Introduction
Triple-negative breast cancer (TNBC), as a molecular
subtype of breast cancer, is susceptible to early recurrence and
metastasis. Patients with TNBC have poor prognosis owing to a lack
of effective clinical treatment measures (1). Tumor cells receive blood supply by
fusion with the host blood vessels, as well as by vascular
formation methods, such as constructing new vascular systems, which
is called vasculogenic mimicry (VM) (2). This phenomenon was discovered by
Maniotis et al (2), who
studied the microcirculation of human uveal melanoma and
demonstrated a novel manner by which tumor cells obtain blood
supply. According to the VM theory, when the tumor diameter is
>2 mm, vascular endothelial cells are activated to construct new
blood vessels to obtain blood supply to prevent tumor cell necrosis
as a result of ischemia and hypoxia (1). The VM theory suggests that tumor cells
form tubular structures, and it has been hypothesized that tumor
cells exhibit stem cell plasticity, similar to the phenotype and
function of endothelial cells, and can differentiate into
endothelial cells in the tumor microenvironment (3,4). VM
has been reported in melanoma, glioma and bidirectional malignancy,
as well as colon, liver, ovarian, bladder and prostate cancer
(5–10). A previous study found that VM also
exists in TNBC, and that VM formation and high expression levels of
CD133+ may be primary reasons for recurrence and
progression of TNBC (11).
Epithelial-to-mesenchymal transition (EMT) refers to
the process by which epithelial cells lose their polarity and
cell-cell contacts to acquire certain characteristics of
mesenchymal cells (12). EMT serves
an important role in the invasion and the metastasis of numerous
types of cancer, including breast and colorectal cancer (13,14).
The expression levels of E-cadherin and occludin are downregulated,
whereas the expression levels of N-cadherin and vimentin are
upregulated during EMT (15).
MicroRNAs (miRNAs or miRs) are non-coding RNAs that
serve important roles in regulating the expression levels of mRNAs.
Aberrant miRNAs are known to act as promoters or inhibitors of EMT
(16). miR-93 is a member of the
miR-106b-25 cluster (17).
Overexpression of miR-93 has been identified in multiple types of
cancer, including hepatocellular, gastric and breast cancer, as
well as glioma and endometrial carcinoma, in which it serves as an
oncogene (17). The expression
levels of miR-93 have been demonstrated to be markedly increased in
TNBC tissue compared with non-TNBC tissue, and are associated with
clinicopathological features, such as increased lymph node
metastasis, higher TNM staging and increased Ki-67 expression
(18), indicating that miR-93
contributes to cell proliferation and metastasis. Moreover, miR-93
is involved in the process of EMT in endometrial carcinoma and
breast cancer cells (19,20). However, the regulatory mechanism of
miR-93 on EMT and VM in TNBC remains unclear. Therefore, the
present study investigated the role and underlying mechanism of
miR-93 in EMT transformation and VM formation to provide a
potential therapeutic target for TNBC.
Materials and methods
Cell culture
Human breast cancer cell lines MDA-MB-231, SK-BR-3
and MCF-7 were donated by The Medical Center Laboratory of Xi'an
Jiaotong University (Xi'an, China) and cultured in RPMI-1640 and
DMEM (Gibco; Thermo Fisher Scientific, Inc.) at 37°C.
Non-tumorigenic MCF-10A cells were purchased from The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences and
cultured in McCoy's 5A medium (Gibco; Thermo Fisher Scientific,
Inc.). All media were supplemented with 10% FBS (Biological
Industries), along with 1% penicillin and streptomycin.
miR-93 RNA interference (RNAi)
MDA-MB-231 cells were seeded in a 6-well plate at
5×104 cells/well for RNAi. Cells were then divided into
lentivirus-short hairpin (sh)RNA and negative control (shNC)
groups, in which cells were infected with the miR-93 knockdown
recombinant (5′-CTACCTGCACGAACAGCACTTTG-3′) and empty green
fluorescent protein (GFP) lentivirus (5′-TTCTCCGAACGTGTCACGT-3′),
respectively. Lentiviral vectors were constructed by Shanghai
GeneChem Co., Ltd. Cells were infected with shNC and shRNA
lentiviral particles (multiplicity of infection, 10) and incubated
for 8 h at 37°C. The medium containing lentiviruses was then
removed, and cells were cultured in normal culture medium for
another 12 h. Subsequently, 72 h after infection, the expression
levels of GFP were observed through a fluorescent microscope
(magnification, ×100; data not shown). Cells with infection
efficiency >80% were selected for subsequent analysis.
MTT assay
Following digestion by trypsin, MDA-MB-231 cells in
logarithmic growth phase were resuspended in complete medium
(RPMI-1640 medium supplemented with 10% FBS and 1% penicillin and
streptomycin). Cells were plated at a density of 5×103
cells/well into 96-well plates. Following incubation at 37°C for 48
h, 20 µl MTT reagent (5 mg/ml) was added to each well and incubated
at 37°C for 4 h. Finally, 100 µl dimethyl sulfoxide was added to
completely dissolve the purple formazan crystals. The absorbance of
each well was measured at 492 nm using a multi-functional
microplate reader for evaluation of cell viability.
Western blot analysis
Total cellular protein was extracted using RIPA
buffer (Beyotime Institute of Biotechnology) and the protein
concentrations were determined by a BCA Protein Assay kit (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Each sample contain 50 µg of protein was separated by
10% SDS-PAGE, the proteins were transferred to PVDF membrane and
blocked with 5% non-fat milk at 37°C for 1 h. The blots were
incubated overnight with specific primary antibodies
(anti-E-cadherin; cat. no. 4065; Cell Signaling Technology, Inc.,
anti-N-cadherin; cat. no. ab76057, Abcam, anti-vimentin; cat. no.
5741, Cell Signaling Technology, Inc. and anti-occludin; cat. no.
