Introduction
The difficulty of early diagnosis, the diversity and
complexity of tumorigenesis and the progression of non-small cell
lung cancer (NSCLC) contribute to the poor prognosis of the disease
(1,2). A number of patients with NSCLC die as
a result of tumor metastasis, and until the last decade, the 5-year
overall survival rate of patients with metastatic NSCLC was <5%
(3,4). Moreover, as the mechanism underlying
the recurrence and metastasis of NSCLC is not completely
understood, exploring the molecular mechanisms underlying the
metastasis and progression of the disease may aid with the
development of therapeutic strategies for NSCLC.
During epithelial-mesenchymal transition (EMT),
epithelial cells transform into cells with a mesenchymal phenotype
via a specific process, which is accompanied by alterations in cell
structure, adhesion, morphology and migration (5). A number of studies have demonstrated
that EMT serves an important role in the invasion and metastasis of
various malignant tumors, such as colorectal (6), pancreatic (7), prostate (8), breast (9) and lung (10) cancer. According to Otsuki et
al (11), EMT signaling
promotes epithelial tumor malignancy, such as lung cancer,
indicating that inhibiting EMT signaling may suppress tumor
metastasis, recurrence or drug resistance. Therefore, developing
EMT-targeted therapy may serve as a promising therapeutic strategy
for lung cancer.
Long non-coding RNAs (lncRNAs) are
non-protein-coding transcripts that are >200 nucleotides in
length (12). Although the
mechanisms of action underlying lncRNAs are not completely
understood, increasing evidence has suggested that lncRNAs are
important regulator in a number of biological processes, such as
cell proliferation and apoptosis (13). Non-coding RNA activated by DNA
damage (NORAD) is an lncRNA that is downregulated in breast and
lung cancer, and low NORAD expression levels in the two types of
cancer are correlated with lymph node metastasis and poor patient
prognosis (14). Moreover, previous
studies have indicated that NORAD acts as a sponge for miRNAs
(miRs) to affect the occurrence and development of NSCLC (15,16).
However, whether NORAD affects EMT during the
development of NSCLC is not completely understood; therefore, the
present study aimed to investigate whether NORAD was involved in
NSCLC EMT and its underlying mechanism, with the aim of identifying
a novel therapeutic target for NSCLC.
Materials and methods
Tissue collection
NSCLC cancer tissues and healthy adjacent tissues
(≥5 cm from the tumor margin) were obtained from 50 patients (age,
32–70 years; mean age, 45±7 years; 32 male patients and 18 female
patients) who were diagnosed with NSCLC in Jingmen No. 1 People's
Hospital between February 2018 and April 2019. In all patients with
NSCLC, diagnosis was histopathologically confirmed. Written
informed consent was obtained from all patients. The present study
was approved by Jingmen No. 1 People's Hospital Ethics Committee
(approval no. JM2018010232).
Cell transfection and grouping
Human bronchial epithelium 16HBE cells were
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. NSCLC lines (A549, SK-MES-1, H1975 and
SK-LU-1) were purchased from American Type Culture Collection.
Cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) in humidified air with 5% CO2 at
37°C.
To explore the effects of NORAD on lung cancer
cells, cells were divided into the following groups: i) Control
(cells without treatment); ii) pc-control (cells transfected with
pc-control); iii) pc-NORAD (cells transfected with pc-NORAD); iv)
small interfering RNA (si)-control (cells transfected with
si-control); and v) si-NORAD (cells transfected with si-NORAD). To
explore the potential mechanisms underlying NORAD in lung cancer
cells, cells were divided into the following groups: i) Control
(cells without treatment); ii) mimic-control (cells transfected
with miR-422a mimic-control); iii) mimic (cells transfected with
miR-422a mimic); iv) pc-NORAD; v) mimic + pc-NORAD; vi)
inhibitor-control (cells transfected with miR-422a
inhibitor-control); vii) inhibitor (cells transfected with miR-422a
inhibitor); viii) si-NORAD; and ix) inhibitor + si-NORAD.
SK-MES-1 cells were transfected with pc-NORAD,
pc-control, miR-422a mimic (5′-ACUGGACUUAGGGUCAGAAGGC-3′; Shanghai
GenePharma Co., Ltd.) or mimic control
(5′-UUUGUACUACACAAAAGUACUG-3′; Shanghai GenePharma Co., Ltd.). A549
cells were transfected with si-NORAD (5′-AAGCCACCTTTGTGAACAGTA-3′),
si-control (5′-TTCTCCGAACGTGTCACGT-3′), miR-422a inhibitor
(5′-GCCUUCUGACCCUAAGUCCAGU-3′) or inhibitor control
(5′-CAGUACUUUUGUGUAGUACAAA-3′). Cell transfection (2×105
cells/well) was performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h
post-transfection, subsequent experiments were performed.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
Total RNA was reverse transcribed into cDNA using a High-Capacity
cDNA Reverse Transcription kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The following temperature protocol was used for
reverse transcription: 37°C for 40 min and 85°C for 5 min.
Subsequently, NORAD expression levels were determined by qPCR using
a 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and SYBR Green PCR Master Mix (Toyobo Life
Science). The sequences of the primers used to measure NORAD
expression levels were as follows: NORAD forward,
5′-CCTGGAAGGTGAGCGAAGT-3′ and reverse, 5′-AGAGGGTGGTGGGCATTT-3′;
and GAPDH forward, 5′-TGGTCACCAGGGCTGCTT-3′ and reverse,
5′-AGCTTCCCGTTCTCAGCC-3′. To detect miR-422a expression levels,
qPCR was performed using the SYBR Premix Ex Taq kit (Takara Bio,
Inc.). The sequences of the primers used to measure miR-422a
expression levels were as follows: miR-422a forward,
5′-ACUGGACUUAGGGUCAGAAGGC-3′ and reverse,
5′-GCCUUCUGACCCUAAGUCCAGU-3′; and U6 forward,
5′-CTTCGGCAGCACATATAC-3′ and reverse, 5′-GAACGCTTCACGAATTTGC-3′.
The following thermocycling conditions were used for qPCR:
Pre-degeneration at 95°C for 30 sec; 39 cycles at 95°C for 10 sec
and 60°C for 30 sec; and final extension at 72°C for 30 sec. mRNA
and miRNA expression levels were quantified using the
2−ΔΔCq method (17) and
normalized to the internal reference genes GAPDH and U6,
respectively.
Cell Counting Kit-8 (CCK-8) assay
Cells (1×103/well) were plated in 96-well
plates for 24, 48 or 72 h at 37°C. Subsequently, 10 µl CCK-8
reagent was added to each well and incubated for 1 h 37°C. Cell
viability was measured by detecting the absorbance at a wavelength
of 450 nm using a microtiter plate.
