Introduction
Cervical cancer (CC) is known as one of the most
common types of gynecological cancers worldwide and it is also the
fourth leading cause of female mortality (1). Advances in early diagnosis, surgical
resection and chemotherapy/radiation enable patients to receive
effective treatments, however, CC prognosis remains poor (2). Causes of death in patients with CC are
mainly cancer progression, metastasis and resistance (3). Therefore, investigations into the
mechanism and progression of tumorigenesis may provide novel
insights for the development of new treatment methods of CC.
MicroRNAs (miRNAs or miRs) are 22nt small non-coding
RNA molecules, which are closely implicated in gene expression
(4). miRNAs either degrade certain
specific genes or inhibit their expression by binding to
3′-untranslated regions (3′-UTRs) of their mRNAs in a complimentary
manner (5,6). miRNAs are widely involved in various
biological processes including proliferation, tumor metastasis and
drug tolerance (7,8). miR-34c-5p has been reported to
function as a tumor suppressor in numerous cancer types (9). However, whether the same miRNA serves
as an oncogene or not is ultimately dependent on the
characteristics of the target genes (10–12).
The present study aimed at investigating the effects and mechanism
of miR-34c-5p in CC.
The Notch signaling pathway is an evolutionarily
conserved signaling pathway that mediates proliferation and
differentiation as well as the survival and apoptosis of cells
(13). The Notch signaling pathway
encompasses Notch transmembrane receptors (Notch1-4) and their
ligands (Delta-like 1, 3 and 4 and Jagged 1 and 2) (14). The Notch receptor features a single
channel transmembrane protein and consists of an extracellular
domain, a transmembrane domain and an intracellular domain. Once
the Notch signaling pathway is activated by the ligand-receptor of
joint cells, the Notch1 receptor is cleaved by γ-secretase, thereby
releasing Notch1 intracellular domain (NICD) from the plasma
membrane (15). Subsequently, NICD
translocates into the nucleus and then participates in the
transcription of other transcription factors to further regulate
its downstream genes including members of Hes and Hey families
(16). The Notch signaling pathway
regulates the growth of numerous tissues and cells in a cellular
context-dependent way, which further affects cell specialization,
proliferation and apoptosis (16).
A dysregulated Notch signaling pathway has been revealed in various
types of cancer including CC (17).
However, the interaction between miR-34c-5p and Notch1 remains to
be elucidated. The present study was designed to explore the
biological functions and mechanism of miR-34c-5p on CC at a
molecular level, hoping to provide a novel approach to the current
treatment of this disease.
Materials and methods
Tissue collection
CC and adjacent tissues were collected from 30
patients aged between 29 and 72 who underwent surgical resections
at the Emergency General Hospital (Beijing, China) between December
2017 and December 2019. One of the patients who received anticancer
treatment was excluded from the study. All tissues were frozen by
liquid nitrogen immediately and stored at −80°C for later use. The
experimental protocol was authorized by the Ethics Committee of
Emergency General Hospital (approval no. 2017SY1503). Prior written
informed consent was obtained from each patient.
Hematoxylin and eosin (H&E)
staining
Tissue samples were fixed in 4% formalin for 48 h at
room temperature. The tissues were then paraffin-embedded, then cut
to 5-µm-thick sections. Slides were subjected to H&E stain
according the manufacturer's instructions (Beyotime Institute of
Biotechnology).
EdU analysis
An EdU detection kit (cat. no. C10310; Guangzhou
RiboBio Co., Ltd.) was used to detect cell proliferation. Briefly,
cells were plated in 24-well plates at a density of
5×104 cells/well and treated with 50 µM EdU solution for
4 h at 37°C. The cells were then fixed with 4% paraformaldehyde for
10 min at room temperature and treated with 0.5% Triton X-100 at
room temperature for 5 min. The nuclei were labeled with DAPI
(Guangzhou RiboBio Co., Ltd.).
Cell culture and transfection
Human CC cell lines (C33A, CaSki, HeLa and SiHa) and
human immortalized normal cervical cell line (Ect1/E6E7) were
acquired from the American Type Culture Collection (ATCC). 293T
cells were purchased from the Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences. All cell lines were cultured in
DMEM (HyClone; Cytiva) supplemented with 10% FBS (HyClone; Cytiva),
100 U/ml penicillin and 100 µg/ml streptomycin (Beyotime Institute
of Biotechnology) in a humidified incubator containing 5%
CO2 at 37°C. The miR-34c-5p mimics
(5′-AGGCAGUGUAGUUAGCUGAUUGC-3′), miR-34c-5p mimics negative control
(5′-ACUACUGAGUGACAGUAGA-3′), miR-34c-5p inhibitors
(5′-GCAAUCAGCUAACUACACUGCCU-3′) and miR-34c-5p inhibitors negative
control (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by Guangzhou
RiboBio, Co., Ltd. Full-length Notch1 from human cDNA library was
inserted a pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific,
Inc.). A pcDNA3.1 vector alone (empty plasmid) served as a negative
control. Cells were transfected using Lipofectamine® LTX
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. HeLa cells were transfected either
with miR-NC (50 nM) or miR-34c-5p mimics (50 nM) and/or
pcDNA3.1/Notch1 vector (100 nM) or pcDNA3.1 (100 nM). In addition,
CaSki cells were transfected with anti-NC (50 nM) or miR-34c-5p
inhibitors (50 nM). Following transfection for 48 h, cells were
harvested for subsequent experiments and repeated 4 times.