71-1500, Invitrogen (Thermo Fisher Scientific, Inc.); all at a
dilution of 1:1,000) at 4°C and thereafter with the HRP-conjugated
secondary antibody (dilution of 1:1,000, goat anti-rabbit, cat. no.
sc-2004; goat anti-mouse; cat. no. sc-2005, Santa Cruz
Biotechnology, Inc.) for 1 h at room temperature. ECL reagent (EMD
Millipore) was used to detect protein expression levels that were
analyzed with Quantity One 6.0 software (Bio-Rad Laboratories,
Inc), and the protein expression levels were normalized to
β-actin.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cell lines using
TRIzol® (Thermo Fisher Scientific, Inc.). The RNA
samples were diluted with sterile water, and the absorbance values
at 260 and 280 nm were detected using a spectrophotometer; a
260/280 value of 1.8–2.0 indicated optimal RNA quality. cDNA was
synthesized using the isolated RNA as a template by PrimeScript RT
reagent kit (Fermentas; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. The SYBR Green PCR kit (Takara
Biotechnology Co., Ltd.) was used to perform qPCR. The
thermocycling conditions were: 95°C for 10 sec, followed by 40
cycles of 95°C for 5 sec, 60°C for 30 sec, one cycle of 95°C for 15
sec, 60°C for 30 sec. Melting curve analysis was used to confirm
specificity of the PCR products. Relative expression levels of the
detected genes were assessed using the 2−ΔΔCq method
(21). GAPDH was used as an
internal control for mRNA, and U6 was an internal control for
miRNA. The primers used are listed in Table I.
 | Table I.Primer sequences used for
RT-quantitative PCR. |
Table I.
Primer sequences used for
RT-quantitative PCR.
| Gene | Primer name | Primer sequence
(5′→3′) |
|---|
| Hsa-miR-93 | miR-93 RT | GTCGTATCCAGTGCA
GGGTCCGAGGTATTCGCACTGGATACGACCTACCT |
|
| Forward |
ATTCGGCAAAGTGCTGTTCGTGC |
|
| Reverse |
ATCCAGTGCAGGGTCCGAGG |
| U6 | U6 RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATA |
|
| Forward |
AGAGAAGATTAGCATGGCCCCTG |
|
| Reverse | GTGCAGGGTCCGAGGT |
| E-cadherin | Forward |
GATTCCTTGCCAGTTGGTGT |
|
| Reverse |
TTCCTGCCTGGATTGGTATC |
| N-cadherin | Forward |
TTCTGGCTGTTGTGTTGAGG |
|
| Reverse |
ATTCCACCGCTACCACTTTG |
| Vimentin | Forward |
TGCCTCAACCTCCCAAGTAG |
|
| Reverse |
GGTCAGGAGTTCGAGACCAG |
| GAPDH | Forward |
ATCTGGCACCACACCTTCTACAATGAGCTGCG |
|
| Reverse |
CGTCATCCCTGCTTGCTGATCCACATC |
Cell migration and invasion assay
For the cell migration assay, equal numbers of cells
(1×105 cells/ml) were seeded in the top compartment of a
24-well chamber in 200 µl serum-free RPMI-1640. Additionally, 600
µl medium containing 10% FBS was added into the bottom chamber of
the system. The cells were incubated at 37°C for 24 h. After