Wound healing assay
Cell migration was assessed by performing a wound
healing assay. During the wound healing assay, cells
(2×105 cells/well) were cultured in RPMI-1640 medium
supplemented with 1% FBS at 37°C with 5% CO2. At 90%
confluence, a single scratch was made in the cell monolayer using a
medium-sized pipette tip. The monolayer was washed with PBS to
remove cell debris. At 0 and 24 h, images of the cells were
captured using a light microscope to measure the scratch width
(magnification, ×100). Migration rate=(scratch width at 0 h-scratch
width at 48 h)/scratch width at 0 h.
Transwell assay
At 48 h post-transfection, cells (3×105)
were cultured in medium without serum. A Transwell assay (pore
size, 8 µm; Corning, Inc.) was performed to detect cell invasion.
Cells (2×105 cells/ml) were plated into the upper
chamber with medium containing 1% FBS, which was pre-coated with
Matrigel® at 37°C for 30 min. Medium supplemented with
10% FBS (500 µl) was plated in the lower chambers. Following
incubation for 24 h at 37°C, cells on the upper chamber surface
were removed. Invading cells were fixed with 50% methanol for 30
min at 4°C and stained with 0.1% crystal violet for 30 min at room
temperature. Stained cells were visualized under a light microscope
(magnification, ×100) and the number of invading cells was
calculated to determine the relative invasion rate.
Dual-luciferase reporter assay
starBase (starbase.sysu.edu.cn) was used to predict the target
gene of NORAD, and the binding sites were verified by performing
adual-luciferase reporter assay. pGL3-NORAD 3′-UTR wild-type (wt)
plasmid and pGL3-NORAD 3′-UTR mutated (mut) plasmid were obtained
from Guangzhou RiboBio Co., Ltd. In brief, the wt 3′-UTR of NORAD
containing the miR-422a binding sequences was obtained by PCR
amplification from human genomic DNA, and mutated (mut) NORAD
obtained using the Quick-Change Site-Directed Mutagenesis kit
(Stratagene). The wt and mut 3′-UTRs of NORAD were inserted into
the pGL3 luciferase reporter plasmids (Promega Corporation) to
obtain pGL3-NORAD 3′-UTR wt plasmid and pGL3-NORAD 3′-UTR mut
plasmid, respectively. 293T cells (2×105 cells; The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences)
were co-transfected with NORAD-wt plasmid (100 ng) or NORAD-mut
plasmid (100 ng), and 50 nM miR-422a mimic or 50 nM miR-422a
mimic-control using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. At 48 h post-transfection, luciferase activity was
measured using the dual-luciferase reporter assay system (Promega
Corporation). Firefly luciferase activities were normalized to
Renilla luciferase activities.
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (Santa Cruz Biotechnology, Inc.) and cell lysates were
centrifuged at 20,000 × g for 10 min at 4°C. Equal amounts of
protein (30 µg) were separated via 10% SDS-PAGE and transferred
onto PVDF membranes at 100 V for 1.5 h. The membranes were blocked
with 5% non-fat milk for 1 h at room temperature and incubated
overnight at 4°C with primary antibodies targeted against: Matrix
metallopeptidase (MMP)2 (1:1,000; 72 kD; cat. no. ab37150; Abcam),
MMP9 (1:1,000; 95 kD; cat. no. ab73734; Abcam), E-cadherin
(1:10,000; 97 kD; cat. no. ab40772; Abcam), N-cadherin (1:1,000;
130 kD; cat. no. ab18203; Abcam) and GAPDH (1:10,000; 36 kD; cat.
no. ab181602; Abcam). Following primary incubation, the membranes
were incubated with a secondary horseradish peroxidase-conjugated
antibody (goat anti-rabbit IgG; 1:10,000; cat. no. ab205718; Abcam)
for 1 h at room temperature. Protein bands were visualized using
enhanced chemiluminescence detection reagent (Thermo Fisher
Scientific, Inc.). Protein expression was quantified using
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.) with
GAPDH as the loading control.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 20.0; IBM Corp). Data are presented as the mean ±
standard deviation. Comparisons between two groups were analyzed
using the paired or unpaired Student's t-test. Comparisons among
multiple groups were analyzed using one-way ANOVA followed by
Tukey's post hoc test. The correlation between continuous variables
was analyzed using Pearson's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were performed at least three times.
Results
NORAD is highly expressed in NSCLC
tissues and cells
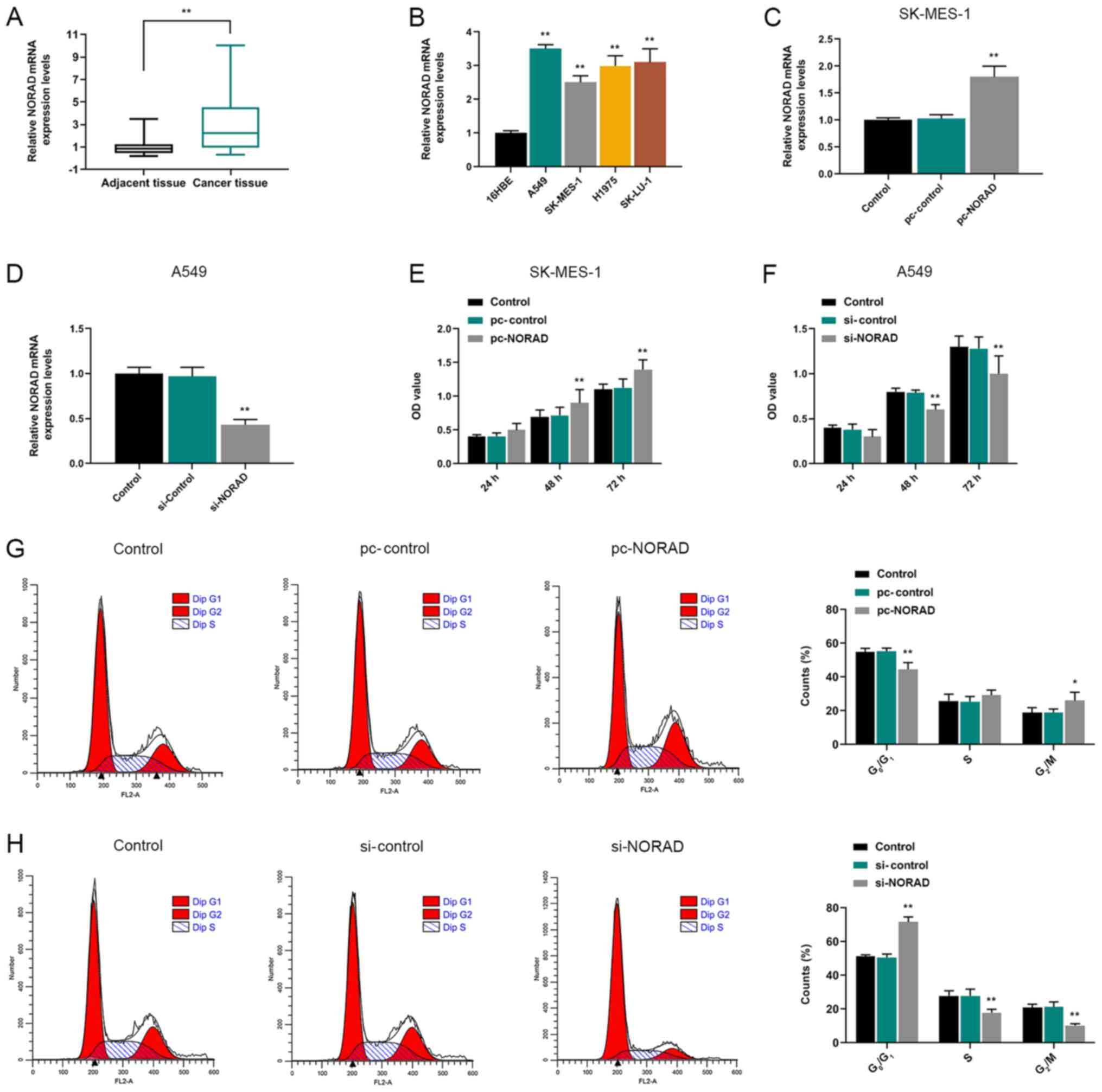
The expression levels of NORAD in NSCLC tissues and
cells were detected via RT-qPCR. The results indicated NORAD
expression was significantly higher in NSCLC tissues compared with
healthy adjacent tissues (Fig. 1A).
