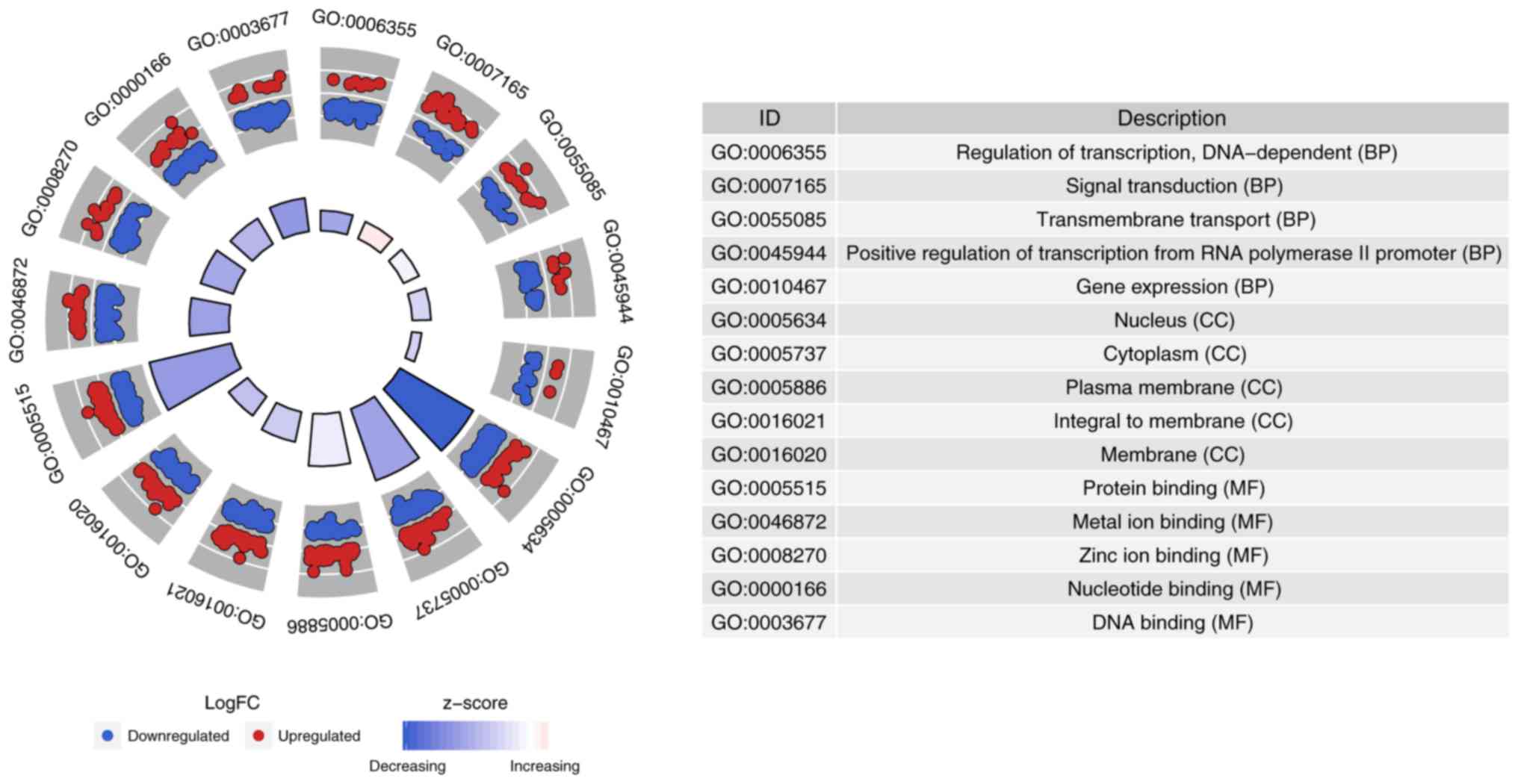
expressed miRNAs, GO and KEGG pathway analysis were performed. The
top 5 significantly enriched GO terms and top 12 KEGG terms are
shown in Figs. 5 and 6, respectively. The biological process,
cell component and molecular function ‘regulation of transcription,
DNA dependent’, ‘nucleus’ and protein binding’ were significantly
enriched, respectively (Fig. 5).
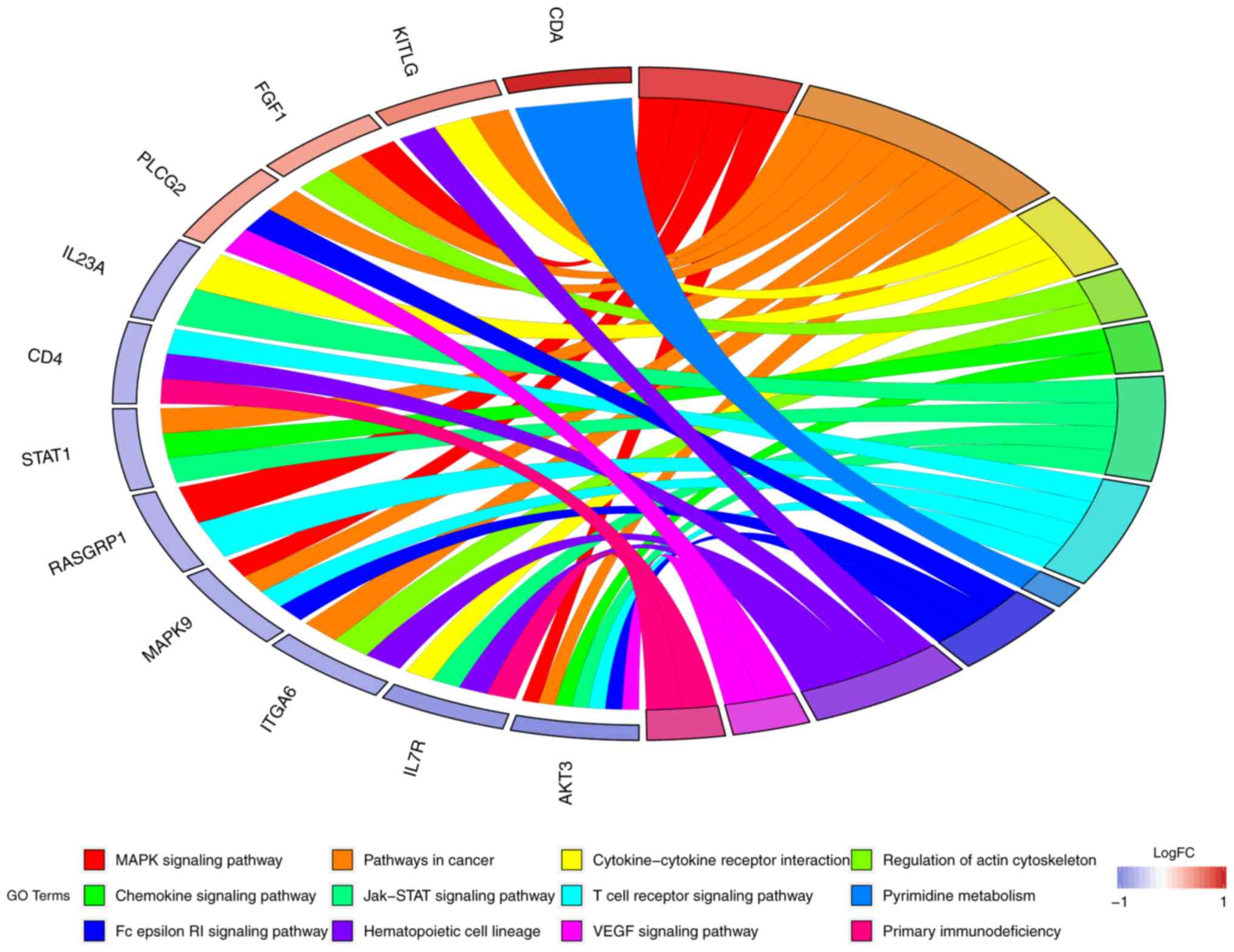
For KEGG terms, it was found that STAT1 was involved in the
‘chemokine signaling pathway’ and ‘Jak-STAT signaling pathway’. In
addition, AKT3 was involved in the ‘MAPK signaling pathway’ and ‘T
cell receptor signaling pathway’ (Fig.
6).
Identification of differentially
expressed target mRNAs associated with prognosis
Through comparative analysis between mRNAs in the
GSE59867 dataset and identified 481 differentially expressed target
mRNAs, it was found that 185 differentially expressed target mRNAs,
including long-chain-fatty-acid-CoA ligase 1 (ACSL1), nicotinamide
phosphoribosyltransferase (NAMPT), regulator of G-protein signaling
2 (RGS2), Jun dimerization protein 2 (JDP2), aquaporin-9 (AQP9),
STAT1 and AKT3, were associated with prognosis. The expression
levels of ACSL1, NAMPT, RGS2, AQP9, JDP2, STAT1 and AKT3 were
measured in the blood of patients with MI at admission, discharge,