carefully removing the non-migrated cells on the upper surface of
the Transwell chamber membrane, the cells that had migrated from
the membrane were fixed with 4% paraformaldehyde for 30 min
followed by staining with 0.1% crystal violet for 10 min, both at
room temperature. The number of migrated cells was counted from
five random fields of view under an inverted phase-contrast
microscope (magnification, ×200). Cell invasion assays were
performed in the same way as migration assays except that the cells
were seeded in the upper compartment of the chamber and the
membrane was pre-coated with diluted Matrigel for 30 min at
37°C.
VM
Matrigel was added into 48-well plates at 100
µl/well, and then incubated at 37°C for 30 min. Cells
(2×104 cells/ml) were added to each well. Following
incubation at 37°C for 12 h, cells with tubular structure formation
were imaged using an inverted microscope, and the number of tubular
cells (completely surrounded by a circle) were counted manually
from five random visual fields (up, down, left, right and center)
to calculate the average. Following incubation at 37°C for 48 h,
cells were digested with dispase and recycled with cell recovery
solution, which enables recovery of cells cultured on Matrigel for
subsequent biochemical analysis of PCR and western blot.
Statistical analysis
All data are presented as the mean ± standard
deviation of at least three independent repeats. SPSS 19.0 software
(IBM Corp.) was used for statistical analysis. Data were analyzed
by unpaired Student's t-test or one-way ANOVA followed by Tukey's
or Fisher's least significant difference post hoc test, as
appropriate. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-93 expression levels in breast
cancer cell lines and knockdown of miR-93 using shRNA
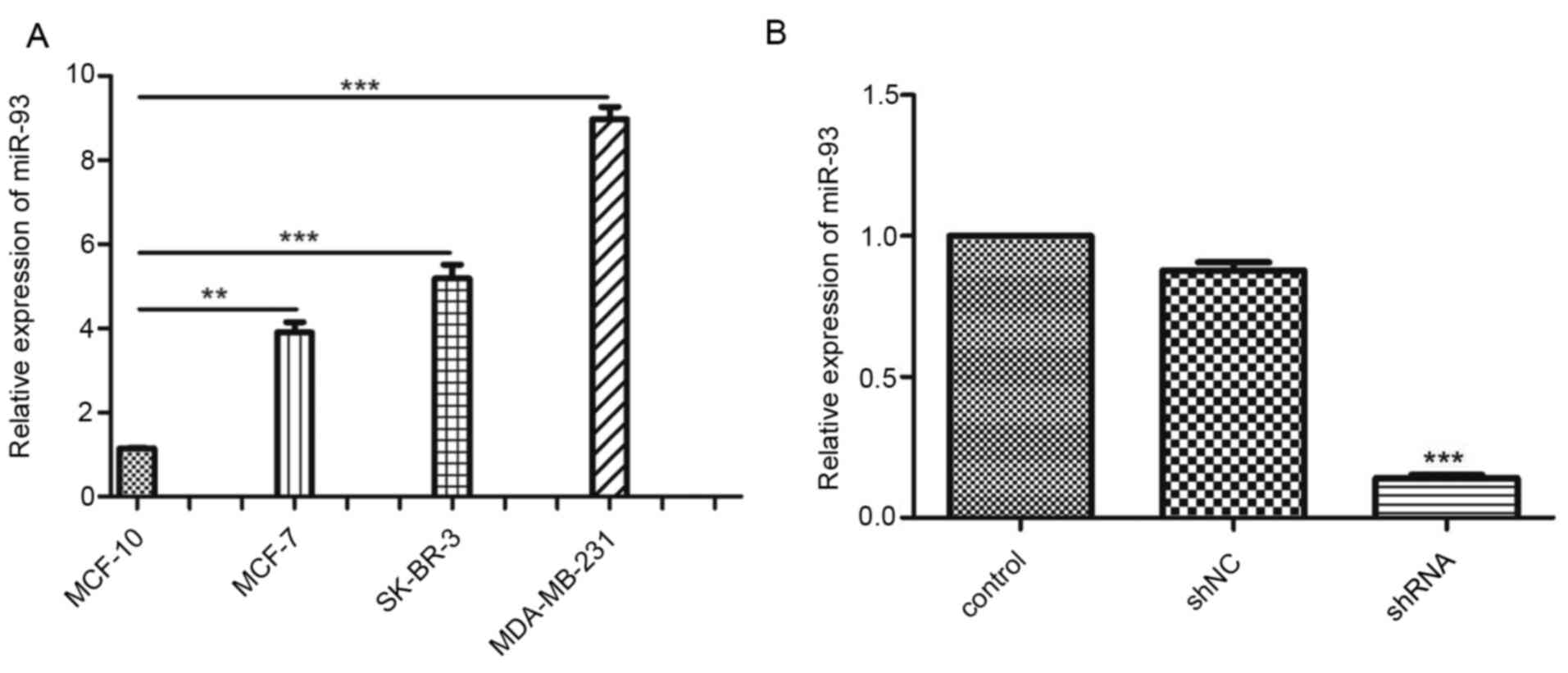
The expression levels of miR-93 in MCF-7, SK-BR3,
MDA-MB-231 and non-tumorigenic MCF-10A cell lines were detected by
RT-qPCR. The results showed that the expression levels of miR-93
were significantly higher in the three breast cancer cell lines
compared with expression in MCF-10A cells (P<0.01 and
P<0.001; Fig. 1A), with the
highest expression in MDA-MB-231 cells. Hence, the MDA-MB-231 cell
line was chosen for subsequent experiments. RT-qPCR confirmed that
expression of miR-93 was significantly inhibited by miR-93 shRNA
(P<0.001; Fig. 1B).
miR-93 depletion decreases MDA-MB-231
breast cancer cell viability
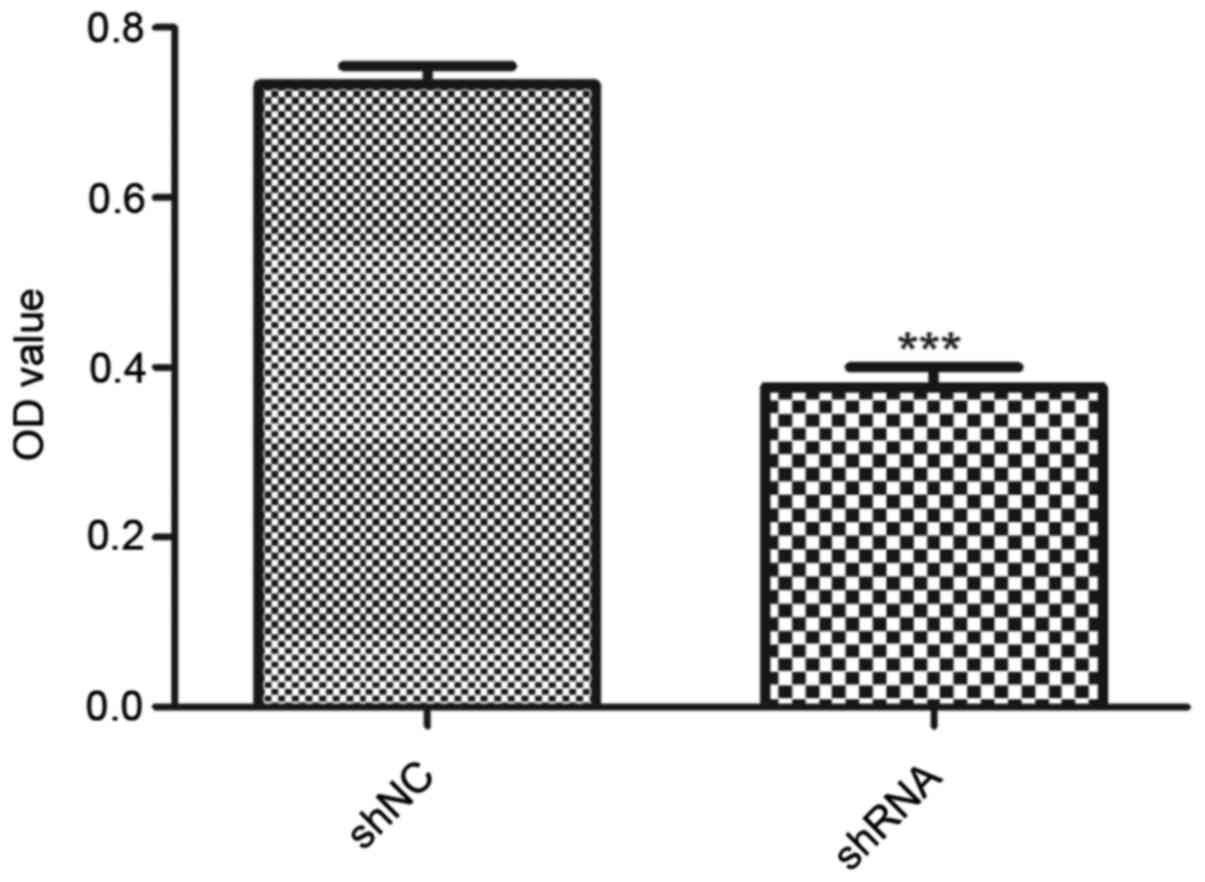
The effect of miR-93 on the viability of MDA-MB-231
cells was determined by MTT assay. Results showed a statistically
significant decrease in optical density in the shRNA group compared
with the shNC group (P<0.001; Fig.