Moreover, NORAD expression levels in NSCLC cell lines (A549,
SK-MES-1, H1975 and SK-LU-1) were significantly higher compared
with the normal human bronchial epithelial 16HBE cell line
(Fig. 1B).
Effects of NORAD on NSCLC cell
biological behaviors
The RT-qPCR results indicated that pc-NORAD and
si-NORAD were successfully transfected into SK-MES-1 and A549
cells, respectively (Fig. 1C and
D). CCK-8, wound healing and Transwell assays were performed to
detect the effects of NORAD on NSCLC cell viability, migration and
invasion, respectively. At 48 and 72 h, pc-NORAD-transfected cells
displayed higher viability compared with the pc-control group,
whereas si-NORAD-transfected cells displayed significantly lower
viability compared with the si-control group (Fig. 1E and F). Moreover, the results
indicated that NORAD overexpression significantly reduced the
number of cells in G1 phase and increased the number of
cells in G2/M phase compared with the pc-control group.
By contrast, NORAD knockdown increased the number of cells in
G1 phase and reduced the number of cells in
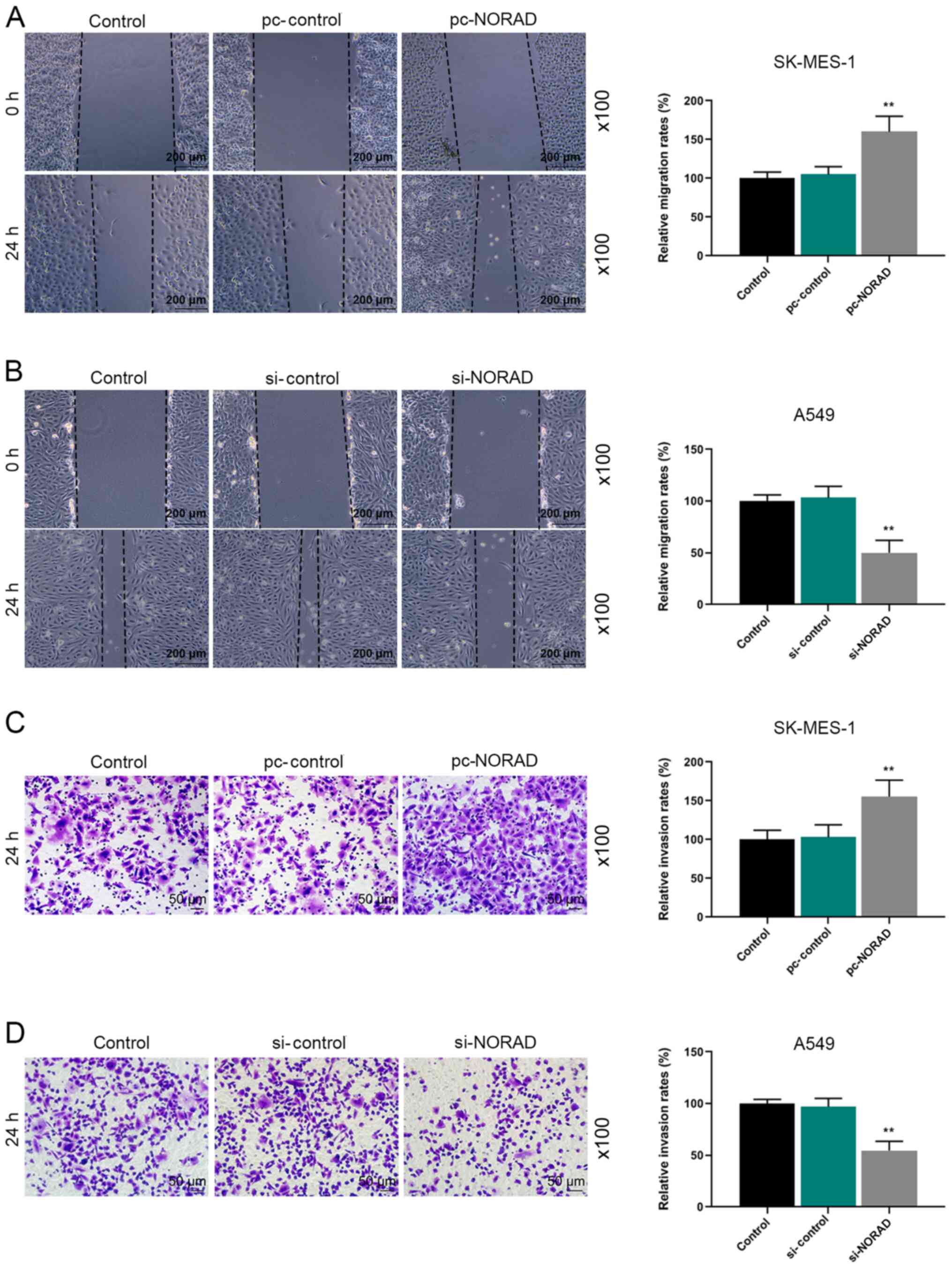
G2/M phase compared with the si-control group (Fig. 1G and H). In addition, the wound
healing assay demonstrated that pc-NORAD significantly increased
cell migration compared with the pc-control group, whereas si-NORAD
displayed the opposite results compared with the si-control group
(Fig. 2A and B). Similarly, the
results of the Transwell assay indicated that NORAD-overexpression
cells displayed a significantly higher invasion rate compared with
the pc-control group, whereas NORAD-knockdown cells displayed a
significantly decreased invasion rate compared with the si-control
group (Fig. 2C and D).
