Introduction
Liver fibrosis is a common pathological alteration
that occurs during the process of tissue repair after chronic liver
injury, and gradually develops into cirrhosis, which eventually
leads to irreversible liver injury (1,2).
Emerging evidence indicates that the abnormal activation and
proliferation of hepatic stellate cells (HSCs) are important
factors leading to liver fibrosis (3) and that epithelial-mesenchymal
transition (EMT) is associated with the abnormal activation of HSCs
(4). Choi et al (5) reported that EMT in HSC activation is
regulated via the hedgehog (Hh) signaling pathway. Therefore,
inhibiting HSC excessive viability and EMT by regulating the Hh
signaling axis may serve as a potential therapeutic strategy to
alleviate liver fibrosis.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
composed of 22–25 nucleotides that restrain the expression of
target mRNAs by binding to their 3′untranslated region (UTR)
(6). Yu et al (7) demonstrated that increasing the
expression of miR-200a regulated the Hh signaling pathway by
targeting the binding to GLI family zinc finger 2 (Gli2), thus
restraining EMT in HSC activation to hinder the progression of
liver fibrosis. The increased expression of miR-152 regulates the
Hh signaling pathway by downregulating DNA methyltransferase 1, and
further restrains HSC activation and EMT to exert its anti-fibrotic
function (8). A study demonstrated
that the abnormal expression of miR-375 has been observed in
various diseases, including inflammatory bowel disease and liver
cancer, suggesting that miR-375 serves important regulatory
functions in various human diseases (9). Besides, Yang et al (10) reported that miR-375 is downregulated
in a fructose-induced liver fibrosis rat model. However, the
molecular mechanism underlying miR-375 in liver fibrosis is not
completely understood.
Rac family small GTPase 1 (RAC1) is an important
member of the Rho family of small GTPase proteins and participates
in the regulatory processes of cell proliferation, differentiation
and EMT (11). RAC1 overexpression
in HSCs leads to abnormal activation, which in turn aggravates
liver fibrosis (12). Another study
demonstrated that RAC1 activation induces HSC activation and
promotes HSC EMT by mediating the Hh signaling pathway, thus
aggravating liver fibrosis (13).
However, the mechanism underlying RAC1 in regulating HSC activation
and EMT in liver fibrosis is not completely understood.
The present study aimed to investigate the
expression of miR-375 in liver fibrosis tissues and cells compared
with healthy control tissues and hepatocytes (HCs), respectively.
Moreover, the mechanisms underlying miR-375 in liver fibrosis were
investigated.
Materials and methods
Tissue samples
In the present study, 15 healthy controls and 15
patients (43.2% male patients and 56.8% female patients; age,
60.1±11.7 years) with liver cirrhosis undergoing partial liver
resection or liver biopsy were recruited from the First Affiliated
Hospital of Zhengzhou University (Zhengzhou, China) between March
2017 and March 2020. Liver cirrhosis was diagnosed by performing a
liver biopsy, abdominal ultrasound or computed tomography scan. The
exclusion criteria were as follows: i) The use of non-protective
liver drugs to treat viral hepatitis or liver fibrosis; and ii)
liver biochemical indicators were normal. The participants were
divided into two groups: i) Healthy controls (n=15); and ii)
cirrhosis (n=15). The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Zhengzhou University
(approval no. 2020-KY-047). Written informed consent was obtained
from all patients before obtaining liver tissues.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
RT-qPCR was performed as previously described
(14). Briefly, total RNA was
extracted from human and mouse liver fibrosis tissues and mouse
liver fibrosis cells using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed
into cDNA using the PrimeScript One-Step RT-PCR kit (Takara
Biotechnology Co., Ltd.) at 42°C. Subsequently, qPCR was performed
using an SYBR Green PCR kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and an ABI 7900HT Fast Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used for qPCR: 95°C for 5 min;
followed by 40 cycles of 95°C for 15 sec and 60°C for 20 sec. The
sequences of the primers used for qPCR are presented in Table I. miRNA and mRNA expression levels
were quantified using the 2−ΔΔCq method (15) and normalized to the internal
reference genes U6 and β-actin, respectively.