2), suggesting that downregulation of miR-93 suppressed the
viability of MDA-MB-231 cells.
miR-93 depletion contributes to cell
migration and invasion
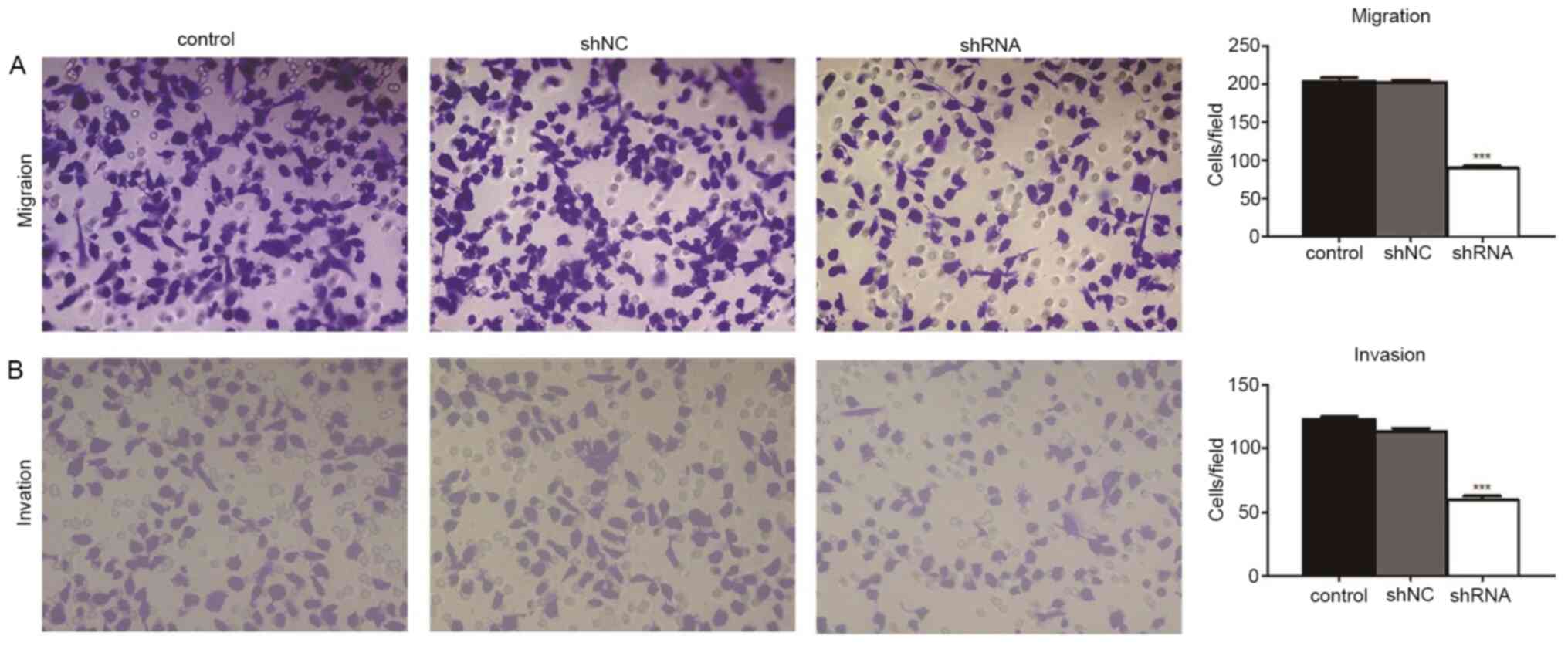
The Transwell assays revealed the effects of miR-93
knockdown on the migratory and invasive activity of MDA-MB-231
cells. Compared with the control (209.3±7.2 cells/field) and shNC
groups (202.0±8.7 cells/field), cell migration was significantly
decreased in the shRNA group (88.8±5.0 cells/field; both
P<0.001; Fig. 3). Compared with
the control (129.6±8.4 cells/field) and shNC groups (112.0±4.7
cells/field), cell invasion was also significantly decreased in the
shRNA group (68.8±7.0 cells/field; both P<0.001; Fig. 3). Cell migration and invasion were
not significantly different between the control and shNC groups
(both P>0.05; Fig. 3). These
results showed that the migratory and invasive ability of
MDA-MB-231 cells decreased following miR-93 depletion.