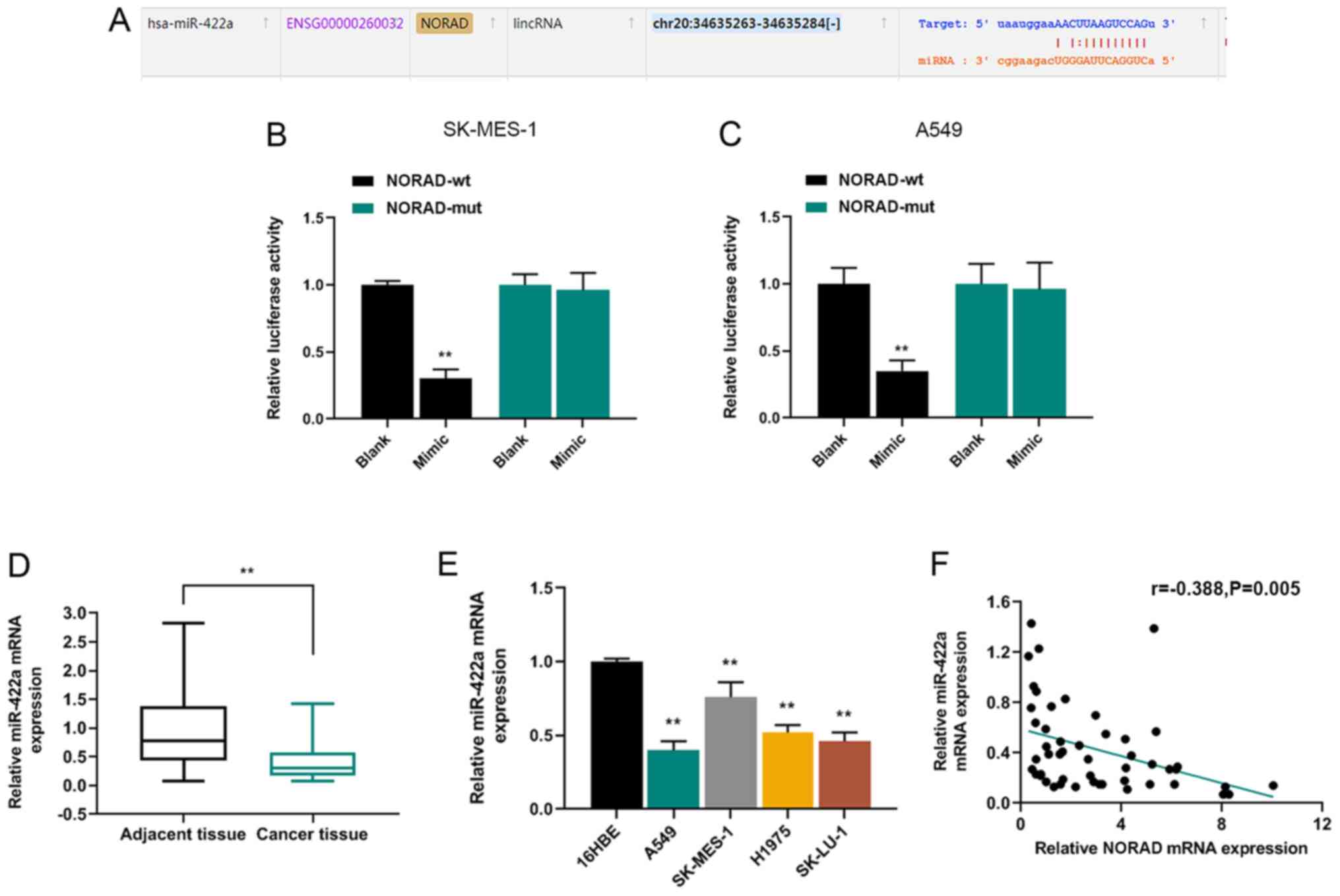
NORAD interacts with miR-422a and the
expression of miR-422a in NSCLC tissues
The potential mechanisms underlying NORAD in NSCLC
cells were explored, and the relationship between NORAD and miRNA
was predicted. starBase was used to identify the binding sites
between NORAD and miR-422a (Fig.
3A). A dual-luciferase reporter assay indicated that the
luciferase activity of NORAD-wt was significantly reduced by
miR-422a mimic compared with the blank group, which verified the
relationship between NORAD and miR-422a (Fig. 3B and C). Furthermore, the expression
of miR-422a in NSCLC tissues was detected via RT-qPCR. The results
demonstrated that miR-422a expression levels were significantly
lower in NSCLC tissues compared with healthy adjacent tissues
(Fig. 3D). In addition, miR-422a
expression levels in the NSCLC cell lines (A549, SK-MES-1, H1975
and SK-LU-1) were significantly reduced compared with 16HBE cells
(Fig. 3E). Pearson's correlation
analysis indicated that miR-422a expression was negatively
correlated with NORAD expression in NSCLC tissues (Fig. 3F).