one month after discharge and six months after discharge (Fig. 7). The results showed that the
expression levels of ACSL1 (P=2.2×10−11;
P<2.22×10−16; P<2.22×10−16), NAMPT
(P=1.1×10−07; P=2.5×10−10;
P=5.6×10−11), RGS2 (P=3.8×10−05;
P=1.6×10−07; P=1.3×10−05), AQP9
(P=9×10−12; P<2.22×10−16;
P<2.22×10−16) and JDP2 (P=0.00031;
P=1.3×10−08; P=1.3×10−07) in MI were
significantly downregulated at discharge, one month after discharge
and six months after discharge compared with that at admission.
Whereas, it was found that the expression levels of STAT1
(P=8.1×10−11; P=2.1×10−13;
P=7×10−10) and AKT3 (P=0.0036; P=5.6×10−06;
P=1×10−09) in MI were significantly upregulated at
discharge, one month after discharge and six months after discharge
compared with that at admission. This suggested that ACSL1, NAMPT,
RGS2, AQP9, JDP2, STAT1 and AKT3 expression could be used to
monitor the recurrence risk of patients with MI.
Diagnosis prediction of differentially
expressed target mRNAs associated with prognosis
ROC curve analysis was performed to assess the
diagnostic ability of ACSL1, NAMPT, RGS2, AQP9, JDP2, STAT1 and
AKT3 (Fig. 8). Of note, AUC values
of the aforementioned differentially expressed target mRNAs were
all >0.6, suggesting that they have a potential diagnostic value
for MI.