Cell Counting Kit-8 (CCK-8) assay
HeLa cell viability was detected by CCK-8 assay
(Beyotime Institute of Biotechnology). HeLa cells (1×103
cells/well) were cultured in 96-well plates for 0, 24, 48 and 72 h,
four times in each time group. At a fixed time, 10 µl of CCK-8 was
added into each well and incubated for 3 h. The optical density and
450 nm of each well was determined in quadruplicate using Multiskan
MK3 (Thermo Fisher Scientific, Inc.).
Transwell assay
Migration and invasion abilities of cells were
determined through Corning Transwell chambers (Corning, Inc.). For
the detection of invasion capability, an 8-µm pore size Transwell
membrane filter was precoated with 30 µl Matrigel™ (BD Biosciences)
at 37°C for 4 h. In the migration and invasion detection, HeLa
cells (5×104 cells) were resuspended in 100 µl DMEM
without the addition of FBS before being transferred to the upper
chamber. A total of 600 µl of DMEM supplemented with 10% FBS was
added to the lower chamber. Cells were incubated for 12 h before
being fixed with 4% paraformaldehyde for 10 min at room temperature
and stained with 0.5% crystal violet at room temperature for 20
min. The number of stained cells randomly selected from six fields
were counted with images captured under a light microscope (Olympus
Corporation; magnification, ×100) and repeated four times.
Western blotting
Tissue samples and treated cells were lysed by
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology). Protein concentrations were determined by BCA
Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). A
quantity of protein extract (30 µg total protein/lane) was resolved
via 10% SDS-PAGE and transferred onto PVDF membranes and blocked
with 5% dried skimmed milk at room temperature for 1 h. The PVDF
membranes were incubated with primary antibody (Notch1; dilution
1:500; cat. no. ab52627; Abcam) and β-actin antibody (dilution
1:1,000; cat. no. ab8227; Abcam) with gentle agitation at 4°C
overnight and then treated with secondary antibody (horseradish
peroxidase-labeled goat anti-rabbit; dilution 1:1,000; cat. no.
ab150077; Abcam) at room temperature for 2 h. β-actin served as a
loading control. Protein bands were visualized via an enhanced
chemiluminescence system (Beyotime Institute of Biotechnology) and
repeated four times. Western blots were quantified by ImageJ
software (V 1.46; National Institutes of Health).
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
A total of 2 µl RNA (at a concentration of 200
ng/µl) was extracted from 2×106 cells and tissues with
TRIzol® reagent (Thermo Fisher Scientific, Inc.). cDNA
was synthesized by TaqMan® MicroRNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. To quantify miRNA and mRNA, a qPCR assay
was performed with iQ™ SYBR® Green Supermix (Bio-Rad
Laboratories, Inc.) on the platform of an iCycler iQ™ qPCR
detection system (Bio-Rad Laboratories, Inc.). Relative levels of
miR-34c-5p and Notch1 were calculated as an inverse log of 2-ΔΔCq
and normalized to the reference gene (18), repeated four times. Conditions of
the thermocycling were as follows: 95°C for 10 min; followed by 40
cycles at 95°C for 15 sec and 60°C for 1 min; annealed at 55°C for
30 sec; and elongated at 72°C for 3 min. β-actin was considered as
an internal reference and employed to analyze the expression of
Notch 1 gene. U6 was regarded as an internal control for the
detection of miR-34c-5p expression. Primers were as follows:
Notch1-forward (F), 5′-GAGGCGTGGCAGACTATGC-3′ and Notch1-reverse
(R), 5′-CTTGTACTCCGTCAGCGTGA-3′; miR-34c-5p RT primer,
5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGCAATC;
miR-34c-5p-F, 5′-CGGAGGCAGTGTAGTTAGCT-3′ and miR-34c-5p-R,
5′-GTGCAGGGTCCGAGGT-3′; U6 RT primer, 5′-AACGCTTCACGAATTTGCGT-3′;
U6-F, 5′-CTCGCTTCGGCAGCACA-3′ and U6-R, 5′-AACGCTTCACGAATTTGCGT-3′;
β-actin-F, 5′-CATGTACGTTGCTATCCAGGC-3′ and β-actin-R,
5′-CTCCTTAATGTCACGCACGAT-3′.
Flow cytometry
According to the manufacturer's protocol, estimation
of the apoptosis rate was performed with Annexin V-PI detection kit
(Beyotime Biotechnology Institute). Cell cycles, proliferation and
apoptosis rate of each sample were analyzed by flow cytometry
(Cytomics Fc500 MPL Flow Cytometer; Beckman Coulter, Inc.) and
repeated four times. All data were analyzed with ModFit LT 3.0
(Verity Software House, Inc.).
Luciferase reporter assay
Wild-type (WT) or mutant (MUT) Notch1-3′UTR with the
miR-34c-5p binding site was loaded into psicheck2 vector (Promega
Corporation). The 293T cells (1×105 cells/well) were
co-transfected with 0.1 mg psiCHECK2-WT Notch1-3′-UTR or 0.1 mg
psiCHECK2-MUT Notch1-3′-UTR and 10 nM miR-34c-5p mimics or 10 nM
miR-34c-5p inhibitors using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Cells were cultured at 37°C for 48 h and
then analyzed for luciferase activity according to the
manufacturer's protocol (GeneCopoeia, Inc.) and the experiment was
repeated four times. Luciferase activity was standardized by
comparison with Renilla luciferase activity.