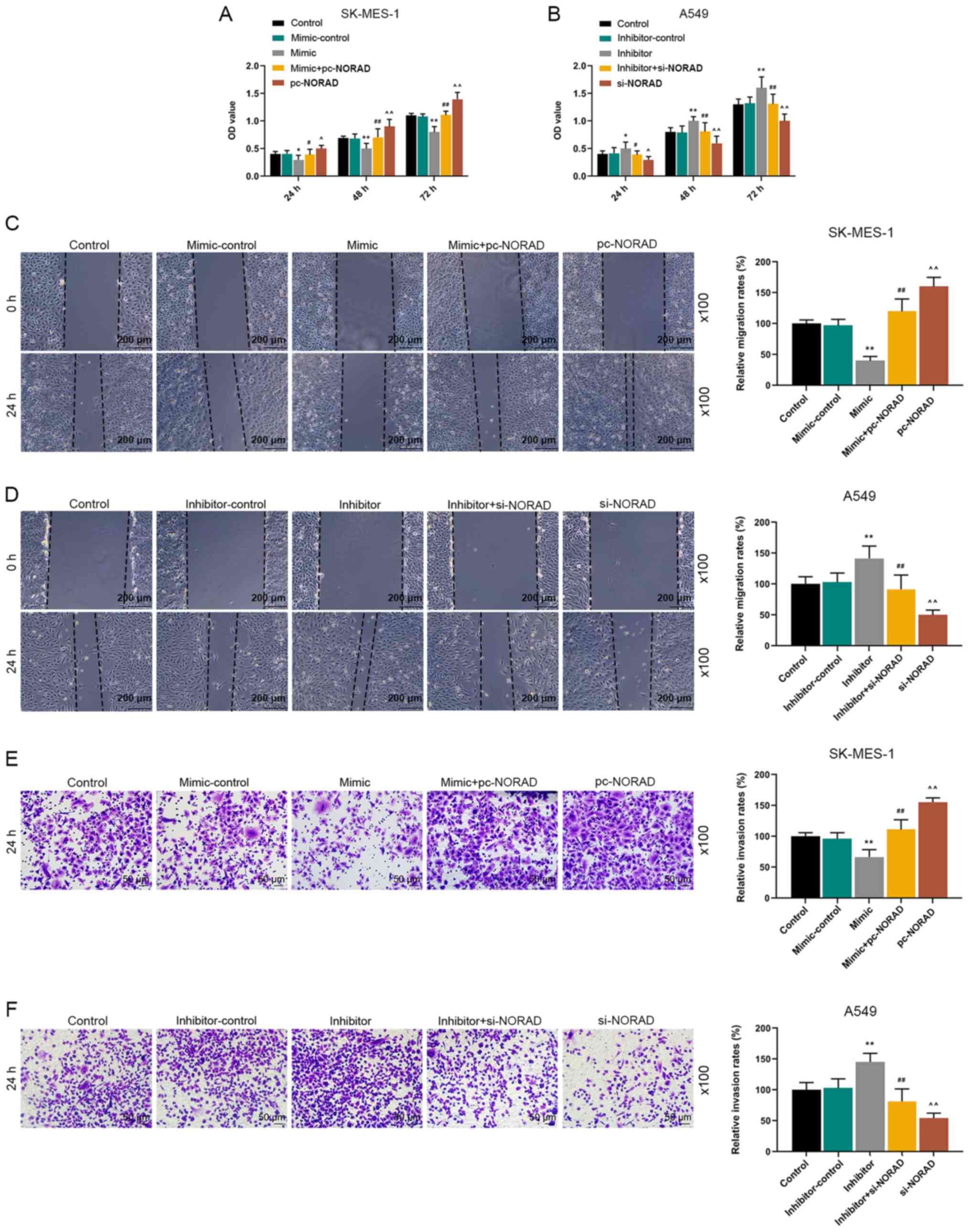
NORAD regulates the biological
behaviors of NSCLC cells via miR-422a
CCK-8, wound healing and Transwell assays were
performed to detect the effects of NORAD and miR-422a on cell
viability, migration and invasion. The CCK-8 assay results
indicated that miR-422a overexpression significantly reduced cell
viability compared with the mimic-control, but cells transfected
with miR-422a mimic and pc-NORAD displayed significantly increased
cell viability compared with the mimic group. However, miR-422a
knockdown significantly increased cell viability compared with the
inhibitor-control, which was reversed by co-transfection with
si-NORAD (Fig. 4A and B). Moreover,
the wound healing assay results suggested that miR-422a mimic
significantly decreased cell migration compared with the
mimic-control group, whereas miR-422a inhibitor displayed the
opposite effect compared with the inhibitor-control group.
Furthermore, cells co-transfected with miR-422a mimic and pc-NORAD
displayed significantly increased cell migration compared with the
mimic group, whereas cells co-transfected with miR-422a inhibitor
and si-NORAD displayed decreased cell migration compared with the
inhibitor group (Fig. 4C and D).
The results of the Transwell assay revealed that miR-422a
overexpression significantly inhibited cell invasion compared with
the mimic-control group, but miR-422a knockdown displayed the
opposite effects compared with the inhibitor-control group.
However, alterations to NORAD expression levels partly reversed the
effects of miR-422a on cell invasion (Fig. 4E and F).
NORAD regulates EMT by regulating
miR-422a expression
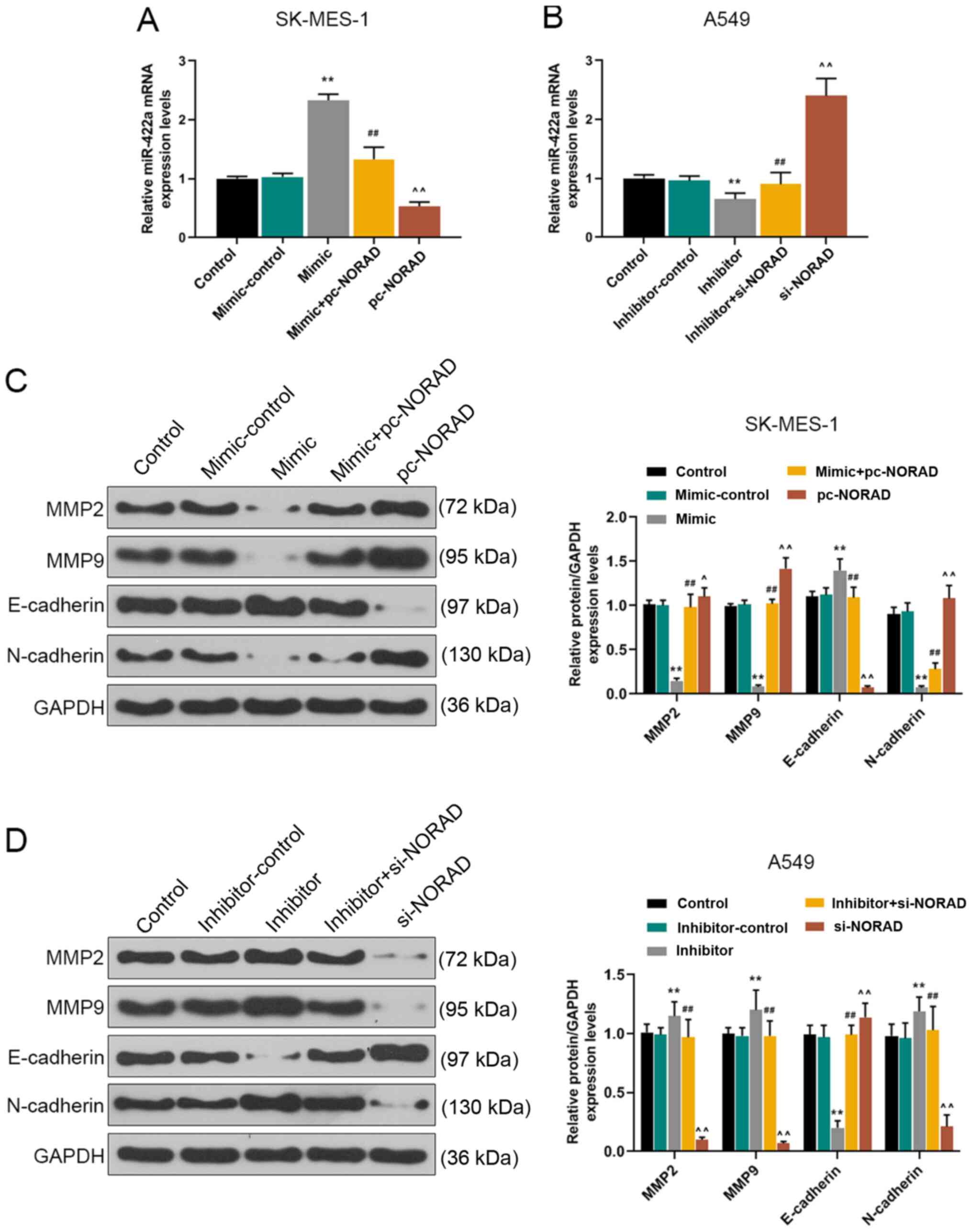
The RT-qPCR results indicated that the expression
levels of miR-422a in SK-MES-1 cells were significantly decreased
by NORAD overexpression compared with the mimic + pc-NORAD group,
whereas NORAD knockdown displayed the opposite effects compared
with the inhibitor + si-NORAD group (Fig. 5A and B). To explore the effect of
NORAD on NSCLC cell EMT, western blotting was performed to measure
the expression levels of EMT-related proteins, such as MMP2, MMP9,
E-cadherin and N-cadherin. The results demonstrated that miR-422a
overexpression significantly reduced the protein expression levels
of MMP2, MMP9 and N-cadherin, but significantly increased the
protein expression levels of E-cadherin compared with the
mimic-control group. NORAD overexpression reversed miR-422a
overexpression-mediated effects on protein expression (Fig. 5C). However, miR-422a knockdown
significantly increased the protein expression levels of MMP2, MMP9
and N-cadherin, and significantly reduced the expression levels of
E-cadherin compared with the inhibitor-control group. NORAD
knockdown reversed miR-422a knockdown-mediated effects on protein
expression (Fig. 5D).
Discussion
NSCLC is one of the most frequently diagnosed types
of cancer, with a poor prognosis and high mortality rate (18). Moreover, developing effective
strategies to improve the prognosis of NSCLC remains a challenge.
NORAD is an important lncRNA that serves a key role in cancer cell
migration and invasion (19,20).
lncRNAs have critical functions during cancer progression (21,22).
For example, lncRNA small nucleolar RNA host gene 1 (SNHG1)
functions as an oncogene in colorectal cancer, promoting colorectal
cancer cell proliferation via epigenetic silencing of Kruppel like
factor 2 and cyclin dependent kinase inhibitor 2B in the nucleus
(23). Knockdown of HOXD antisense
growth-associated long non-coding RNA inhibited cell proliferation
and migration, and promoted cell apoptosis in bladder cancer cells
(24). The present study