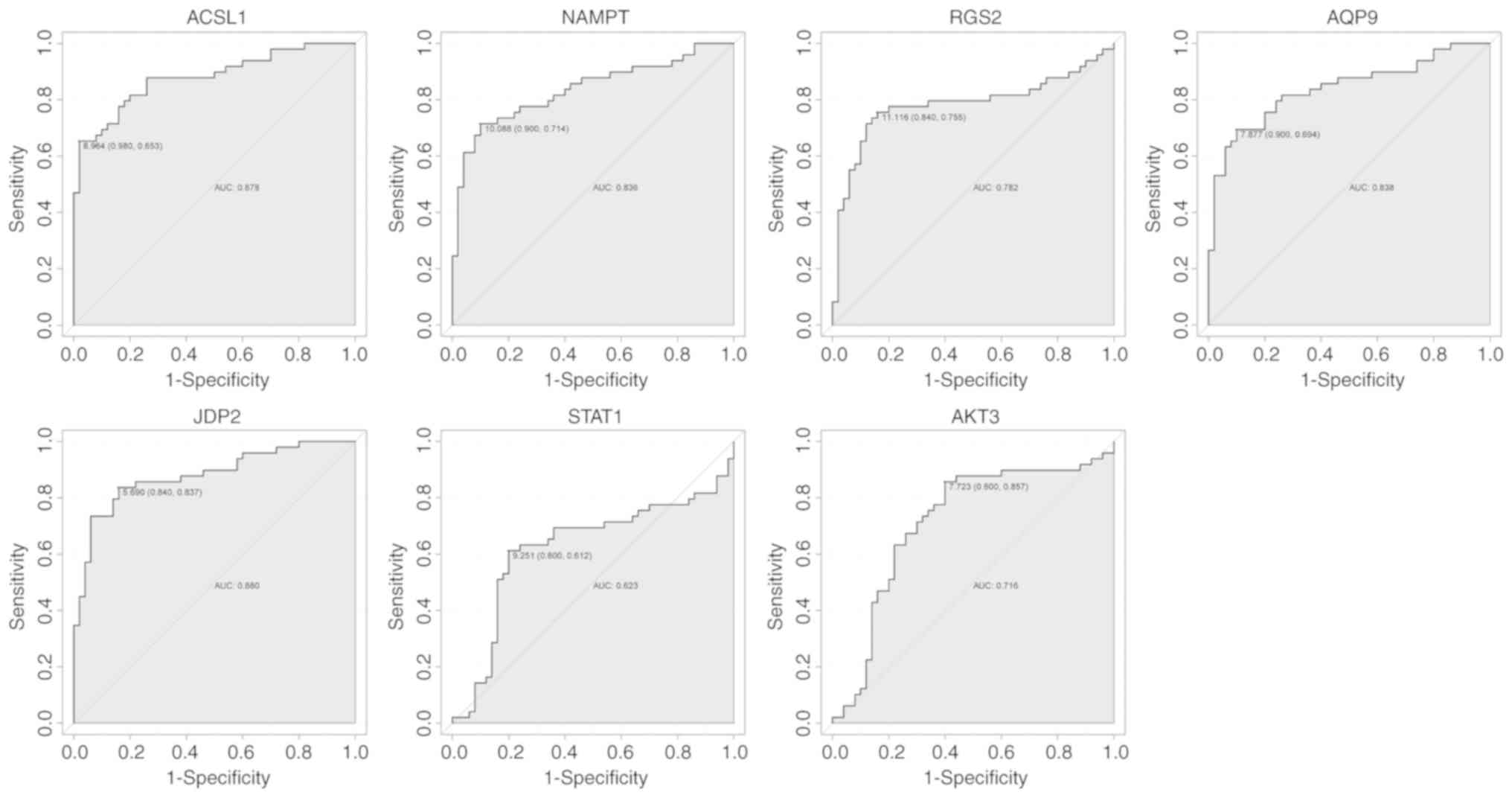 | Figure 8.ROC curves of ACSL1, NAMPT, RGS2,
AQP9, JDP2, STAT1 and AKT3 expression in myocardial infarction. The
ROC curves were used to show the diagnostic value of these genes
with 1-specificity and sensitivity. ACSL1,
long-chain-fatty-acid-CoA ligase 1; NAMPT, nicotinamide
phosphoribosyltransferase; RGS2, regulator of G-protein signaling
2; JDP2, Jun dimerization protein 2; AQP9, aquaporin-9; ROC,
receiver operating characteristic; AUC, area under curve. |
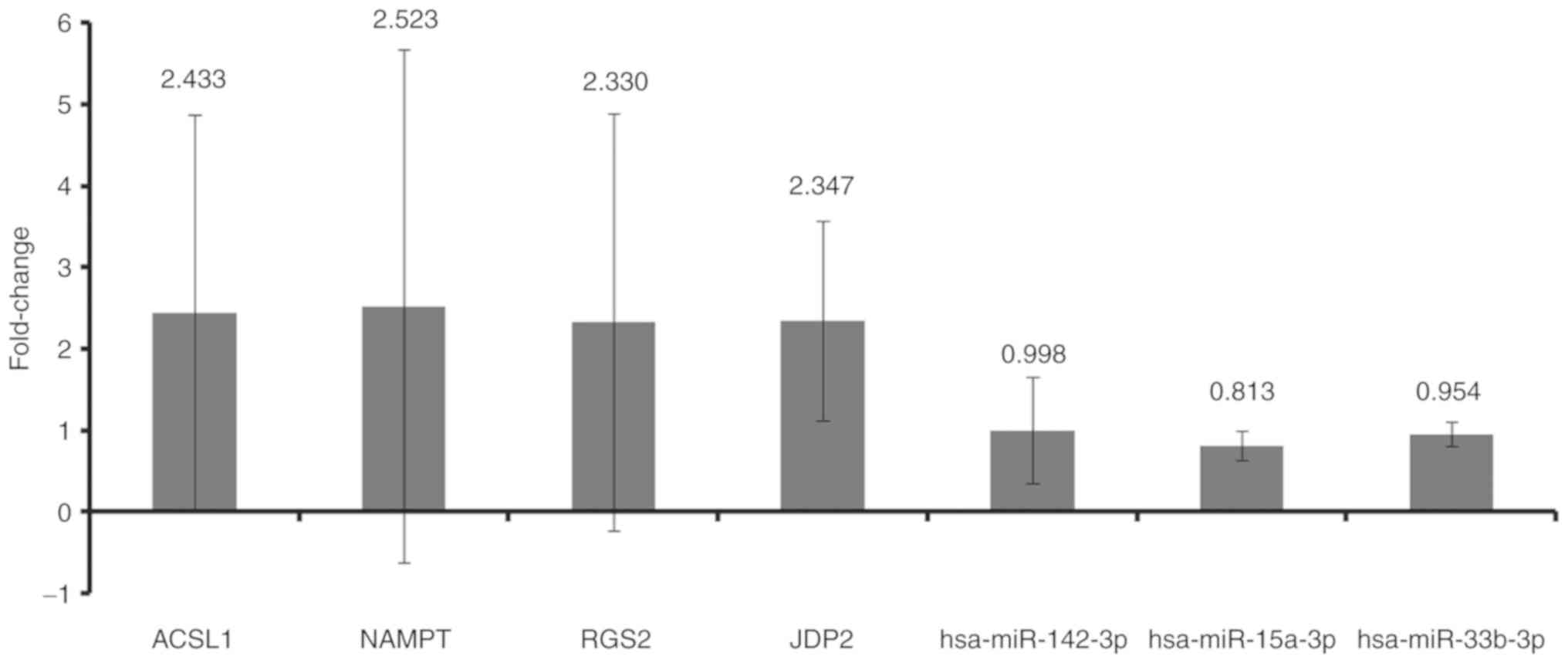
RT-qPCR
A total of five patients with MI and five normal
controls were included in the present study. Clinical information
of these individuals is presented in Table IV. In addition, three
differentially expressed miRNAs (hsa-miR-142-3p, hsa-miR-15a-3p and
hsa-miR-33b-5p) and their target mRNAs (ACSL1, NAMPT, RGS2 and
JDP2) were randomly selected for validation via RT-qPCR (Fig. 9). The relative expression levels of
hsa-miR-142-3p, hsa-miR-15a-3p and hsa-miR-33b-5p were
downregulated in the blood of patients with MI. The relative
expression levels of ACSL1, NAMPT, RGS2 and JDP2 were upregulated
in the blood of patients with MI. These results were consistent
with the bioinformatics analysis.
 | Table IV.Clinical information of patients with
MI and healthy controls. |
Table IV.