Bioinformatics prediction
Potential target genes of miR-34c-5p were identified
by using the online prediction system TargetScan 7.1 (http://www.targetscan.org).
Statistical analysis
Data were presented as the mean ± standard error of
the mean. SPSS 13.0 software (SPSS, Inc.) was used for statistical
processing. Unpaired Student's t-test or one-way ANOVA was used for
data analysis. Assays of significant difference were subjected to
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
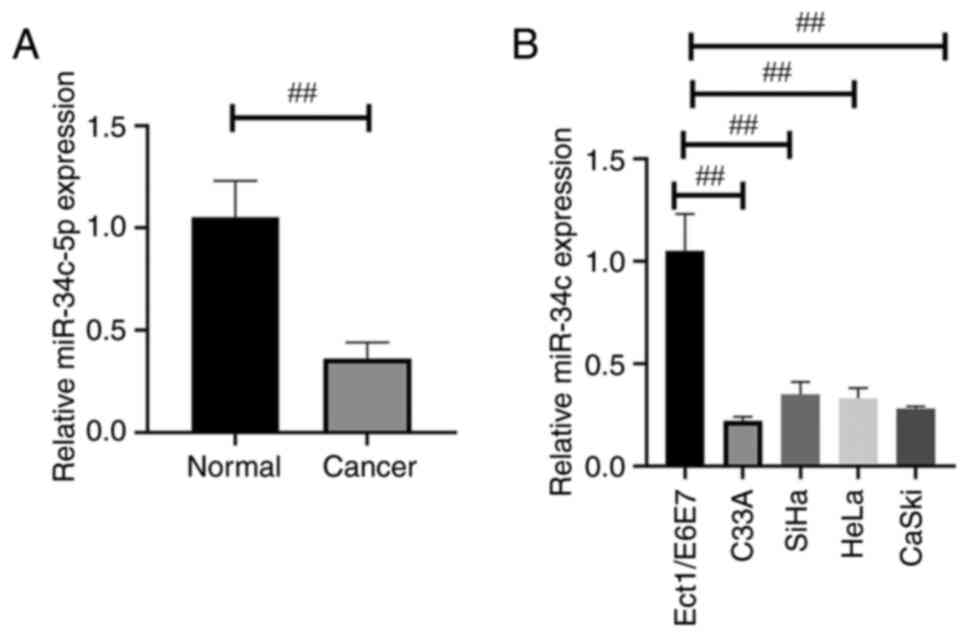
miR-34c-5p expression is downregulated
in human CC tissues and related cells
The morphology of CC and adjacent tissues (control
group) was observed using H&E staining (Fig. S1). To verify the role of miR-34c-5p
in CC, miR-34c-5p expression was initially determined in 30 pairs
of CC tissues and adjacent tissues by RT-qPCR, which indicated that
the miR-34c-5p expression was significantly reduced in tumor
tissues compared with normal tissues (Fig. 1A). The level of miR-34c-5p