miR-93 depletion alters expression
levels of EMT-associated markers in MDA-MB-231 breast cancer
cells
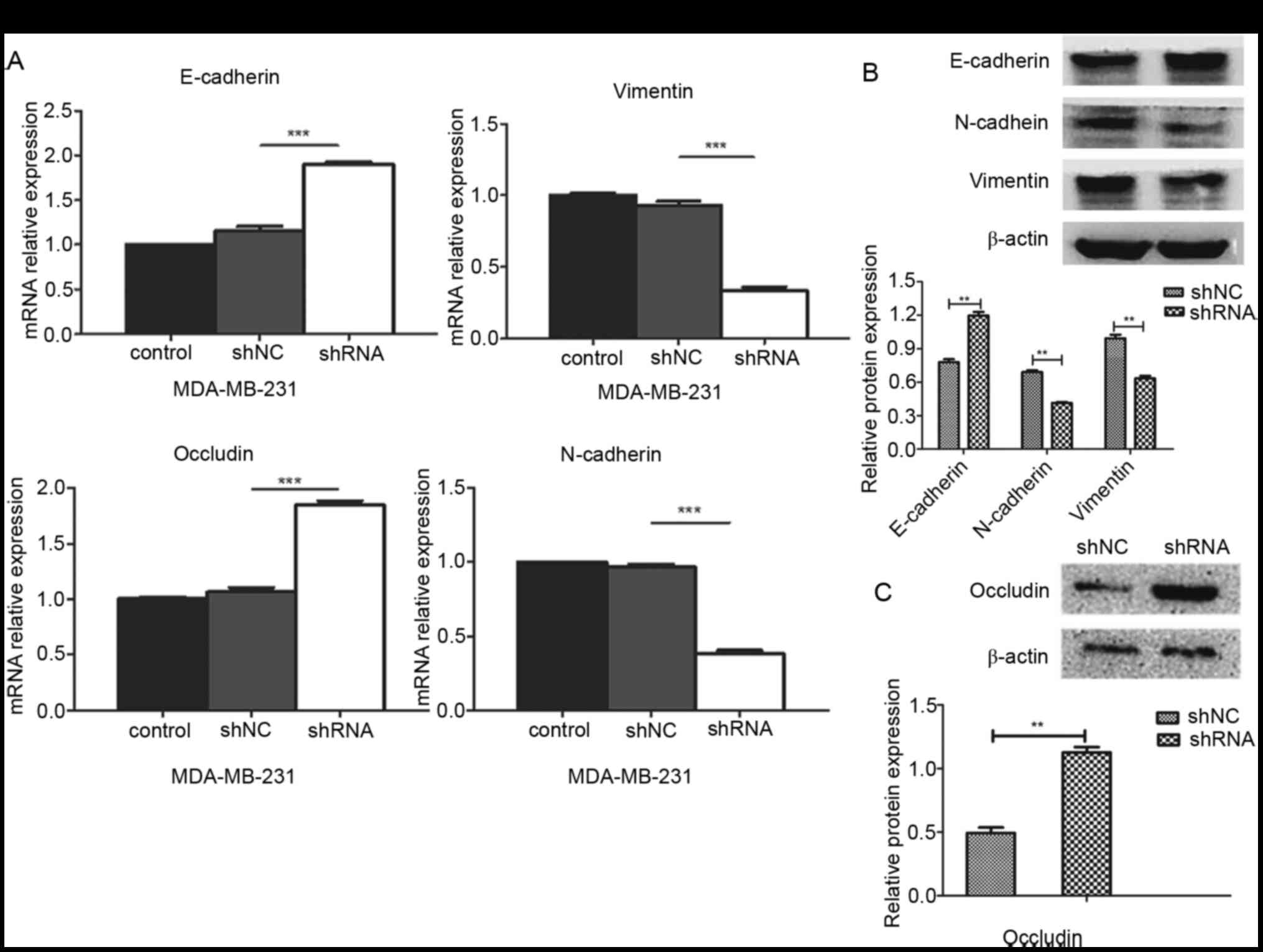
The association between miR-93 and EMT markers was
analyzed. RT-qPCR and western blotting were used to detect changes
in the expression levels of EMT-associated markers (E-cadherin,
N-cadherin, vimentin and occludin) following miR-93 depletion.
RT-qPCR results showed that miR-93-knockdown cells displayed
increased mRNA expression levels of E-cadherin and occludin and
decreased mRNA expression levels of N-cadherin and vimentin
(P<0.001; Fig. 4A). Western blot
assay results were consistent with the RT-qPCR and showed that the
protein expression levels of E-cadherin and occludin were
significantly upregulated, whereas the protein expression levels of
N-cadherin and vimentin were significantly downregulated in the
shRNA group compared with the respective expression levels in the
shNC group (P<0.01; Fig. 4B and
C). These results suggested that knockdown of miR-93 may
inhibit the EMT process in MDA-MB-231 cells.
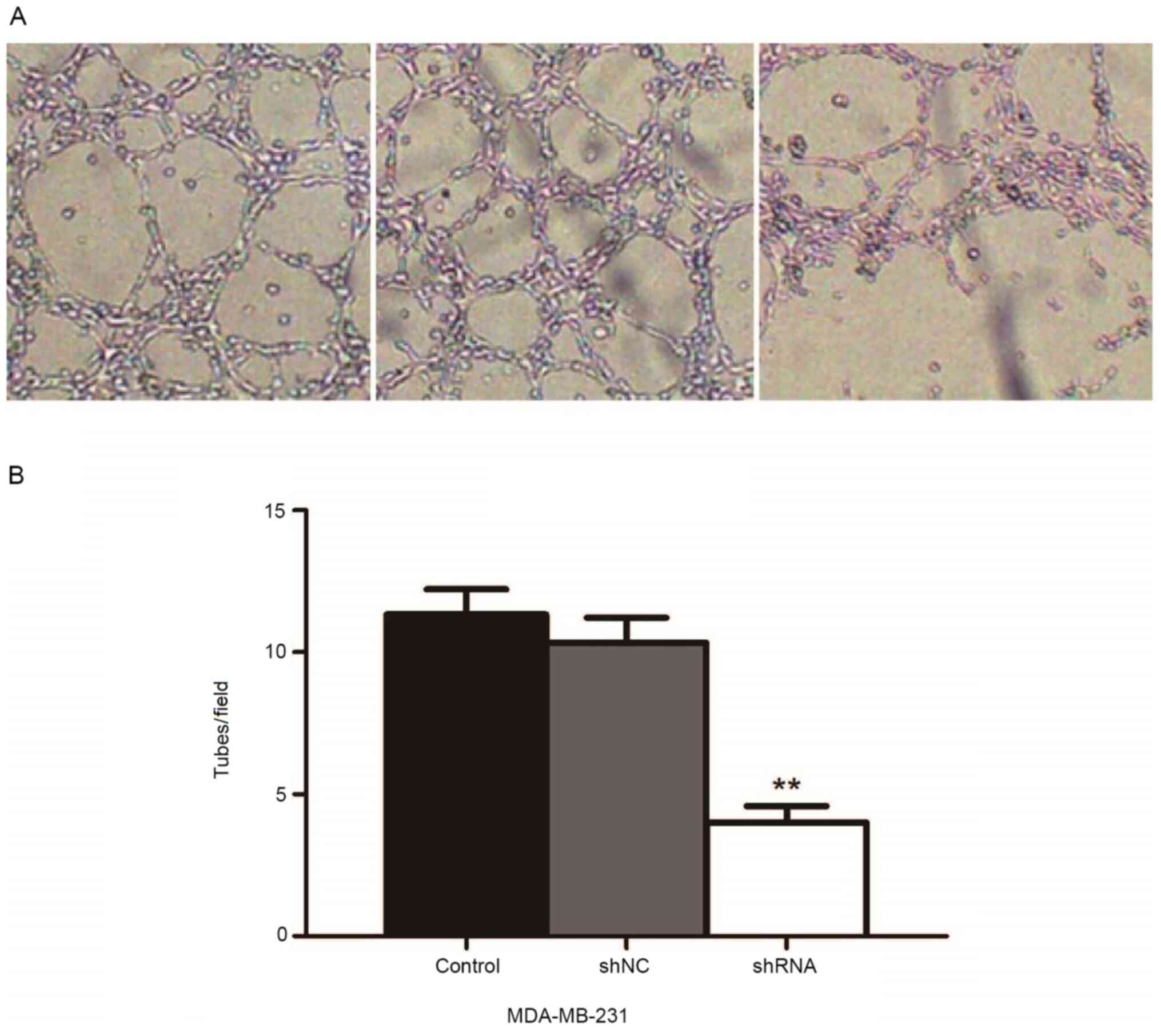
miR-93 depletion suppresses VM
formation in MDA-MB-231 breast cancer cells
Vascularization was observed with an inverted
microscope in three-dimensional culture of MDA-MB-231 cells
following miR-93 knockdown. VM was observed in all three groups
(Fig. 5A). The results showed that
the microtubule-forming ability of cells was significantly
decreased in the shRNA group compared with the control and the shNC
groups (P<0.01; Fig. 5B),
suggesting that miR-93 may serve as a promoter of VM formation in
MDA-MB-231 cells.