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence (5′→3′) |
|---|
| miR-375 | F:
CACAAAATTTGTTCGTTCGGCT |
|
| R:
GTGCAGGGTCCGAGGT |
| U6 | F:
CAAATTCGTGAAGCGTTCCATAT |
|
| R:
GCTTCACGAATTTGCGTGTCATCCTTGC |
| RAC1 | F:
GAGCAGAAGCTGATCTCCGAGGAG |
|
| R:
TTACAACAGCAGGCATTTTCTCTT |
| α-SMA | F:
CTGACAGAGGCACCACTGAA |
|
| R:
CATCTCCAGAGTCCAGCACA |
| Col1A1 | F:
GATTGAGAACATCCGCAGC |
|
| R:
CATCTTGAGGTCACGGCAT |
| β-actin | F:
CAGGAGGCATTGCTGATGAT |
|
| R:
GAAGGCTGGGGCTCATTT |
Establishment of liver fibrosis mouse
model
A total of 12 male C57BL/6J mice (age, 8 weeks;
weight, 20–22 g; Experimental Animal Center of Zhengzhou
University) were used to establish a liver fibrosis mouse model.
The mice were housed in a temperature (23±3°C) and humidity
(40–70%) controlled environment with 12-h light/dark cycles, and
free access to food and water. Animal health and behavior were
assessed every 24 h. Mice were divided into the following two
groups: i) Sham (n=6); and ii) CCl4 (n=6). Mice in the
CCl4 group were intraperitoneally injected with 7 ml/kg
CCl4 (diluted 1:9 in olive oil; Sinopharm Chemical
Reagent Co., Ltd.; cat. no. XW00562352) twice a week for six weeks.
Mice in the sham group were intraperitoneally injected with the
same volume of olive oil twice a week for six weeks. At the end of
the six weeks, mice were sacrificed and the liver tissues were
isolated for subsequent experiments. The animal experimental
protocol was approved by the Ethics Committee of the First
Affiliated Hospital of Zhengzhou University (approval no.
2020-KY-047).
Masson and hematoxylin and eosin
(H&E) staining
Mouse liver tissue samples were fixed with 10%
formalin at room temperature for 48 h, embedded in paraffin and cut
into 5-µm thick sections. According to the standard protocols,
Masson staining was performed to assess collagen deposition and
H&E staining was performed to assess the degree of liver
injury. For Masson staining, sections were stained with hematoxylin
for 5–10 min at room temperature, followed by Masson staining for
5–10 min at room temperature. For H&E staining, sections were
stained with hematoxylin for 5–10 min at room temperature, followed
by eosin staining for 1 min at room temperature. Stained sections
were observed using a light microscope (Nikon Corporation;
magnification, ×200).
Western blotting
Total protein was extracted from human and mouse
liver fibrosis tissues and mouse liver fibrosis cells using RIPA
(Beijing Solarbio Science & Technology Co., Ltd.). Total
protein was quantified using the BCA Protein Assay kit (Beijing
Solarbio Science & Technology Co., Ltd.). Proteins (20 µg/lane)
were separated via 10% SDS-PAGE and transferred onto PVDF
membranes. After blocking with 5% skim milk at room temperature for
1 h, the membranes were incubated overnight at 4°C with the
following primary antibodies: Anti-α-smooth muscle actin (α-SMA;
cat. no. ab5694; 1:1,000; Abcam), anti-collagen type I-α1 (Col1A1;
cat. no. ab34710; 1:1,000; Abcam), anti-RAC1 (cat. no. ab155938;
1:1,000; Abcam), anti-E-cadherin (cat. no. ab227639; 1:25; Abcam),
anti-Snail (cat. no. ab216347; 1:1,000; Abcam), anti-Vimentin (cat.
no. sc-6260; 1:1,000; Santa Cruz Biotechnology, Inc.),
anti-hedgehog interacting protein (Hhip; cat. no. ab86450; 1:1,000;
Abcam), anti-sonic hedgehog signaling molecule (Shh; cat. no.
sc-365112; 1:1,000; Santa Cruz Biotechnology, Inc.), anti-Gli2
(cat. no. sc-271786; 1:1,000; Santa Cruz Biotechnology, Inc.) and
anti-β-actin (cat. no. sc-8432; 1:1,000; Santa Cruz Biotechnology,
Inc.). Subsequently, the membranes were incubated with a
HRP-conjugated specific secondary antibody (cat. no. ab205718;
1:2,000; Abcam) at room temperature for 1 h. Protein bands were
visualized using the enhanced chemiluminescence kit (Shanghai XP
Biomed Ltd.). β-actin was used as the loading control.