investigated the role of NORAD in NSCLC cell viability, migration,
invasion, as well as the underlying mechanisms. The results
revealed that NORAD expression was significantly higher in NSCLC
tissues and cells compared with adjacent healthy tissues and cells.
In addition, compared with the control groups, NORAD overexpression
promoted NSCLC cell viability, migration and invasion, whereas
NORAD knockdown displayed the opposite effects. Furthermore, the
results indicated that NORAD regulated NSCLC cell EMT by serving as
a sponge for miR-422a.
Previous studies have indicated that NORAD serves a
vital role in promoting the development of a number of different
types of cancer. For instance, Zhou et al (25) reported that NORAD expression is
increased in breast cancer tissues, which promotes cell
proliferation, migration and invasion, and is related to a poor
prognosis. According to Tong et al (26), NORAD knockdown can inhibit cell
proliferation, reduce bufalin's chemical resistance, and inhibit
cell cycle transition and xenograft growth, suggesting that it
displays an anticancerous effect on epithelial ovarian cancer.
Another study demonstrated that low expression of NORAD in
osteosarcoma significantly inhibits cell proliferation and invasion
in vivo (27). Moreover, it
has been reported that NORAD promotes the development of other
types of cancer, such as gastric (28), prostate (29) and ovarian cancer (26,30),
as well as osteosarcoma (27).
Similarly, Chen et al (15)
indicated that NORAD increases the expression of AKT1 by adsorbing
miR-656-3p to promote NSCLC cell proliferation and migration. The
results of the aforementioned studies were consistent with the
results of the present study. The present study indicated that
NORAD was associated with the occurrence and development of NSCLC,
and that the expression of NORAD was increased in NSCLC tissues and
cell lines compared with adjacent healthy tissues and cells. In
addition, NORAD expression was associated with A549 and SK-MES-1
cell viability, migration and invasion. Based on the results, it
was hypothesized that NORAD may serve as a novel therapeutic target
for NSCLC.
lncRNAs can regulate cell biological behaviors via
miRNAs (31). In the present study,
the relationship between NORAD and miR-422a was investigated using
a dual-luciferase reporter assay. In addition, a negative
correlation between NORAD expression and miR-422a expression was
identified in NSCLC. Moreover, compared with the control group,
miR-422a overexpression reduced NSCLC cell viability, migration and
invasion, which was reversed by NORAD overexpression. By contrast,
compared with the inhibitor-control group, miR-422a knockdown
increased NSCLC cell viability, migration and invasion, which was
reversed by NORAD knockdown. Therefore, the results suggested that
NORAD served as an oncogene during NSCLC progression by regulating
miR-422a. A previous study reported that miR-422a regulates
metabolism and malignant tumors of gastric cancer cells by
targeting pyruvate dehydrogenase kinase 2 (32). In addition, it was also reported
that LINC00313, an oncogene in papillary thyroid cancer, regulates
cell proliferation, invasion, migration and EMT by regulating
miR-422a expression (33). Another
previous study demonstrated that miR-422a also functions as a tumor
suppressor in lung cancer cells by regulating sulfatase 2-mediated
transforming growth factor-β/SMAD signaling pathway (34). LINC00858 facilitates NSCLC cell
proliferation by sponging miR-422a and activating kallikrein
related peptidase 4 (35). However,
the specific signaling pathways involved in the procancer effects
of NORAD in NSCLC require further investigation.