expression was detected in four types of CC cells (C33A, SiHa, HeLa
and CaSki) by RT-qPCR and the results indicated that the expression
of miR-34c-5p was significantly decreased in CC cell lines when
compared with that in Ect1/E6E7 cells (Fig. 1B).
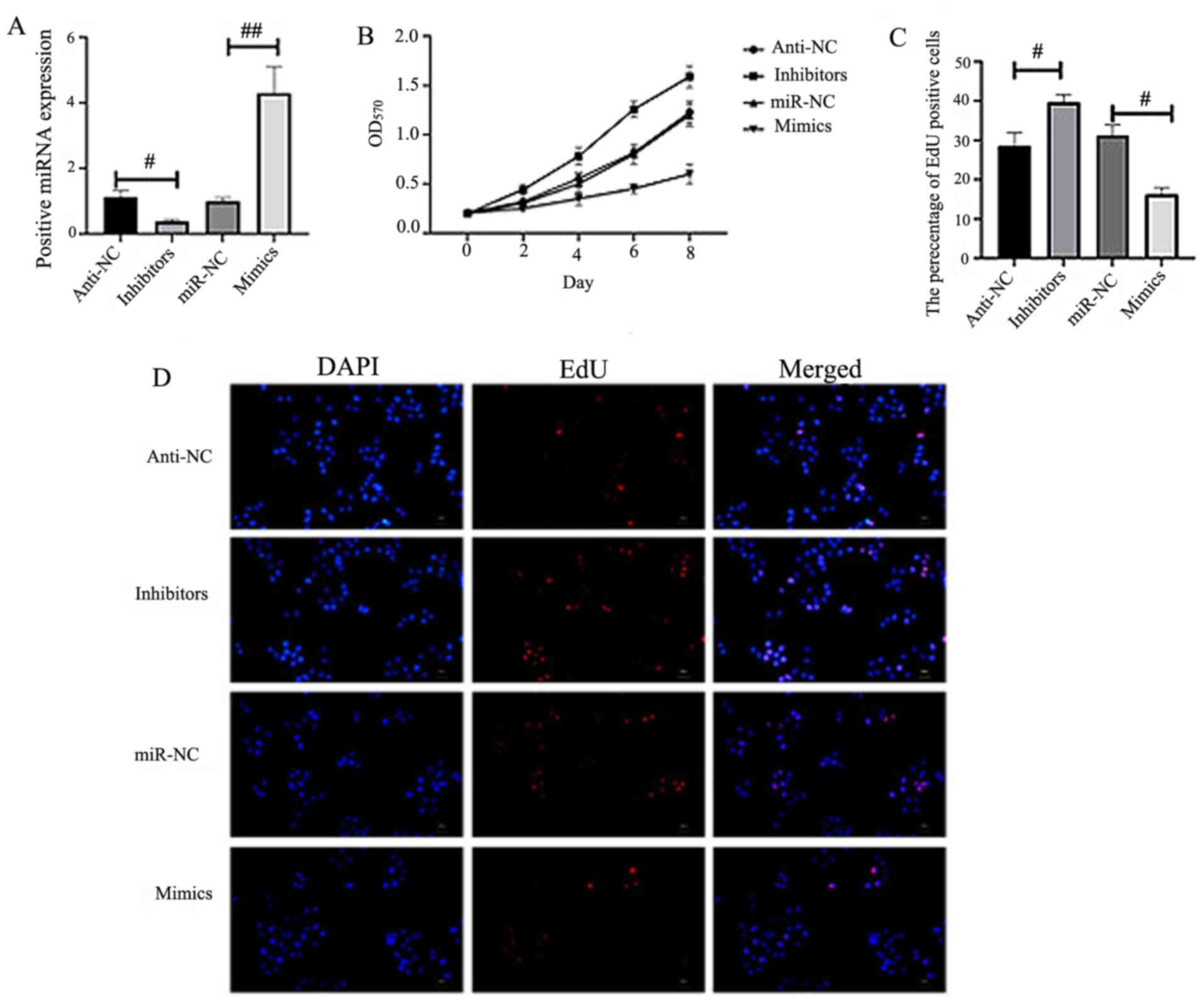
miR-34c-5p inhibits CC cell
proliferation and enhances cell apoptosis
miR-34c-5p was overexpressed or silenced in HeLa
cells to ascertain its role in the development of CC (Fig. 2A). CCK-8 (Fig. 2B) and EdU analysis (Fig. 2C-D) indicated that the proliferation
of HeLa cells was inhibited in the miR-34c-5p-mimics group, while
an increase was revealed in miR-34c-5p-inhibitor-transfected HeLa
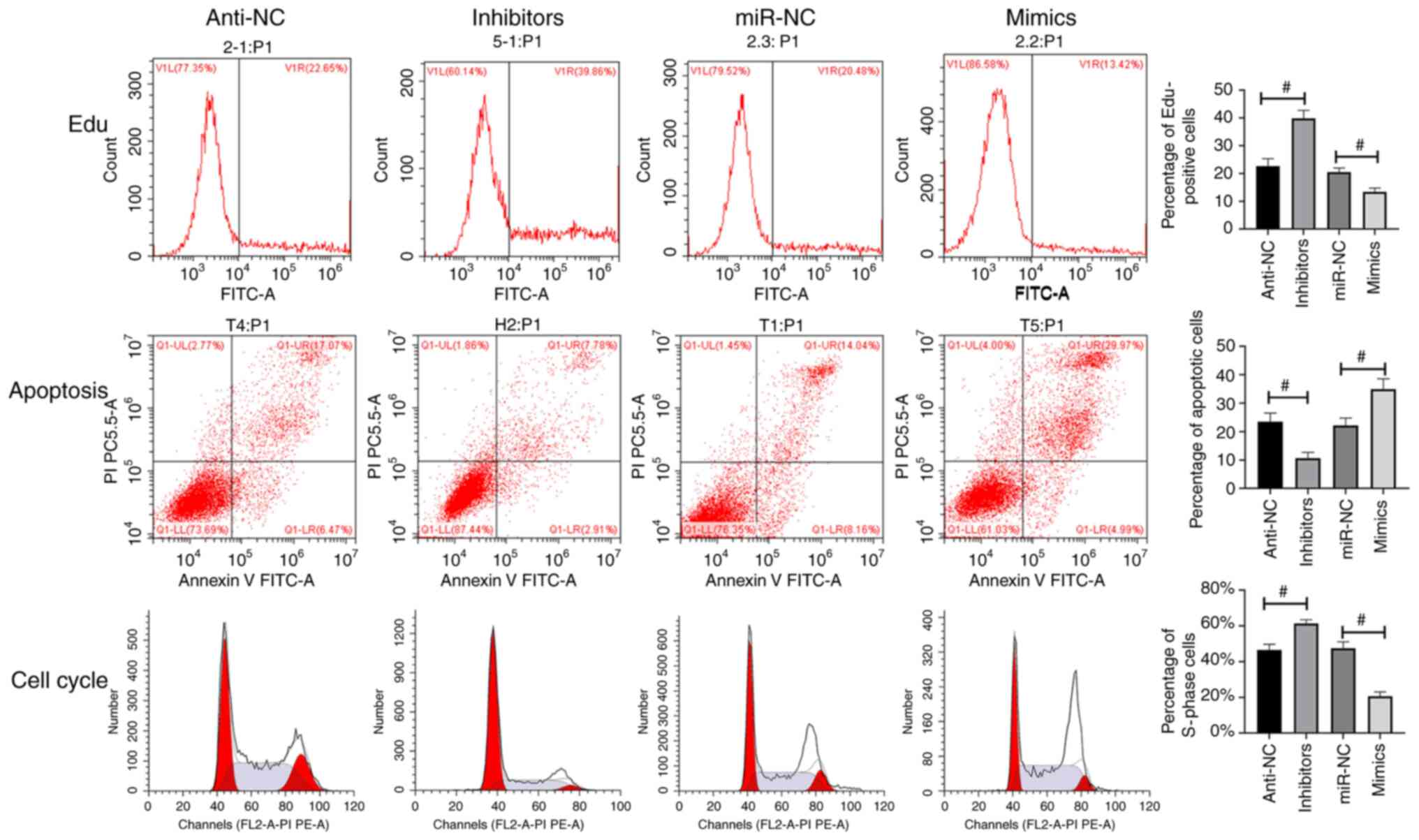
cells (Fig. 2B-D). Flow cytometry
was employed to detect cell proliferation, cell cycle and apoptosis
with results revealing that miR-34c-5p mimics inhibited cell
proliferation and promoted cell apoptosis (Fig. 3).