Cell isolation and culture
Mouse primary HCs and primary HSCs were isolated as
previously described (16). Cells
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 µg/ml streptomycin at 37°C with 5%
CO2. Moreover, HSCs were isolated from healthy control
mice and cultured in DMEM at 37°C, 5% CO2 in
vitro for 1, 3 and 5 days, respectively.
Cell transfection and treatment
HSCs were isolated from healthy control mice (Sham
group) and cultured for 1 day in vitro to obtain
primary-1-day HSCs. Primary-1-Day HSCs were seeded
(1×105 cells/well) into 24-well plates and cultured for
~24 h. At 80% confluence, cells were transfected with 10 nM miR-375
mimic, 10 nM pre-NC, 10 nM miR-375 mimic + 10 nM pcDNA-RAC1, 10 nM
miR-375 inhibitor or 10 nM NC at 37°C for 48 h using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. miR-375 mimic, miR-375 inhibitor, pre-NC, inhibitor NC,
pcDNA and pcDNA-RAC1 were purchased from Shanghai GenePharma Co.,
Ltd. The specific sequences were as follows: miR-375 mimic,
5′-UUUGUUCGUUCGGCUCGCGUGA-3′; miR-375 inhibitor,
5′-UCACGCGAGCCGAACGAACAAA-3′; pre-NC, 5′-CAGUACUUUUGUGUAGUACAA-3′;
and NC, 5′-CAGUACUUUUGUGUAGUACAA-3′. At 48 h post-transfection,
cells were used for subsequent experiments.
To investigate whether miR-375 inhibited mouse HSC
viability and EMT via the Hh signaling pathway, cells
(1×105) were cultured in DMEM supplemented with 0.3 µM
smoothened agonist (SAG; an exogenous Hh agonist; Beyotime
Institute of Biotechnology) at 37°C for 24 h.
Cell viability
The MTT assay was performed to assess mouse HSC
viability. HSC cells were seeded (1×104 cells/well) into
96-well plates and cultured at 37°C for 48 h. Subsequently, 20 µl
MTT solution was added to each well and incubated at 37°C for 2 h.
Then, 150 µl DMSO was added to dissolve the purple formazan
crystals. The optical density was measured at a wavelength of 570
nm to determine HSC viability.
Cell cycle assay
The cell cycle distribution of mouse HSCs was
assessed via flow cytometry. Following fixation with 70% ethanol
for at 37°C for 24 h, cells (1×106) were resuspended in
PI/RNase staining buffer (BD Pharmingen; BD Biosciences)
supplemented with 50 ng/µl RNase A and incubated at 37°C for 30
min. The cell cycle distribution was analyzed via flow cytometry
using an Attune™ NxT Flow Cytometer flow cytometer (Invitrogen;
Thermo Fisher Scientific, Inc.) and FlowJo software (version 10;
Flow Jo LLC).
Dual-luciferase reporter gene
assay
Online bioinformatics software TargetScan (version
3.1; www.targetscan.org/mamm_31) was used to predict that
miR-375 contained the binding sites in the 3′UTR region of
RAC1.
The RAC1 3′-UTR-wild-type (WT) and RAC1 3′UTR-mutant
(Mut) sequences were inserted into the pmirGLO luciferase reporter
vector (Shanghai GenePharma Co., Ltd.). Subsequently, HSCs
(1×106) were co-transfected with 1 µg miR-375 mimic,
pre-NC, miR-375 inhibitor or NC and 1 µg RAC1 3′UTR-WT or RAC1
3′UTR-Mut using Lipofectamine 2000 transfection reagent. At 48 h
post-transfection, luciferase activity was measured using a
Dual-Luciferase Reporter Assay system (Promega Corporation).
Firefly luciferase activity was normalized to Renilla
luciferase activity.
Statistical analysis
Experiments were repeated at least three times.
Statistical analyses were performed using SPSS software (version
17.0; SPSS, Inc.). Comparisons between two groups were analyzed
using the unpaired Student's t-test. Comparisons among multiple
groups were analyzed using one-way ANOVA followed by Tukey's post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-375 is downregulated in liver
fibrosis tissues and cells
The abnormal expression of miRNAs is associated with
the occurrence of liver fibrosis (17,18).
In the present study, the expression levels of miR-375 and Col1A1
(a liver fibrosis molecular marker) in the tissue samples of
patients with cirrhosis were assessed. Compared with the healthy
control group, miR-375 expression was significantly decreased in
the cirrhosis group, whereas Col1A1 expression was significantly
increased (Fig. 1A). Pearson's
correlation analysis demonstrated that miR-375 expression was
negatively correlated with Col1A1 expression and RAC1 expression in
human cirrhosis tissue samples (Fig.