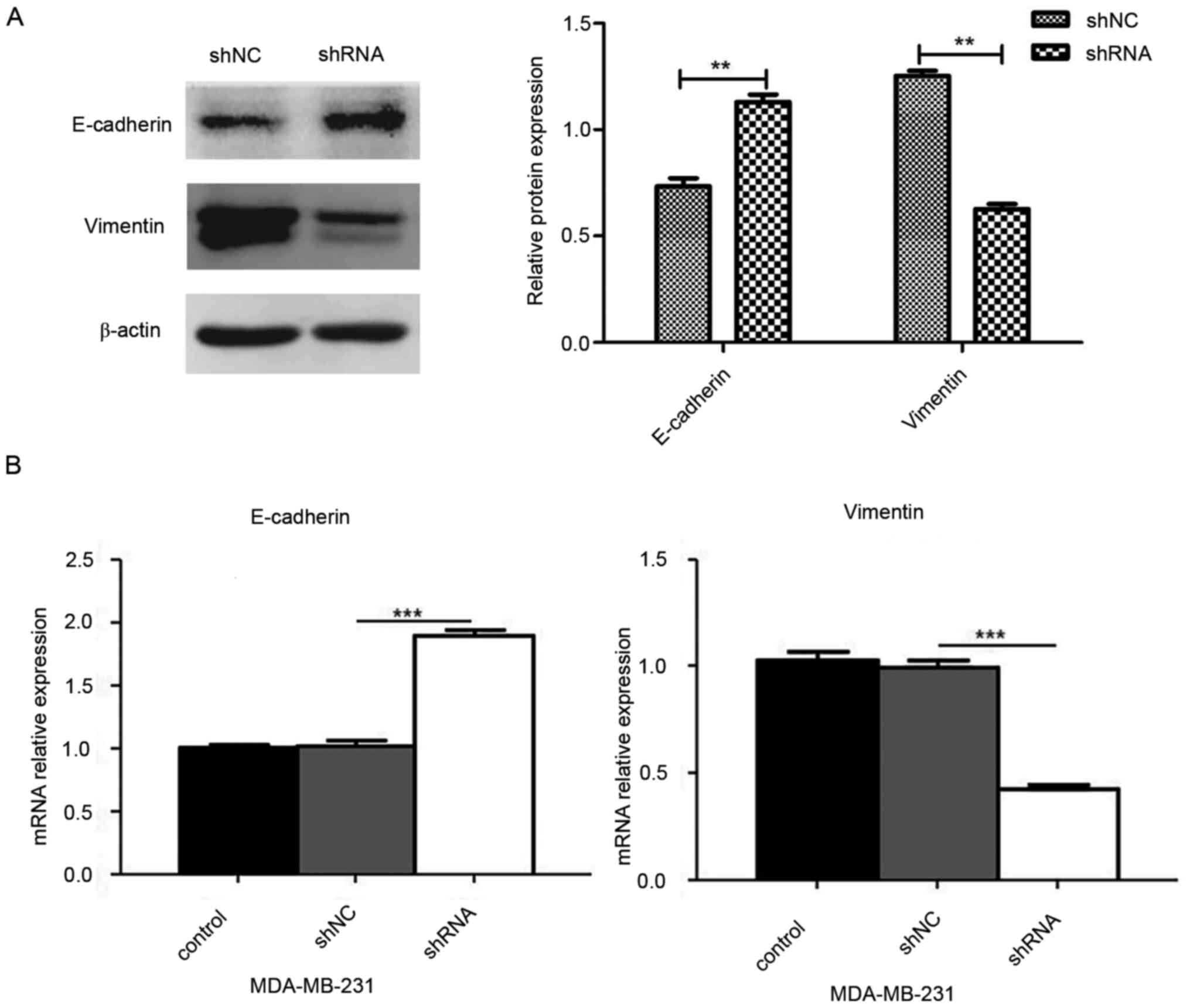
miR-93 depletion inhibits expression
levels of EMT-associated markers in three-dimensional cell
culture
To determine whether knockdown of miR-93 affects the
EMT process in three-dimensional culture of MDA-MB-231 cells,
western blotting (Fig. 6A) and
RT-qPCR (Fig. 6B) were performed.
The results indicated that the mRNA and protein expression levels
of E-cadherin increased, whereas those of vimentin decreased in the
shRNA group compared with the shNC group, suggesting that miR-93
may promote VM formation through the EMT process (P<0.01 and
P<0.001).