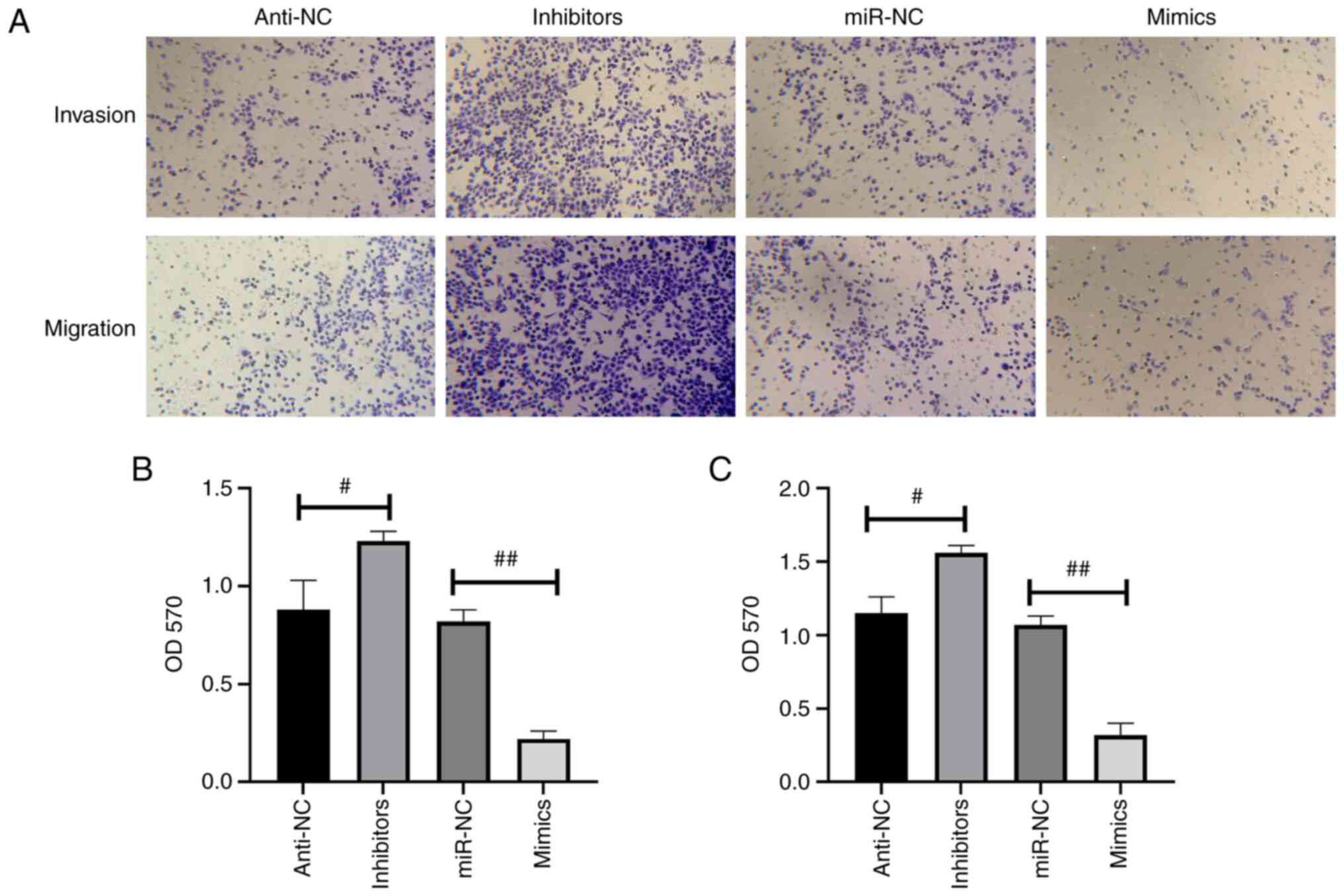
miR-34c-5p inhibits the migration and
invasion capacities of CC cells
Transwell assays demonstrated that the both the
migration and invasion abilities in miR-34c-5p-mimic-transfected
HeLa cells were significantly reduced compared with the miR-NC
group; however, that of miR-34c-5p-silenced HeLa cells exhibited an
increase in migration and invasion abilities (Fig. 4).