1B). Subsequently, a liver fibrosis mouse model was established
via intraperitoneal injection of CCl4. The Masson and
H&E staining results indicated that the liver fibrosis tissues
of mice in the CCl4 group displayed increased collagen
deposition, inflammation, infiltration, steatosis and fibrosis
compared with the sham group (Fig.
1C). α-SMA and Col1A1 are common molecular marker of liver
fibrosis (19). The mRNA and
protein expression levels of α-SMA and Col1A1 were dramatically
increased in the CCl4 group compared with the sham group
(Fig. 1D). By contrast, miR-375
expression was significantly decreased in the CCl4 group
compared with the sham group (Fig.
1E). Subsequently, mouse primary HCs and primary HSCs were
isolated from liver fibrosis model mice. miR-375 expression was
significantly decreased in the HSC group compared with the HC group
(Fig. 1F). Moreover, HSCs were
isolated from healthy control mice and cultured in vitro for
1, 3 and 5 days, respectively. The mRNA and protein expression
levels of α-SMA and Col1A1 were markedly increased in the day 3 and
5 groups compared with the day 1 group (Fig. 1G). By contrast, miR-375 expression
was significantly decreased in the day 3 and 5 groups compared with
the day 1 group (Fig. 1H). The
aforementioned results indicated that miR-375 was downregulated in
human and mouse liver fibrosis tissues and mouse liver fibrosis
cells compared with healthy control tissues and HCs, respectively,
suggesting that miR-375 may serve a regulatory function in the
process of liver fibrosis.
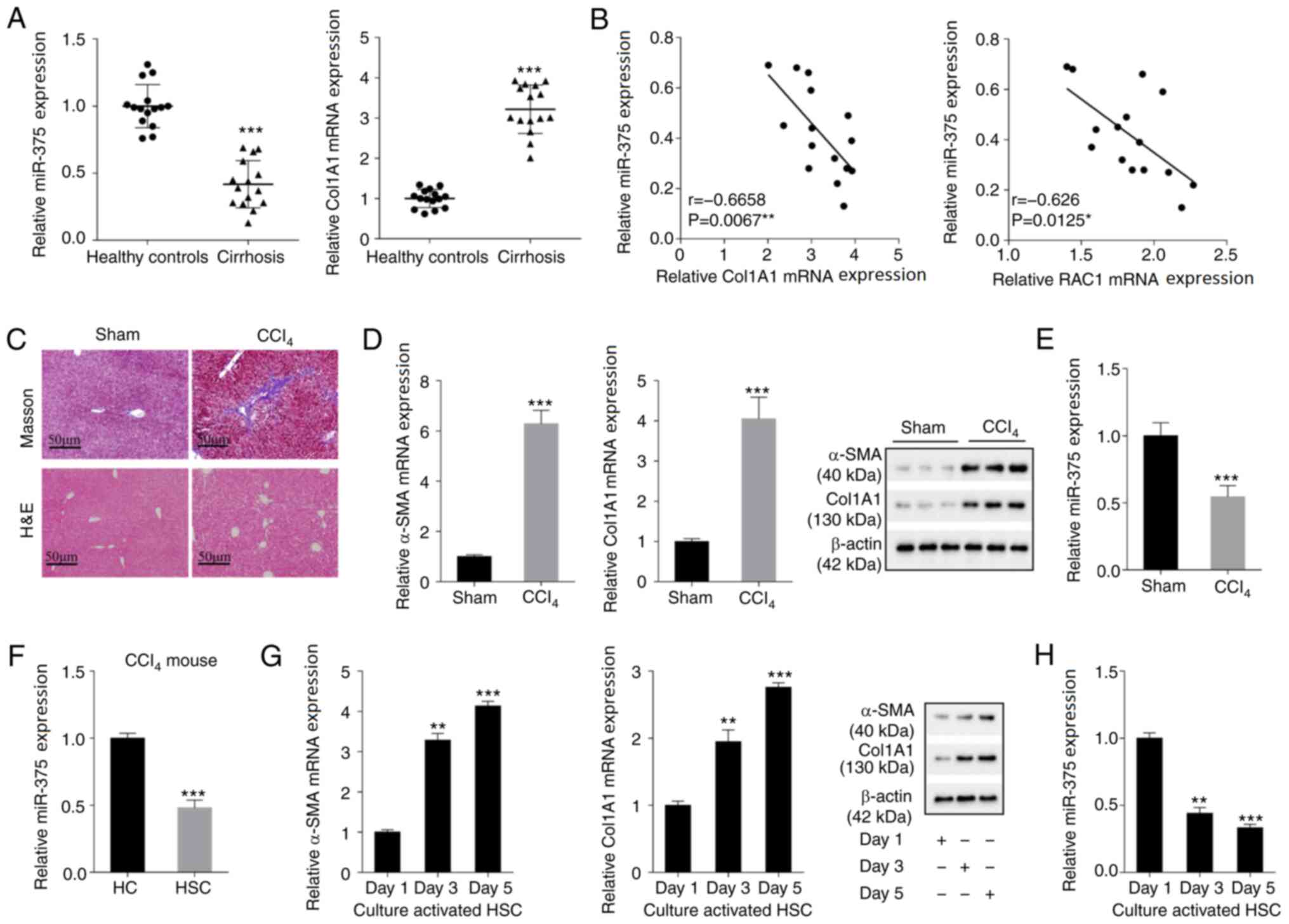 | Figure 1.miR-375 expression in liver fibrosis
tissues and cells. Liver tissues were isolated from healthy
controls (n=15) and patients with cirrhosis (n=15) who underwent
partial liver resection or liver biopsy. (A) miR-375 and Col1A1 (a
liver fibrosis molecular marker) expression levels in the tissue
samples of healthy controls and patients with cirrhosis. (B)
Pearson's correlation analysis was performed to analyze the
correlation between miR-375 expression and Col1A1 or RAC1
expression in tissue samples isolated from patients with cirrhosis.
A liver fibrosis mouse model of liver fibrosis was established by
an intraperitoneal injection of CCl4. (C) Pathological
alterations in tissue samples isolated from liver fibrosis mouse