Discussion
miR-93, a member of the miR-106b-25 cluster, is
located on intron 13 of the MCM 7 gene on chromosome 7q22. Studies
have previously reported that the miR-106b-25 cluster is highly
expressed in esophageal, gastrointestinal, liver, prostate and
endometrial cancer, suggesting that it may be a potential
proto-oncogene (22–24). Lowery et al (25) showed that the expression profile of
miRNAs is associated with the hormone receptor status in breast
cancer and may be an important indicator for the molecular
classification, clinical diagnosis, treatment choice and prognosis.
Cascione et al (26) found
that the expression profiles of miRNAs and mRNAs are associated
with the survival of patients with TNBC; miR-16, miR-155, miR-125b
and miR-374a are associated with survival, and miR-16, miR-125b,
miR-374a, miR-374b, miR-421, miR-655 and miR-497 are associated
with disease-free survival. Multi-factor analysis has suggested
that the aforementioned indicators are independent prognostic
factors that affect the survival of patients. Nam et al
(27) found that compared with
normal ovarian tissue, miR-93 expression levels are significantly
increased in ovarian cancer tissue, and that this is associated
with worse prognosis. In our previous study (28) gene chip technology was used to
investigate the differential expression profiles of miRNAs in TNBC
compared with corresponding paracarcinoma tissue; the results
demonstrated a significant upregulation of miR-93. The target genes
of miR-93 were predicted using bioinformatics and found that these
target genes promoted cell viability, invasion and migration
(28). Our earlier work confirmed
that miR-93 expression is increased in clinical specimens of
patients with TNBC and that this is associated with poor prognosis
(high TNM staging and lymph node metastasis) (29). The present study confirmed at the
cytology level that miR-93 depletion inhibited the viability of
TNBC cells, and decreased cell invasion and migration, which was
consistent with the results of previous experiments using clinical
specimens.
miRNAs are involved in the formation of VM. Weng
et al (30) found that
miR-409-3p inhibits VM formation in human fibrosarcoma cells. Wu
et al (31) demonstrated
that miR-26b directly regulates the expression levels of Ephrin
type-A receptor 2 to inhibit VM formation in glioma. Shevde et
al (32) confirmed that
miR-299-5p is associated with VM formation in breast cancer. In the
present study, miR-93 depletion was found to inhibit the formation
of VM in three-dimensional culture of MDA-MB-231 cells, which was
consistent with the results of the aforementioned studies.
EMT refers to the process by which epithelial cells
acquire specific characteristics of mesenchymal cells. Mani et
al (13) found that mammary
epithelial cells that underwent the EMT process acquired certain
characteristics of stem cells, such as increased tumorigenesis. Liu
et al (14) reported high
expression levels of zinc-finger E-box binding homeobox 1 (ZEB1) in
VM-positive colon cancer tissue and that expression levels of
E-cadherin are decreased, whereas those of vimentin are increased
in ZEB1-positive tissue. Moreover, the formation of VM in colon
cancer significantly decreases following downregulation of ZEB1
(14). Sun et al (33) observed a significant decrease in VM
formation following downregulation of Twist-related protein 1 in
liver cancer. The present study showed that miR-93 depletion
significantly increased the protein expression levels of E-cadherin
and occludin and decreased those of N-cadherin and vimentin in
monolayer cell culture, suggesting that miR-93 may promote the
occurrence of EMT in TNBC cells. Moreover, in the shRNA group VM
was inhibited and the expression levels of E-cadherin were higher,
whereas the expression levels of vimentin were lower in
three-dimensional cell culture, suggesting that miR-93 may promote
the formation of VM through the EMT process.
VM determines the ability of tumor cells to form
blood cavities, which is associated with poor prognosis in patients
with TNBC (34). Lack of
experimentation on non-TNBC cell lines is a limitation of the
present study; further investigations involving both TNBC and
non-TNBC cell lines are required to determine if the effects of
miR-93 specifically act on the TNBC subtype. In summary, the
present study demonstrated that knockdown of miR-93 inhibited the
formation of VM in MDA-MB-231 TNBC cells and this was associated
with the expression levels of EMT markers. Therefore, targeting
miR-93 may provide a novel option for the anti-angiogenic treatment
of TNBC.
Acknowledgements
The authors would like to thank Professor Xinhan
Zhao (Department of Oncology, The First Affiliated Hospital of
Xi'an Jiaotong University, Shaanxi, China) for insightful
discussion and modification of the paper.
Funding
The present study was supported by grants from the
Project of Shaanxi Provincial Health commission fund: Effects and
mechanism of miR-93 on EMT and VM in triple-negative breast cancer
cells (grant no. 2016D037).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GA, FL, SH, YL and LHo performed all the
experiments. GA, LHe, JB and LHo designed the experiments. GA and
FL analyzed the data. GA, FL and LHo wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Anders C and Carey LA: Understanding and
treating triple-negative breast cancer. Oncology (Williston Park).
22:1233–1239; discussion 1239–1240, 1243. 2008.PubMed/NCBI
|
|
2
|
Maniotis AJ, Folberg R, Hess A, Seftor EA,
Gardner LM, Pe'er J, Trent JM, Meltzer PS and Hendrix MJ: Vascular
channel formation by human melanoma cells in vivo and in vitro:
Vasculogenic mimicry. Am J Pathol. 155:739–752. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hendrix MJ, Seftor EA, Hess AR and Seftor
RE: Vasculogenic mimicry and tumour-cell plasticity: Lessons from
melanoma. Nat Rev Cancer. 3:411–421. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paulis YWJ, Soetekouw PMMB, Verheul HMW,
Tjan-Heijnen VC and Griffioen AW: Signalling pathways in
vasculogenic mimicry. Biochim Biophys Acta. 1806:18–28.
2010.PubMed/NCBI
|
|
5
|
McDonald DM and Foss AJ: Endothelial cells
of tumor vessels: Abnormal but not absent. Cancer Metastasis Rev.