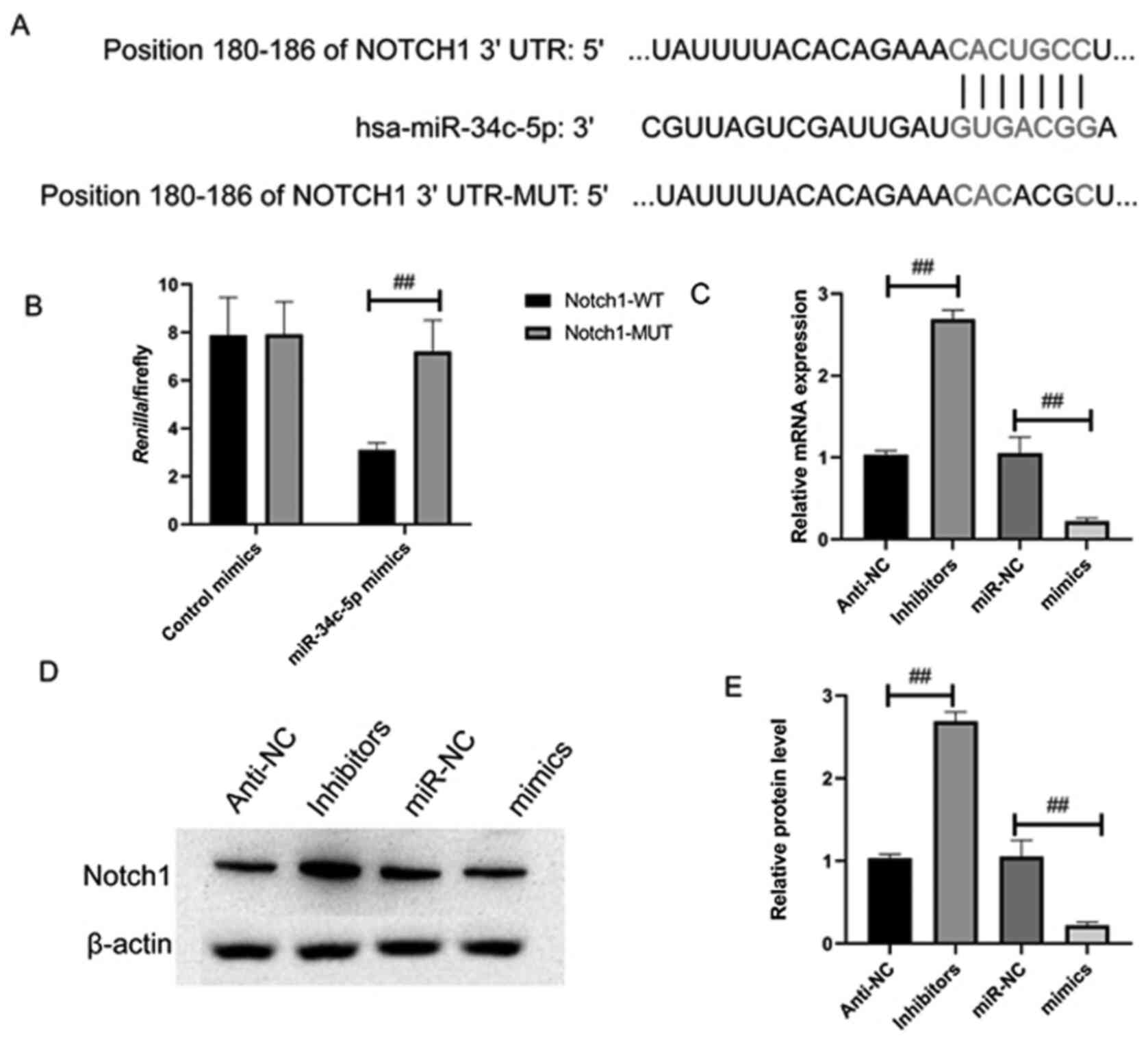
Notch1 acts as a target of
miR-34c-5p
The underlying mechanism of miR-34c-5p was
investigated in CC progression, the potential targets of which were
predicted using TargetScan. The results demonstrated that Notch1
mRNA 3′-UTR possesses highly conserved binding sites for miR-34c-5p
to bind with (Fig. 5A).
Correlations between Notch1 and miR-34c-5p were analyzed through
luciferase reporter assay. Luciferase reporter plasmid containing
wt/mut 3′-UTR human Notch1 binding site was co-transfected with
miR-34c-5p mimics into the 293T cells. miR-34c-5p mimics
efficiently reduced the luciferase activity of Notch1 wt 3′-UTR in
the 293T cells but no effects were observed in the cells
transfected with the mut Notch1 3′-UTR (Fig. 5B). As it was identified as a
specific target of miR-34c-5p (Fig.
3B), Notch1was highly expressed in CC (Fig. S2). The results of RT-qPCR and
western blot analysis demonstrated that overexpression of
miR-34c-5p disabled Notch1 expression in HeLa cells at mRNA and
protein levels (Fig. 5C-E) in
contrast to the miR-NC cells; the opposite results obtained from
miR-34c-5p silencing in HeLa cells further demonstrated Notch 1 to
be the target of miR-34c-5p.
Discussion
Previous studies have validated the relationship
between cancer progression and dysregulated miRNA expression.
Several miRNAs have been demonstrated to serve as either tumor
suppressors or oncogenes in the development of CC (19–21).
Despite numerous miRNAs acting as tumor suppressors and inhibitors
of their target gene expression at a low level, they may also
elicit an intense oncogene translation, thereby facilitating the
development of tumors (22).
Overexpressed oncogenic miRNAs may also result in an inhibitory
effect on tumor suppressor genes (23).
miR-34c-5p has been revealed to be downregulated in
types of cancer (10–12), although the mechanism in CC remains
unclear. The present study revealed that the miR-34c-5p expression
exhibited a significant decrease in CC and relevant cell lines,
indicating that miR-34c-5p served an essential role in cell
proliferation, metastasis and apoptosis of CC. The aforementioned
findings suggested that miR-34c-5p overexpression hindered the
viability, proliferation, migration and invasion of different types
of cancer cells and accelerated apoptosis in vitro.
miRNAs inhibit the expression of specific target
genes, resulting in regulation of a variety of biological changes
(24). miR-34c-5p is expressed at a
low level in gliomas compared with normal brain tissues and normal
glial cell lines (12).