Clinical information of patients with
MI and healthy controls.
| A, Patients with
MI |
|---|
| Sex | Age, years | Height, m | Weight, kg | Hypertension
history | Diabetes
history | Smoking
history | Drinking
history | LDL | HDL | TC | TG | CTnT, ng/ml | CK-MB, µg/l |
|---|
| Female | 75 | 1.56 | 50 | Yes | No | No | No | 3.34 | 1.20 | 5.20 | 1.46 | 1.52 | 87 |
| Male | 84 | 1.70 | 70 | Yes | No | No | No | 3.05 | 1.17 | 4.56 | 0.75 | 0.07 | 46 |
| Male | 31 | 1.73 | 75 | Yes | Yes | No | Yes | 4.04 | 1.12 | 6.12 | 0.86 | 1.35 | 82 |
| Female | 65 | 1.61 | 61 | Yes | No | Yes | No | 3.51 | 1.31 | 5.69 | 1.24 | 1.21 | 49 |
| Male | 56 | 1.58 | 54 | Yes | Yes | Yes | No | 3.45 | 1.07 | 6.31 | 1.36 | 0.07 | 65 |
|
| B, Healthy
controls |
|
| Female | 83 | 1.55 | 80 | Yes | No | No | No | 1.26 | 1.03 | 3.12 | 1.83 |
<0.02 | <1l |
|
| Male | 82 | 170 | 81 | Yes | Yes | No | Yes | 2.74 | 1.01 | 4.40 | 1.42 | 0.33 | 2.48 |
| Female | 67 | 1.58 | 60 | No | No | No | No | 2.20 | 0.61 | 3.42 | 1.35 | 0.01 | 1.00 |
| Male | 59 | 1.75 | 85 | Yes | No | Yes | No | 3.45 | 1.14 | 5.21 | 1.89 | 0.01 | 2.23 |
| Male | 41 | 1.73 | 68 | No | No | No | No | 3.89 | 1.23 | 6.01 | 0.75 | 0.02 | 2.45 |
Discussion
Decreased expression of hsa-miR-142-3p was found in
patients with heart failure (34).
Su et al (35) demonstrated
that upregulating hsa-miR-142-3p could ameliorate myocardial
ischemic injury. Of note, hsa-miR-142-3p is considered to be a
biomarker for acute MI (36). In
the present study, it was found that hsa-miR-142-3p was
downregulated in the blood of patients with MI. Additionally, ACSL1
was found to be one of the target mRNAs of hsa-miR-142-3p and the
most upregulated gene. Moreover, ACSL1 was shown to have a
diagnostic and prognostic value for patients with MI. ACSL1
catalyzes the synthesis of acyl-CoA from fatty acid synthase
(37). Overexpression of ACSL1 can
lead to severe cardiomyopathy (38). The expression of ACSL1 is
significantly related to the degree of atherosclerosis in the
coronary artery (39). It has been
observed that ACSL1 is upregulated in peripheral blood leukocytes
of patients with acute MI (39).
The present study further demonstrated the important roles of
hsa-miR-142-3p and ACSL1 in MI.
Mature hsa-miR-15a-3p influences vascular function,
including the expression of vascular endothelial growth factor
receptor 2 (40–43). hsa-miR-15a-3p is notably
downregulated in the atrial myocardial tissue of patients with
congenital heart defect (44). The
present study found decreased expression of hsa-miR-15a-3p in the
blood of patients with MI; NAMPT was shown to be regulated by
hsa-miR-15a-3p. Furthermore, NAMPT could be regarded as a
diagnostic and prognostic biomarker for patients with MI. NAMPT is
involved in atherosclerosis and myocardial failure (45). Increased expression of NAMPT has
been found in MI (46). In
addition, FK866 (a specific inhibitor of NAMPT) reduced infarct
size following MI (47). The
present results suggested that hsa-miR-15a-3p may play a critical
role in the process of MI by targeting NAMPT.
Upregulation of hsa-miR-17-3p was demonstrated to
promote the viability of cardiomyocytes (48). Overexpression of hsa-miR-17-3p can
inhibit fibrosis of cardiac fibroblasts (49). In addition, hsa-miR-17-3p is
related to atherosclerosis and angiogenesis (50–52).
In the present study, it was found that hsa-miR-17-3p was
downregulated in the blood of patients with MI. In addition, JDP2
was shown to be the target of hsa-miR-17-3p. It is worth mentioning
that JDP2 was associated with diagnosis and prognosis. The
expression of JDP2 was revealed to be increased in patients with
heart failure and MI (46,53). Of note, JDP2 is considered to be a
candidate marker for the risk stratification of patients who may
develop heart failure following MI (54). The present results indicated that
JDP2 may be involved in the development of MI under the regulation
of hsa-miR-17-3p.
hsa-miR-33b-5p is a hypoxia-regulated miRNA. The
methylation level of the encoding region of hsa-miR-33b-5p is
related to lipid levels (55). The
present study found that hsa-miR-33b-5p was downregulated in the
blood of patients with MI. Moreover, upregulated RGS2 with
diagnostic and prognostic value was shown to be regulated by
hsa-miR-33b-5p. RGS2 is involved in regulating vascular smooth
muscle cell contraction and blood pressure (56,57).