19:109–120. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sood AK, Seftor EA, Fletcher MS, Gardner
LM, Heidger PM, Buller RE, Seftor RE and Hendrix MJ: Molecular
determinants of ovarian cancer plasticity. Am J Pathol.
158:1279–1288. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shirakawa K, Kobayashi H, Heike Y,
Kawamoto S, Brechbiel MW, Kasumi F, Iwanaga T, Konishi F, Terada M
and Wakasugi H: Hemodynamics in vasculogenic mimicry and
angiogenesis of inflammatory breast cancer xenograft. Cancer Res.
62:560–566. 2002.PubMed/NCBI
|
|
8
|
Sharma N, Seftor RE, Seftor EA, Gruman LM,
Heidger PM Jr, Cohen MB, Lubaroff DM and Hendrix MJ: Prostatic
tumor cell plasticity involves cooperative interactions of distinct
phenotypic subpopulations: Role in vasculogenic mimicry. Prostate.
50:189–201. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hao X, Sun B, Zhang S and Zhao X:
Microarray study of vasculogenic mimicry in bi-directional
differentiation malignant tumor. Zhonghua Yi Xue Za Zhi.
82:1298–1302. 2002.(In Chinese). PubMed/NCBI
|
|
10
|
Dupuy E, Hainaud P, Villemain A,
Bodevin-Phèdre E, Brouland JP, Briand P and Tobelem G: Tumoral
angiogenesis and tissue factor expression during hepatocellular
carcinoma progression in a transgenic mouse model. J Hepatol.
38:793–802. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang D, Sun B, Zhao X, Ma Y, Ji R, Gu Q,
Dong X, Li J, Liu F, Jia X, et al: Twist1 expression induced by
sunitinib accelerates tumor cell vasculogenic mimicry by increasing
the population of CD133+ cells in triple-negative breast cancer.
Mol Cancer. 13:2072014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shook D and Keller R: Mechanisms,
mechanics and function of epithelial-mesenchymal transitions in
early development. Mech Dev. 120:1351–1383. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Sun B, Qi L, Li H, Gao J and Leng
X: Zinc finger E-box binding homeobox 1 promotes vasculogenic
mimicry in colorectal cancer through induction of
epithelial-to-mesenchymal transition. Cancer Sci. 103:813–820.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang L, Du WW, Yang W, Rutnam ZJ, Peng C,
Li H, O'Malley YQ, Askeland RW, Sugg S, Liu M, et al: MiR-93
enhances angiogenesis and metastasis by targeting LATS2. Cell
Cycle. 11:4352–4365. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu J, Xu J, Wu Y, Chen Q, Zheng W, Lu X,
Zhou C and Jiao D: Identification of microRNA-93 as a functional
dysregulated miRNA in triple-negative breast cancer. Tumour Biol.
36:251–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen S, Chen X, Sun KX, Xiu YL, Liu BL,
Feng MX, Sang XB and Zhao Y: MicroRNA-93 promotes
epithelial-mesenchymal transition of endometrial carcinoma cells.
PLoS One. 11:e01657762016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shyamasundar S, Lim JP, Bay BH and SUKANYA
S: miR-93 inhibits the invasive potential of triple-negative breast
cancer cells in vitro via protein kinase WNK1. Int J Oncol.
49:2629–2636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, et al: Functional links
between clustered microRNAs: Suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li W, Liang L, He X, Wan D and Gu J: Study
on the mechanism of hsa-miR93 in liver cancer. Weichangbingxue he
Ganbingxue Zazhi. 17:478–480. 2008.(in Chinese).
|
|
24
|
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe
TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al:
Genomic profiling of microRNA and messenger RNA reveals deregulated
microRNA expression in prostate cancer. Cancer Res. 68:6162–6170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G, et
al: MicroRNA signatures predict oestrogen receptor, progesterone
receptor and HER2/neu receptor status in breast cancer. Breast
Cancer Res. 11:478–480. 2009. View
Article : Google Scholar
|
|
26
|
Cascione L, Gasparini P, Lovat F, Carasi
S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, et
al: Integrated microRNA and mRNA signatures associated with
survival in triple negative breast cancer. PLoS One. 8:e559102013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao X, Hu J and Zhao X: Screening of
differentially expressed miRNA in triple negative breast cancer.
Jiaotong University Xuebao. 5:569–571. 2012.(in Chinese).
|
|
29
|
Zhao X, Hu J and Zhao X: Expression and
significance of hsa-miR-93 in triple negative breast cancer.
Zhonghua Linchuang Yishi Zazhi. 6:3914–3916. 2012.
|
|
30
|
Weng C, Dong H, Chen G, Zhai Y, Bai R, Hu
H, Lu L and Xu Z: miR-409-3p inhibits HT1080 cell proliferation,
vascularization and metastasis by targeting angiogenin. Cancer
Lett. 323:171–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu N, Zhao X, Liu M, Liu H, Yao W, Zhang
Y, Cao S and Lin X: Role of microRNA-26b in glioma development and
its mediated regulation on EphA2. PLoS One. 6:e162642011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shevde LA, Metge BJ, Mitra A, Xi Y, Ju J,
King JA and Samant RS: Spheroid-forming subpopulation of breast
cancer cells demonstrates vasculogenic mimicry via hsa-miR-299-5p
regulated de novo expression of osteopontin. J Cell Mol Med.
14((6B)): 1693–1706. 2010.PubMed/NCBI
|
|
33
|
Sun T, Zhao N, Zhao XL, Gu Q, Zhang SW,
Che N, Wang XH, Du J, Liu YX and Sun BC: Expression and functional
significance of Twist1 in hepatocellular carcinoma: Its role in
vasculogenic mimicry. Hepatology. 51:545–556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Plantamura I, Casalini P, Dugnani E, Sasso
M, D'Ippolito E, Tortoreto M, Cacciatore M, Guarnotta C, Ghirelli
C, Barajon I, et al: PDGFRβ and FGFR2 mediate endothelial cell
differentiation capability of triple negative breast carcinoma
cells. Mol Oncol. 8:968–981. 2014. View Article : Google Scholar : PubMed/NCBI
|