Overexpression of miR-34c-5p was revealed to inhibit U251 cell
proliferation and result in S-phase arrest, G0/G1 reduction and
cell apoptosis (12). In gastric
cancer, miR-34c-5p was revealed to directly bind to the 3′UTR of
the microtubule-associated protein tau (MAPT) and inhibit the
expression of MAPT (25). The
present study determined that miR-34c-5p directly targeted Notch1
mRNA and downregulated Notch1 expression, thereby inhibiting the
progression of CC. Notch1 has also been reported to dissociate from
the fused negative inhibitor in the primary cilium and then
converts into an activated form and migrates to the nucleus
(26,27). Notch1 translocation enhances the
expression of downstream target oncogenes including cyclin D1 and
homeobox protein NANOG (28,29).
Notch1 has been revealed to be highly expressed as an oncogene in
various cancers including non-small cell lung, breast, liver and
gastric cancer (30–33). The present study confirmed that
miR-34c-5p impaired Notch1 expression by directly binding to the
3′-UTR of Notch1 mRNA. It also demonstrated a negative correlation
between the level of miR-34c-5p and Notch1 mRNA in CC. In addition,
it was observed that the overexpression of Notch1 in CC hindered
the effect of miR-34c-5p on cell survival and metastasis. All the
aforementioned underpin the hypothesis that Notch1 is the direct
target of miR-34c-5p.
In conclusion, the present study demonstrated that
miR-34c-5p was significantly downregulated in human CC and related
cell lines and that overexpressed miR-34c-5p accelerated cell
apoptosis and inhibited proliferation, invasion and migration of CC
cells. The present study elucidated the biological traits of
miR-34c-5p and Notch1 so as to further study their correlation,
which may provide novel approaches for the treatment of CC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW, HW and XW conceived and designed the
experiments. HW, RJ, XW, XN and XL conducted all the experiments.
SW, HW and XW wrote and revised the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Emergency General Hospital (Beijing, China). Prior
written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu M, Wang Z, Liu Q, Zhu H and Xu N:
Expression of Micro-RNA-492 (MiR-492) in human cervical cancer cell
lines is upregulated by transfection with wild-type P53,
irradiation, and 5-fluorouracil treatment in vitro. Med Sci Monit.
24:7750–7758. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Song TT, Xu F and Wang W: Inhibiting
ubiquitin conjugating enzyme E2 N by microRNA-590-3p reduced cell
growth of cervical carcinoma. Kaohsiung J Med Sci. 36:501–507.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang T, Feng J and Zhang A: miR-584
inhibits cell proliferation, migration and invasion in vitro and
enhances the sensitivity to cisplatin in human cervical cancer by
negatively targeting GLI1. Exp Ther Med. 19:2059–2066.
2020.PubMed/NCBI
|
|
4
|
Jones RA, Franks SE and Moorehead RA:
Comparative mRNA and miRNA transcriptome analysis of a mouse model
of IGFIR-driven lung cancer. PLoS One. 13:e2069482018. View Article : Google Scholar
|
|
5
|
Chen Z, Zhang M, Qiao Y, Yang J and Yin Q:
MicroRNA-1297 contributes to the progression of human cervical
carcinoma through PTEN. Artif Cells Nanomed Biotechnol.
46:1120–1126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu HJ, Jin PY, Tang Y, Fan SH, Zhang XF,
Wang F, Wu DM, Lu J and Zheng YL: microRNA-136 inhibits
proliferation and promotes apoptosis and radiosensitivity of
cervical carcinoma through the NF-κB pathway by targeting E2F1.
Life Sci. 199:167–178. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yin Z and Ren W: MicroRNA-217 acts as a
tumor suppressor and correlates with the chemoresistance of
cervical carcinoma to cisplatin. Onco Targets Ther. 12:759–771.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu LM, Wang WW, Qi R, Leng TG and Zhang
XL: MicroRNA-224 inhibition prevents progression of cervical
carcinoma by targeting PTX3. J Cell Biochem. 119:10278–10290. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding W, Xin J, Jiang L, Zhou Q, Wu T, Shi
D, Lin B, Li L and Li J: Characterisation of peripheral blood
mononuclear cell microRNA in hepatitis B-related acute-on-chronic
liver failure. Sci Rep. 5:130982015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng D, Wang H, Li L, Ma X, Chen Y, Zhou
H, Luo Y, Xiao Y and Liu L: miR-34c-5p promotes eradication of
acute myeloid leukemia stem cells by inducing senescence through
selective RAB27B targeting to inhibit exosome shedding. Leukemia.