Dysfunction of RGS2 is associated with cardiac hypertrophy
(58). The present results
suggested that hsa-miR-33b-5p could be associated with MI pathology
by targeting RGS2.
A previous report found that hsa-miR-24-1-5p reduced
cardiac fibrosis following MI (59). In the current study, decreased
expression of hsa-miR-24-1-5p was found in the blood of patients
with MI. In addition, upregulated AQP9 was one of the targets of
hsa-miR-24-1-5p. Moreover, AQP9 had a diagnostic and prognostic
value for patients with MI. It has been reported that increased
AQP9 is associated with the development of atherosclerotic lesions
(60). It has been demonstrated
that AQP9 is notably upregulated in patients with heart failure and
MI (46,61). In addition, silencing of the AQP9
gene was revealed to inhibit apoptosis of myocardial cells and
improve cardiac function following MI (62). Thus, it has been demonstrated that
hsa-miR-24-1-5p and AQP9 may play a significant role in cardiac
function following MI.
has-miR-34a-5p, which is associated with cardiac
fibrosis and regeneration, is upregulated during atherosclerosis
progression (63,64). Increased expression of
hsa-miR-34a-5p in circulation is related to cardiac remodeling and
heart failure following MI (65–67).
In the present study, it was found that hsa-miR-34a-5p was
upregulated in the blood of patients with MI. In addition,
downregulation of STAT1 and AKT3 was shown to be regulated by
hsa-miR-34a-5p. Furthermore, STAT1 and AKT3 were associated with
the diagnosis and prognosis of patients with MI. Activation of
STAT1 has been demonstrated to induce cardiomyocyte apoptosis
following ischemia/reperfusion injury by enhancing the expression
of pro-apoptotic proteins, which suggests that STAT1 plays a key
role in the pathology of MI (68–70).
AKT3 is involved in cardiomyocyte cell death (71). In rats, the expression of AKT3 is
decreased in the spinal cord following myocardial
ischemia-reperfusion injury (72).
In addition, AKT3 deficiency is related to an increase in
atherosclerosis (73).
A side from being a target of has-miR-34a-5p, it was
also found that STAT1 was involved in the ‘chemokine signaling
pathway’ and ‘Jak-STAT signaling pathway’ according to KEGG. AKT3
was found to be involved in the ‘MAPK signaling pathway’ and ‘T
cell receptor signaling pathway’ according to KEGG. Previous
studies revealed that chemokines were expressed in the infarcted
myocardium and play an important role in myocardial inflammation
and healing (74,75). The Jak/STAT signaling pathway plays
a key role in the modulation of cardioprotection against
ischemia/reperfusion injury (76,77).
Activation of p38 MAPK has been observed in cardiac hypertrophy
induced by MI, and inhibition of the signing pathway protects the
heart against MI (78). In
addition, the p38-JNK-MAPK signaling pathway is associated with the
inflammatory response in MI (79–81).
T cells, the main components of cell-mediated inflammation, are
associated with the development of atherosclerotic plaques
(82). Notably, Abbate et
al (83) detected activated T
cells in the infarct and remote areas of the heart tissue of
patients with MI.
In conclusion, the bioinformatics analysis from the
GEO database led to the finding of several potential differentially
expressed miRNAs and mRNAs. The epigenetic modifications of
hsa-miR-142-2p-ACSL1, hsa-miR-15a-3p-NAMPT, hsa-miR-17-3p-JDP2,
hsa-miR-33-5p-RGS2, hsa-miR- 24-1-5p-AQP9 and
hsa-miR-34a-5p-STAT1/AKT3 and four signaling pathways (‘chemokine
signaling pathway’, ‘Jak-STAT signaling pathway’, ‘MAPK signaling
pathway’ and ‘T cell receptor signaling pathway’) may be associated
with the development of MI. In addition, ACSL1, NAMPT, RGS2, JDP2,
AQP9, STAT1 and AKT3 could monitor the recurrence risk of patients
with MI. However, there is a limitation of the present study. The
underlying mechanism of the identified differentially expressed
miRNAs and mRNAs in MI requires further study in vivo and
in vitro.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO repository under accession
nos. GSE24548 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24548),
GSE97320 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97320),
GSE24519 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24519),
GSE66360 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66360)
and GSE60993 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60993).
Authors' contributions
SW drafted the manuscript. SW and NC carried out the
experiments and analyzed and interpreted the data. SW designed the
study. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the Ethics
Committee of Jinan Jigang Hospital (approval no. JG-001) and
informed written consent was obtained from all individuals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bainey KR, Fresco C, Zheng Y, Halvorsen S,
Carvalho A, Ostojic M, Goldstein P, Gershlick AH, Westerhout CM,
Van de Werf F, et al: Implications of ischaemic area at risk and
mode of reperfusion in ST-elevation myocardial infarction. Heart.
102:527–533. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goto Y, Masaki I, Yamada A, Uno M,
Nakamori S, Nagata M, Ichikawa Y, Kitagawa K, Ito M and Sakuma H:
Native T1 mapping allows for the accurate detection of the segments
with chronic myocardial infarction in patients with known or
suspected coronary artery disease. J Cardiovasc Magn Reson.
18:P702016. View Article : Google Scholar
|
|
3
|
Orlic D, Ostojic M, Beleslin B,
Milasinovic D, Tesic M, Borovic M, Vukcevic V, Stojkovic S,
Nedeljkovic M and Stankovic G: The randomized physiologic
assessment of thrombus aspiration in patients with acute ST-segment
elevation myocardial infarction trial (PATA STEMI): Study rationale
and design. J Interv Cardiol. 27:341–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Furtado MB, Costa MW and Rosenthal NA: The
cardiac fibroblast: Origin, identity and role in homeostasis and
disease. Differentiation. 92:93–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kurose H and Mangmool S: Myofibroblasts
and inflammatory cells as players of cardiac fibrosis. Arch Pharm
Res. 39:1100–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao M, Yin D, Chen J and Qu X: Activating
the interleukin-6-Gp130-STAT3 pathway ameliorates ventricular
electrical stability in myocardial infarction rats by modulating
neurotransmitters in the paraventricular nucleus. BMC Cardiovasc
Disord. 20:602020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ouweneel DM, Eriksen E, Sjauw KD, van
Dongen IM, Hirsch A, Packer EJ, Vis MM, Wykrzykowska JJ, Koch KT,
Baan J, et al: Percutaneous mechanical circulatory support versus
intra-aortic balloon pump in cardiogenic shock after acute
myocardial infarction. J Am Coll Cardiol. 69:278–287. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rodríguez-Jiménez AE, Cruz-Inerarity H,
Negrín-Valdés T, Fardales-Rodríguez R and Chávez-González E:
Corrected QT-Interval Dispersion: An Electrocardiographic Tool to
Predict Recurrence of Myocardial Infarction. MEDICC Rev. 21:22–25.