EMT is associated with cancer metastasis and drug
resistance to treatment (36).
Various mechanisms, especially EMT, which is related to
pathogenesis of chronic obstructive pulmonary disease, are involved
in the progression and metastasis of NSCLC (37). EMT can increase cell migration and
invasion to induce cancer metastasis, and MMPs such as MMP2 and
MMP9 can promote the carcinogenesis of EMT by degrading barriers
such as the extracellular matrix (38). E-cadherin and N-cadherin are two
representative EMT markers (39).
Noh et al (40) indicated
that N-cadherin expression may contribute to the biological
aggression of glioma. Liao et al (41) indicated that lncRNA H19 imprinted
maternally expressed transcript overexpression mediates lung cancer
cell proliferation and invasion by blocking the expression of
N-cadherin and inducing the expression of E-cadherin. A recent
study demonstrated that NORAD knockdown inhibits gastric cancer
cell migration and invasion by regulating EMT-related genes
(42). Wu et al (43) reported that miR-422a is associated
with lymphatic metastasis in lung cancer. It has also been reported
that miR-422a overexpression inhibited NSCLC cell EMT progression
(34), which is consistent with the
results of the present study. The present study indicated that
miR-422a overexpression reduced the expression levels of MMP2, MMP9
and N-cadherin, and increased the expression levels of E-cadherin
compared with the mimic-control group, but NORAD overexpression
reversed miR-422a overexpression-mediated effects. By contrast,
miR-422a knockdown increased the expression levels of MMP2, MMP9
and N-cadherin and reduced the expression levels of E-cadherin
compared with the inhibitor-control group, whereas NORAD knockdown
reversed miR-422a knockdown-mediated effects. Therefore, it was
speculated that NORAD may serve as a novel therapeutic target for
NSCLC. However, the present study had a number of limitations. For
example, in vivo experiments need to be conducted to confirm
the results of the present study. In addition, the target genes and
signaling pathways associated with miR-422a-mediated regulation of
NSCLC development, and other potential mechanisms underlying the
role of NORAD in NSCLC cells require further investigation.
In conclusion, the present study indicated that
NORAD displayed a procancer role in NSCLC by sponging miR-422a.
NORAD promoted NSCLC cell viability, migration and invasion via
downregulating miR-422a. Therefore, NORAD may serve as a novel
therapeutic target for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC and QC contributed to the conception and design
of the study. ZC, QC and CX acquired, analyzed and interpreted the
data. ZC and QC drafted and revised the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients. The present study was approved by Jingmen No. 1 People's
Hospital Ethics Committee (approval no. JM2018010232).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Xu-Welliver M and Carbone DP: Blood-based
biomarkers in lung cancer: Prognosis and treatment decisions.
Transl Lung Cancer Res. 6:708–712. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cha G, Xu J, Xu X, Li B, Lu S, Nanding A,
Hu S and Liu S: High expression of CIP2A protein is associated with
tumor aggressiveness in stage I–III NSCLC and correlates with poor
prognosis. Onco Targets Ther. 10:5907–5914. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arbour KC and Riely GJ: Systemic therapy
for locally advanced and metastatic non-small cell lung cancer: A
review. JAMA. 322:764–774. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Z, Wang H, Xia L, Oyang L, Zhou Y,
Zhang B, Chen X, Luo X, Liao Q and Liang J: Overexpression of PAK1
correlates with aberrant expression of EMT markers and poor
prognosis in non-small cell lung cancer. J Cancer. 8:1484–1491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rokavec M, Kaller M, Horst D and Hermeking
H: Pan-cancer EMT-signature identifies RBM47 down-regulation during
colorectal cancer progression. Sci Rep. 7:46872017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krebs AM, Mitschke J, Lasierra Losada M,
Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D,
Reichardt W, Bronsert P, et al: The EMT-activator Zeb1 is a key
factor for cell plasticity and promotes metastasis in pancreatic
cancer. Nat Cell Biol. 19:518–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tong D, Liu Q, Liu G, Xu J, Lan W, Jiang
Y, Xiao H, Zhang D and Jiang J: Metformin inhibits
castration-induced EMT in prostate cancer by repressing
COX2/PGE2/STAT3 axis. Cancer Lett. 389:23–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye X, Brabletz T, Kang Y, Longmore GD,
Nieto MA, Stanger BZ, Yang J and Weinberg RA: Upholding a role for
EMT in breast cancer metastasis. Nature. 547:E1–E3. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam
D, Lee J, Lee SG, Yang WM, Um JY, et al: Ginkgolic acid inhibits
invasion and migration and TGF-β-induced EMT of lung cancer cells
through PI3K/Akt/mTOR inactivation. J Cell Physiol. 232:346–354.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Otsuki Y, Saya H and Arima Y: Prospects
for new lung cancer treatments that target EMT signaling. Dev Dyn.
247:462–472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ricciuti B, Mencaroni C, Paglialunga L,
Paciullo F, Crinò L, Chiari R and Metro G: Long noncoding RNAs: New
insights into non-small cell lung cancer biology, diagnosis and
therapy. Med Oncol. 33:182016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong WJ, Peng JB, Yin JY, Li XP, Zheng W,
Xiao L, Tan LM, Xiao D, Chen YX, Li X, et al: Association between
well-characterized lung cancer lncRNA polymorphisms and
platinum-based chemotherapy toxicity in Chinese patients with lung
cancer. Acta Pharmacol Sin. 38:581–590. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan BS, Yang MC, Singh S, Chou YC, Chen
HY, Wang MY, Wang YC and Chen RH: LncRNA NORAD is repressed by the
YAP pathway and suppresses lung and breast cancer metastasis by
sequestering S100P. Oncogene. 38:5612–5626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen T, Qin S, Gu Y, Pan H and Bian D:
Long non-coding RNA NORAD promotes the occurrence and development
of non-small cell lung cancer by adsorbing MiR-656-3p. Mol Genet
Genomic Med. 7:e7572019. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao W, Weng T, Wang L, Shi B, Meng W, Wang
X, Wu Y, Jin L and Fei L: Long noncoding RNA NORAD promotes cell
proliferation and glycolysis in nonsmall cell lung cancer by acting
as a sponge for miR1365p. Mol Med Rep. 19:5397–5405.
2019.PubMed/NCBI
|
|
17
|
Zou Y, Chen Y, Yao S, Deng G, Liu D, Yuan
X, Liu S, Rao J, Xiong H, Yuan X, et al: MiR-422a weakened breast
cancer stem cells properties by targeting PLP2. Cancer Biol Ther.