32:1180–1188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Re M, Magliulo G, Gioacchini FM,
Bajraktari A, Bertini A, Çeka A, Rubini C, Ferrante L, Procopio AD
and Olivieri F: Expression levels and clinical significance of
miR-21-5p, miR-let-7a, and miR-34c-5p in laryngeal squamous cell
carcinoma. Biomed Res Int. 2017:39212582017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Z, Wu Y, Tian Y, Sun X, Liu J, Ren H,
Liang C, Song L, Hu H, Wang L and Jiao B: Differential effects of
miR-34c-3p and miR-34c-5p on the proliferation, apoptosis and
invasion of glioma cells. Oncol Lett. 6:1447–1452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang JY and Xu ZS: Notch signal pathway
and chronic lymphocytic leukemia. Zhongguo Shi Yan Xue Ye Xue Za
Zhi. 22:1472–1475. 2014.(In Chinese). PubMed/NCBI
|
|
14
|
Yang H, Li Y, Li T, Xu M, Chen Y, Wu C,
Dang X and Liu Y: Multifunctional core/shell nanoparticles
cross-linked polyetherimide-folic acid as efficient Notch-1 siRNA
carrier for targeted killing of breast cancer. Sci Rep. 4:70722014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lähdeniemi I, Misiorek JO, Antila CJM,
Landor SKJ, Stenvall CGA, Fortelius LE, Bergström LK, Sahlgren C
and Toivola DM: Keratins regulate colonic epithelial cell
differentiation through the Notch1 signalling pathway. Cell Death
Differ. 24:984–996. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sharif A, Shaji A, Chammaa M, Pawlik E and
Fernandez-Valdivia R: Notch transduction in non-small cell lung
cancer. Int J Mol Sci. 21:56912020. View Article : Google Scholar
|
|
17
|
Sui C, Zhuang C, Sun D, Yang L, Zhang L
and Song L: Notch1 regulates the JNK signaling pathway and
increases apoptosis in hepatocellular carcinoma. Oncotarget.
8:45837–45847. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Ding Y, Ding N, Zhang H, Lu M, Cui X
and Yu X: MicroRNA-625-5p sponges lncRNA MALAT1 to inhibit cervical
carcinoma cell growth by suppressing NF-κB signaling. Cell Biochem
Biophys. 78:217–225. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li B, Wu N, Zhang XJ, Wei ZL and Shang LX:
MicroRNA-409 inhibits the proliferative ability of cervical
carcinoma cells by regulating AKT. Eur Rev Med Pharmacol Sci.
22:936–942. 2018.PubMed/NCBI
|
|
21
|
Ma C, Xu B, Husaiyin S, Wang L,
Wusainahong K, Ma J, Zhu K and Niyazi M: MicroRNA-505 predicts
prognosis and acts as tumor inhibitor in cervical carcinoma with
inverse association with FZD4. Biomed Pharmacother. 92:586–594.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei S, Zhang ZY, Fu SL, Xie JG, Liu XS, Xu
YJ, Zhao JP and Xiong WN: Hsa-miR-623 suppresses tumor progression
in human lung adenocarcinoma. Cell Death Dis. 8:e28292017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang F, Lin C, Shi YH and Kuerban G:
MicroRNA-101 inhibits cell proliferation, invasion, and promotes
apoptosis by regulating cyclooxygenase-2 in HeLa cervical carcinoma
cells. Asian Pac J Cancer Prev. 14:5915–5920. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ham O, Lee CY, Kim R, Lee J, Oh S, Lee MY,
Kim J, Hwang KC, Maeng LS and Chang W: Therapeutic potential of
differentiated mesenchymal stem cells for treatment of
osteoarthritis. Int J Mol Sci. 16:14961–14978. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu H, Huang M, Lu M, Zhu W, Shu Y, Cao P
and Liu P: Regulation of microtubule-associated protein tau (MAPT)
by miR-34c-5p determines the chemosensitivity of gastric cancer to
paclitaxel. Cancer Chemother Pharmacol. 71:1159–1171. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grisanti L, Revenkova E, Gordon RE and
Iomini C: Primary cilia maintain corneal epithelial homeostasis by
regulation of the notch signaling pathway. Development.
143:2160–2171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eberhart C: Multiple cilia suppress tumour
formation. Nat Cell Biol. 18:368–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Giannone G, Attademo L, Scotto G, Genta S,
Ghisoni E, Tuninetti V, Aglietta M, Pignata S and Valabrega G:
Endometrial cancer stem cells: Role, characterization and
therapeutic implications. Cancers (Basel). 11:18202019. View Article : Google Scholar
|
|
29
|
Shen M, Dong C, Ruan X, Yan W, Cao M,
Pizzo D, Wu X, Yang L, Liu L, Ren X and Wang SE:
Chemotherapy-induced extracellular vesicle miRNAs promote breast
cancer stemness by targeting ONECUT2. Cancer Res. 79:3608–3621.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang S, Cai L, Zhang F, Shang X, Xiao R
and Zhou H: Inhibition of EZH2 attenuates sorafenib resistance by
targeting NOTCH1 activation-dependent liver cancer stem cells via
NOTCH1-related MicroRNAs in hepatocellular carcinoma. Transl Oncol.
13:1007412020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu C, Ren C, Yang T, Sun Y, Qiao P, Wang
D, Lv S and Yu Z: A noncanonical role of fructose-1,
6-bisphosphatase 1 is essential for inhibition of Notch1 in breast
cancer. Mol Cancer Res. 18:787–796. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang LZ, Lei CC, Zhao YP, Sun HW, Yu QH,
Yang EJ and Zhan X: MicroRNA-34c-3p target inhibiting NOTCH1
suppresses chemosensitivity and metastasis of non-small cell lung
cancer. J Int Med Res. 48:3000605209048472020.PubMed/NCBI
|
|
33
|
Hu J, Yu J, Gan J, Song N, Shi L, Liu J,
Zhang Z and Du J: Notch1/2/3/4 are prognostic biomarker and
correlated with immune infiltrates in gastric cancer. Aging (Albany
NY). 12:2595–2609. 2020. View Article : Google Scholar : PubMed/NCBI
|