2019.PubMed/NCBI
|
|
9
|
Frangogiannis NG, Smith CW and Entman ML:
The inflammatory response in myocardial infarction. Cardiovasc Res.
53:31–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nian M, Lee P, Khaper N and Liu P:
Inflammatory cytokines and postmyocardial infarction remodeling.
Circ Res. 94:1543–1553. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Landmesser U, Wollert KC and Drexler H:
Potential novel pharmacological therapies for myocardial
remodelling. Cardiovasc Res. 81:519–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamada Y, Ichihara S and Nishida T:
Molecular genetics of myocardial infarction. Genomic Med. 2:7–22.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Swirski FK and Nahrendorf M: Leukocyte
behavior in atherosclerosis, myocardial infarction, and heart
failure. Science. 339:161–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng Y and Zhang C: MicroRNA-21 in
cardiovascular disease. J Cardiovasc Transl Res. 3:251–255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marques FZ, Vizi D, Khammy O, Mariani JA
and Kaye DM: The transcardiac gradient of cardio-microRNAs in the
failing heart. Eur J Heart Fail. 18:1000–1008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oliveira-Carvalho V, da Silva MM,
Guimarães GV, Bacal F and Bocchi EA: MicroRNAs: new players in
heart failure. Mol Biol Rep. 40:2663–2670. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang JB, Mei J, Jiang LY, Jiang ZL, Liu
H, Zhang JW and Ding FB: miR-196a2 rs11614913 T>C polymorphism
is associated with an increased risk of tetralogy of fallot in a
Chinese population. Acta Cardiol Sin. 31:18–23. 2015.PubMed/NCBI
|
|
18
|
Meder B, Keller A, Vogel B, Haas J,
Sedaghat-Hamedani F, Kayvanpour E, Just S, Borries A, Rudloff J,
Leidinger P, et al: MicroRNA signatures in total peripheral blood
as novel biomarkers for acute myocardial infarction. Basic Res
Cardiol. 106:13–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bostjancic E, Zidar N and Glavac D:
MicroRNA microarray expression profiling in human myocardial
infarction. Dis Markers. 27:255–268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang H, Zhang C, Ban T, Liu Y, Mei L,
Piao X, Zhao D, Lu Y, Chu W and Yang B: A novel reciprocal loop
between microRNA-21 and TGFβRIII is involved in cardiac fibrosis.
Int J Biochem Cell Biol. 44:2152–2160. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL,
Zhu JN, Liu XY, Zhang YY, Li Y, Lin SG and Yu XY: Upregulated
expression of miR-1/miR-206 in a rat model of myocardial
infarction. Biochem Biophys Res Commun. 381:597–601. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi B, Guo Y, Wang J and Gao W: Altered
expression of microRNAs in the myocardium of rats with acute
myocardial infarction. BMC Cardiovasc Disord. 10:112010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reiner-Benaim A: FDR control by the BH
procedure for two-sided correlated tests with implications to gene
expression data analysis. Biom J. 49:107–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
1995.
|
|
27
|
Wu Y, Zhang L, Zhang Y, Zhen Y and Liu S:
Bioinformatics analysis to screen for critical genes between
survived and non-survived patients with sepsis. Mol Med Rep.
18:3737–3743. 2018.PubMed/NCBI
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
The Gene Ontology Consortium, . The Gene
Ontology Resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ellis KL, Cameron VA, Troughton RW,
Frampton CM, Ellmers LJ and Richards AM: Circulating microRNAs as
candidate markers to distinguish heart failure in breathless
patients. Eur J Heart Fail. 15:1138–1147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Su Q, Liu Y, Lv XW, Ye ZL, Sun YH, Kong BH
and Qin ZB: Inhibition of lncRNA TUG1 upregulates miR-142-3p to
ameliorate myocardial injury during ischemia and reperfusion via
targeting HMGB1- and Rac1-induced autophagy. Mol Cell Cardiol.
133:12–25. 2019. View Article : Google Scholar
|
|
36
|
Zhu Y, Lin Y, Yan W, Sun Z, Jiang Z, Shen
B, Jiang X and Shi J: Novel biomarker MicroRNAs for subtyping of
acute coronary syndrome: A Bioinformatics Approach. Biomed Res Int.
2016:46183232016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Durgan DJ, Smith JK, Hotze MA, Egbejimi O,
Cuthbert KD, Zaha VG, Dyck JR, Abel ED and Young ME: Distinct
transcriptional regulation of long-chain acyl-CoA synthetase
isoforms and cytosolic thioesterase 1 in the rodent heart by fatty
acids and insulin. Am J Physiol Heart Circ Physiol.
290:H2480–H2497. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chiu HC, Kovacs A, Ford DA, Hsu FF, Garcia
R, Herrero P, Saffitz JE and Schaffer JE: A novel mouse model of
lipotoxic cardiomyopathy. J Clin Invest. 107:813–822. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang L, Yang Y, Si D, Shi K, Liu D, Meng H
and Meng F: High expression of long chain acyl-coenzyme A
synthetase 1 in peripheral blood may be a molecular marker for
assessing the risk of acute myocardial infarction. Exp Ther Med.
14:4065–4072. 2017.PubMed/NCBI
|
|
40
|
Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai
LS, Zhang L and Hu Y: miR-15a and miR-16 affect the angiogenesis of
multiple myeloma by targeting VEGF. Carcinogenesis. 34:426–435.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jackstadt R and Hermeking H: MicroRNAs as
regulators and mediators of c-MYC function. Biochim Biophys Acta.