19:436–444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin X and Xie C: MA02. 07 Evaluation of
exosomal miRNAs from plasma as potential biomarker for NSCLC. J
Thoracic Oncol. 12 (Suppl):S350–S351. 2017. View Article : Google Scholar
|
|
19
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C and Shen B: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang X, Cai JB, Peng R, Wei CY, Lu JC, Gao
C, Shen ZZ, Zhang PF, Huang XY, Ke AW, et al: The long noncoding
RNA NORAD enhances the TGF-β pathway to promote hepatocellular
carcinoma progression by targeting miR-202-5p. J Cell Physiol.
234:12051–12060. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zuo ZK, Gong Y, Chen XH, Ye F, Yin ZM,
Gong QN and Huang JS: TGFβ1-induced LncRNA UCA1 upregulation
promotes gastric cancer invasion and migration. DNA Cell Biol.
36:159–167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng SQ, Zhang XY, Fan HT, Sun QJ and
Zhang M: Up-regulation of LncRNA MEG3 inhibits cell migration and
invasion and enhances cisplatin chemosensitivity in bladder cancer
cells. Neoplasma. 65:925–932. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B,
Xu X, Xu T, Hu X, Sun L, et al: The long noncoding RNA SNHG1
regulates colorectal cancer cell growth through interactions with
EZH2 and miR-154-5p. Mol Cancer. 17:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Wang Y, Zhang X, Huang Q, Diao Y,
Yin H and Liu H: Long non-coding RNA HOXD-AS1 in cancer. Clin Chim
Acta. 487:197–201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou K, Ou Q, Wang G, Zhang W, Hao Y and
Li W: High long non-coding RNA NORAD expression predicts poor
prognosis and promotes breast cancer progression by regulating
TGF-β pathway. Cancer Cell Int. 19:632019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tong L, Ao Y, Zhang H, Wang K, Wang Y and
Ma Q: Long noncoding RNA NORAD is upregulated in epithelial ovarian
cancer and its downregulation suppressed cancer cell functions by
competing with miR-155-5p. Cancer Med. 8:4782–4791. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Zou J, Chen H, Zhang P, Lu Z, You
Z and Sun J: Long noncoding RNA NORAD regulates cancer cell
proliferation and migration in human osteosarcoma by endogenously
competing with miR-199a-3p. IUBMB Life. 71:1482–1491. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tao W, Li Y, Zhu M, Li C and Li P: LncRNA
NORAD promotes proliferation and inhibits apoptosis of gastric
cancer by regulating miR-214/Akt/mTOR axis. Onco Targets Ther.
12:8841–8851. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang H and Guo H: Long non-coding RNA
NORAD induces cell proliferation and migration in prostate cancer.
J Int Med Res. 47:3898–3904. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang X, Yan Y, Chen Y, Li J and Yang J:
Involvement of NORAD/miR-608/STAT3 axis in carcinostasis effects of
physcion 8-O-β-glucopyranoside on ovarian cancer cells. Artif Cells
Nanomed Biotechnol. 47:2855–2865. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu J, Li H, Zheng B, Sun L, Yuan Y and
Xing C: Competitive endogenous RNA (ceRNA) regulation network of
lncRNA-miRNA-mRNA in colorectal carcinogenesis. Dig Dis Sci.
64:1868–1877. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He Z, Li Z, Zhang X, Yin K, Wang W, Xu Z,
Li B, Zhang L, Xu J, Sun G, et al: MiR-422a regulates cellular
metabolism and malignancy by targeting pyruvate dehydrogenase
kinase 2 in gastric cancer. Cell Death Dis. 9:5052018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan DG, Liu N, Chao M, Tu YY and Liu WS:
SP1-induced upregulation of long noncoding RNA LINC00313
contributes to papillary thyroid cancer progression via the
miR-422a. Eur Rev Med Pharmacol Sci. 23:1134–1144. 2019.PubMed/NCBI
|
|
34
|
Li WQ, Zhang JP, Wang YY, Li XZ and Sun L:
MicroRNA-422a functions as a tumor suppressor in non-small cell
lung cancer through SULF2-mediated TGF-β/SMAD signaling pathway.
Cell Cycle. 18:1727–1744. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu SP, Wang JY, Wang XG and Zhao JP: Long
intergenic non-protein coding RNA 00858 functions as a competing
endogenous RNA for miR-422a to facilitate the cell growth in
non-small cell lung cancer. Aging. 9:475–486. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pastushenko I, Brisebarre A, Sifrim A,
Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D,
Moers V, Lemaire S, et al: Identification of the tumour transition
states occurring during EMT. Nature. 556:463–468. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mahmood MQ, Ward C, Muller HK, Sohal SS
and Walters EH: Epithelial mesenchymal transition (EMT) and
non-small cell lung cancer (NSCLC): A mutual association with
airway disease. Med Oncol. 34:452017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang F, Yu N, Wang H, Zhang C, Zhang Z, Li
Y, Li D, Yan L, Liu H and Xu Z: Downregulated expression of
hepatoma-derived growth factor inhibits migration and invasion of
prostate cancer cells by suppressing epithelial-mesenchymal
transition and MMP2, MMP9. PLoS One. 13:e01907252018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Paolillo M and Schinelli S: Extracellular
matrix alterations in metastatic processes. Int J Mol Sci.
20:49472019. View Article : Google Scholar
|
|
40
|
Noh MG, Oh SJ, Ahn EJ, Kim YJ, Jung TY,
Jung S, Kim KK, Lee JH, Lee KH and Moon KS: Prognostic significance
of E-cadherin and N-cadherin expression in Gliomas. BMC Cancer.
17:5832017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liao S, Yu C, Liu H, Zhang C, Li Y and
Zhong X: Long non-coding RNA H19 promotes the proliferation and
invasion of lung cancer cells and regulates the expression of
E-cadherin, N-cadherin, and vimentin. Onco Targets Ther.
12:4099–4107. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu SY, Peng H, Zhu Q, Wu YX, Wu F, Han CR,
Yan B, Li Q and Xiang HG: Silencing the long noncoding RNA NORAD
inhibits gastric cancer cell proliferation and invasion by the
RhoA/ROCK1 pathway. Eur Rev Med Pharmacol Sci. 23:3760–3770.
2019.PubMed/NCBI
|
|
43
|
Wu L, Hu B, Zhao B, Liu Y, Yang Y, Zhang L
and Chen J: Circulating microRNA-422a is associated with lymphatic
metastasis in lung cancer. Oncotarget. 8:42173–42188. 2017.
View Article : Google Scholar : PubMed/NCBI
|