1849:544–553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chamorro-Jorganes A, Araldi E, Penalva LO,
Sandhu D, Fernández-Hernando C and Suárez Y: MicroRNA-16 and
microRNA-424 regulate cell-autonomous angiogenic functions in
endothelial cells via targeting vascular endothelial growth factor
receptor-2 and fibroblast growth factor receptor-1. Arterioscler
Thromb Vasc Biol. 31:2595–2606. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chan LS, Yue PY, Wong YY and Wong RN:
MicroRNA-15b contributes to ginsenoside-Rg1-induced angiogenesis
through increased expression of VEGFR-2. Biochem Pharmacol.
86:392–400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Abu-Halima M, Poryo M, Ludwig N, Mark J,
Marsollek I, Giebels C, Petersen J, Schäfers HJ, Grundmann U,
Pickardt T, et al: Differential expression of microRNAs following
cardiopulmonary bypass in children with congenital heart diseases.
J Transl Med. 15:1172017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dahl TB, Holm S, Aukrust P and Halvorsen
B: Visfatin/NAMPT: A multifaceted molecule with diverse roles in
physiology and pathophysiology. Annu Rev Nutr. 32:229–243. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qiu L and Liu X: Identification of key
genes involved in myocardial infarction. Eur J Med Res. 24:222019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Montecucco F, Bauer I, Braunersreuther V,
Bruzzone S, Akhmedov A, Lüscher TF, Speer T, Poggi A, Mannino E,
Pelli G, et al: Inhibition of nicotinamide
phosphoribosyltransferase reduces neutrophil-mediated injury in
myocardial infarction. Antioxid Redox Signal. 18:630–641. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang X, Chen J and Huang X: Rosuvastatin
attenuates myocardial ischemia-reperfusion injury via upregulating
miR-17-3p-mediated autophagy. Cell Reprogram. 21:323–330. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tao H, Chen ZW, Yang JJ and Shi KH:
MicroRNA-29a suppresses cardiac fibroblasts proliferation via
targeting VEGF-A/MAPK signal pathway. Int J Biol Macromol.
88:414–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Romaine SP, Tomaszewski M, Condorelli G
and Samani NJ: MicroRNAs in cardiovascular disease: An introduction
for clinicians. Heart. 101:921–928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dews M, Homayouni A, Yu D, Murphy D,
Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT and
Thomas-Tikhonenko A: Augmentation of tumor angiogenesis by a
Myc-activated microRNA cluster. Nat Genet. 38:1060–1065. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Landskroner-Eiger S, Qiu C, Perrotta P,
Siragusa M, Lee MY, Ulrich V, Luciano AK, Zhuang ZW, Corti F,
Simons M, et al: Endothelial miR-17~92 cluster negatively regulates
arteriogenesis via miRNA-19 repression of WNT signaling. Proc Natl
Acad Sci USA. 112:12812–12817. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Maciejak A, Kiliszek M, Michalak M, Tulacz
D, Opolski G, Matlak K, Dobrzycki S, Segiet A, Gora M and Burzynska
B: Gene expression profiling reveals potential prognostic
biomarkers associated with the progression of heart failure. Genome
Med. 7:262015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Heger J, Bornbaum J, Würfel A, Hill C,
Brockmann N, Gáspár R, Pálóczi J, Varga ZV, Sárközy M, Bencsik P,
et al: JDP2 overexpression provokes cardiac dysfunction in mice.
Sci Rep. 8:76472018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pfeiffer L, Wahl S, Pilling LC, Reischl E,
Sandling JK, Kunze S, Holdt LM, Kretschmer A, Schramm K, Adamski J,
et al: DNA methylation of lipid-related genes affects blood lipid
levels. Circ Cardiovasc Genet. 8:334–342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Heximer SP, Knutsen RH, Sun X, Kaltenbronn
KM, Rhee MH, Peng N, Oliveira-dos-Santos A, Penninger JM, Muslin
AJ, Steinberg TH, et al: Hypertension and prolonged vasoconstrictor
signaling in RGS2-deficient mice. J Clin Invest. 111:445–452. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang KM, Wang GR, Lu P, Karas RH,
Aronovitz M, Heximer SP, Kaltenbronn KM, Blumer KJ, Siderovski DP,
Zhu Y and Mendelsohn ME: Regulator of G-protein signaling-2
mediates vascular smooth muscle relaxation and blood pressure. Nat
Med. 9:1506–1512. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wieland T, Lutz S and Chidiac P:
Regulators of G protein signalling: A spotlight on emerging
functions in the cardiovascular system. Curr Opin Pharmacol.
7:201–207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang J, Huang W, Xu R, Nie Y, Cao X, Meng
J, Xu X, Hu S and Zheng Z: MicroRNA-24 regulates cardiac fibrosis
after myocardial infarction. J Cell Mol Med. 16:2150–2160. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Inouye M, Ripatti S, Kettunen J,
Lyytikäinen LP, Oksala N, Laurila PP, Kangas AJ, Soininen P,
Savolainen MJ, Viikari J, et al: Novel Loci for metabolic networks
and multi-tissue expression studies reveal genes for
atherosclerosis. PLoS Genet. 8:e10029072012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Thuny F, Textoris J, Amara AB, Filali AE,
Capo C, Habib G, Raoult D and Mege JL: The gene expression analysis
of blood reveals S100A11 and AQP9 as potential biomarkers of
infective endocarditis. PLoS One. 7:e314902012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Huang X, Yu X, Li H, Han L and Yang X:
Regulation mechanism of aquaporin 9 gene on inflammatory response
and cardiac function in rats with myocardial infarction through
extracellular signal-regulated kinase1/2 pathway. Heart Vessels.
34:2041–2051. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan
M, Lin YD, Fisch S, Unno K, Sereti KI and Liao R: MicroRNA-34a
plays a key role in cardiac repair and regeneration following
myocardial infarction. Circ Res. 117:450–459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huang Y, Qi Y, Du JQ and Zhang DF:
MicroRNA-34a regulates cardiac fibrosis after myocardial infarction
by targeting Smad4. Expert Opin Ther Targets. 18:1355–1365.
2014.PubMed/NCBI
|
|
65
|
Matsumoto S, Sakata Y, Suna S, Nakatani D,
Usami M, Hara M, Kitamura T, Hamasaki T, Nanto S, Kawahara Y and
Komuro I: Circulating p53-responsive microRNAs are predictive
indicators of heart failure after acute myocardial infarction. Circ
Res. 113:322–326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bernardo BC, Gao XM, Winbanks CE, Boey EJ,
Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, et
al: Therapeutic inhibition of the miR-34 family attenuates
pathological cardiac remodeling and improves heart function. Proc
Natl Acad Sci USA. 109:17615–17620. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bernardo BC, Gao XM, Tham YK, Kiriazis H,
Winbanks CE, Ooi JY, Boey EJ, Obad S, Kauppinen S, Gregorevic P, et
al: Silencing of miR-34a attenuates cardiac dysfunction in a
setting of moderate, but not severe, hypertrophic cardiomyopathy.
PLoS One. 9:e903372014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Stephanou A, Brar BK, Scarabelli TM,
Jonassen AK, Yellon DM, Marber MS, Knight RA and Latchman DS:
Ischemia-induced STAT-1 expression and activation play a critical
role in cardiomyocyte apoptosis. J Biol Chem. 275:10002–10008.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Stephanou A: Role of STAT-1 and STAT-3 in
ischaemia/reperfusion injury. J Cell Mol Med. 8:519–525. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang S, Liu W, Liu X, Qi J and Deng C:
Biomarkers identification for acute myocardial infarction detection
via weighted gene co-expression network analysis. Medicine
(Baltimore). 96:e83752017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chang PC, Lin SF, Chu Y, Wo HT, Lee HL,
Huang YC, Wen MS and Chou CC: LCZ696 therapy reduces ventricular
tachyarrhythmia inducibility in a myocardial infarction-induced
heart failure rat model. Cardiovasc Ther. 2019:60326312019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Li SY, Li ZX, He ZG, Wang Q, Li YJ, Yang
Q, Wu DZ, Zeng HL and Xiang HB: Quantitative proteomics reveal the
alterations in the spinal cord after myocardial
ischemia-reperfusion injury in rats. Int J Mol Med. 44:1877–1887.
2019.PubMed/NCBI
|
|
73
|
Getz GS and Reardon CA: Do the
Apoe−/- and Ldlr−/−Mice yield the same
insight on atherogenesis? Arterioscler Thromb Vasc Biol.
36:1734–1741. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Frangogiannis NG: Chemokines in the
ischemic myocardium: From inflammation to fibrosis. Inflamm Res.
53:585–595. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Frangogiannis NG and Entman ML: Chemokines
in myocardial ischemia. Trends Cardiovasc Med. 15:163–169. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Boengler K, Hilfiker-Kleiner D, Drexler H,
Heusch G and Schulz R: The myocardial JAK/STAT pathway: From
protection to failure. Pharmacol Ther. 120:172–185. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Fuglesteg BN, Suleman N, Tiron C, Kanhema
T, Lacerda L, Andreasen TV, Sack MN, Jonassen AK, Mjøs OD, Opie LH
and Lecour S: Signal transducer and activator of transcription 3 is
involved in the cardioprotective signalling pathway activated by
insulin therapy at reperfusion. Basic Res Cardiol. 103:444–453.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liu YH, Wang D, Rhaleb NE, Yang XP, Xu J,
Sankey SS, Rudolph AE and Carretero OA: Inhibition of p38
mitogen-activated protein kinase protects the heart against cardiac
remodeling in mice with heart failure resulting from myocardial
infarction. J Card Fail. 11:74–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ge ZW, Zhu XL, Wang BC, Hu JL, Sun JJ,
Wang S, Chen XJ, Meng SP, Liu L and Cheng ZY: MicroRNA-26b relieves
inflammatory response and myocardial remodeling of mice with
myocardial infarction by suppression of MAPK pathway through
binding to PTGS2. Int J Cardiol. 280:152–159. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu X, Chen K, Zhuang Y, Huang Y, Sui Y,
Zhang Y, Lv L and Zhang G: Paeoniflorin improves pressure
overload-induced cardiac remodeling by modulating the MAPK
signaling pathway in spontaneously hypertensive rats. Biomed
Pharmacother. 111:695–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li C, Li J, Xue K, Zhang J, Wang C, Zhang
Q, Chen X, Gao C, Yu X and Sun L: MicroRNA-143-3p promotes human
cardiac fibrosis via targeting sprouty3 after myocardial
infarction. J Mol Cell Cardiol. 129:281–292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Jia L, Zhu L, Wang JZ, Wang XJ, Chen JZ,
Song L, Wu YJ, Sun K, Yuan ZY and Hui R: Methylation of FOXP3 in
regulatory T cells is related to the severity of coronary artery
disease. Atherosclerosis. 228:346–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Abbate A, Bonanno E, Mauriello A, Bussani
R, Biondi-Zoccai GG, Liuzzo G, Leone AM, Silvestri F, Dobrina A,
Baldi F, et al: Widespread myocardial inflammation and
infarct-related artery patency. Circulation. 110:46–50. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Fan L, Meng H, Guo X, Li X and Meng F:
Differential gene expression profiles in peripheral blood in
northeast chinese han people with acute myocardial infarction.
Genet Mol Biol. 41:59–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Muse ED, Kramer ER, Wang H, Barrett P,
Parviz F, Novotny MA, Lasken RS, Jatkoe TA, Oliveira G, Peng H, et
al: A whole blood molecular signature for acute myocardial
infarction. Sci Rep. 7:122682017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Park HJ, Noh JH, Eun JW, Koh YS, Seo SM,
Park WS, Lee JY, Chang K, Seung KB, Kim PJ and Nam SW: Assessment
and diagnostic relevance of novel serum biomarkers for early
decision of ST-elevation myocardial infarction. Oncotarget.
6:12970–12983. 2015. View Article : Google Scholar : PubMed/NCBI
|