models were observed by performing Masson and H&E staining
(scale bar, 50 µm). (D) α-SMA and Col1A1 mRNA and protein
expression levels in the liver fibrosis mouse model. (E) miR-375
expression in the liver fibrosis mouse model. Primary HCs and HSCs
were isolated from liver fibrosis model mice. (F) miR-375
expression in primary HCs and HSCs. HSCs isolated from healthy
control mice were cultured in vitro for 1, 3 and 5 days. (G)
α-SMA and Col1A1 mRNA and protein expression levels in HSCs. (H)
miR-375 expression in HSCs. *P<0.05, **P<0.01 and
***P<0.001 vs. healthy control, Sham, HC or day 1. miR,
microRNA; Col1A1, collagen type I-α1; RAC1, Rac family small GTPase
1; H&E, hematoxylin and eosin; α-SMA, α-smooth muscle actin;
HC, hepatocyte; HSC, hepatic stellate cell. |
miR-375 overexpression inhibits mouse
HSC viability and EMT, and alleviates liver fibrosis
To further investigate the function of miR-375 in
liver fibrosis, HSCs isolated from healthy control mice were
cultured for 1 day in vitro to obtain mouse primary-1-day
HSC. Subsequently, primary-1-day HSCs were transfected with miR-375
mimic or pre-NC. miR-375 mimic significantly increased miR-375
expression compared with pre-NC (Fig.
2A), indicating successful overexpression of miR-375 in mouse
primary-1-day HSC. miR-375 overexpression notably decreased the
mRNA and protein expression levels of α-SMA, Col1A1 and RAC1
compared with the pre-NC group (Fig.
2B). Moreover, the MTT assay results indicated that miR-375
overexpression significantly inhibited HSC viability compared with
the pre-NC group (Fig. 2C). The
cell cycle analysis results suggested that miR-375 overexpression
significantly inhibited the cell cycle progression of HSCs compared
with the pre-NC group (Figs. 2D and
S1A). The western blotting results
indicated that compared with the pre-NC group, miR-375 mimic
markedly increased the protein expression levels of the epithelial
marker E-cadherin and the Hh signaling pathway-related molecule
Hhip. However, compared with the pre-NC group, miR-375 mimic
notably decreased the protein expression levels of the mesenchymal
markers Snail and Vimentin, and the Hh signaling pathway-related
molecules Shh and Gli2 (Fig. 2E).
The aforementioned results indicated that miR-375 overexpression
inhibited mouse HSC viability and EMT, and alleviated liver
fibrosis.
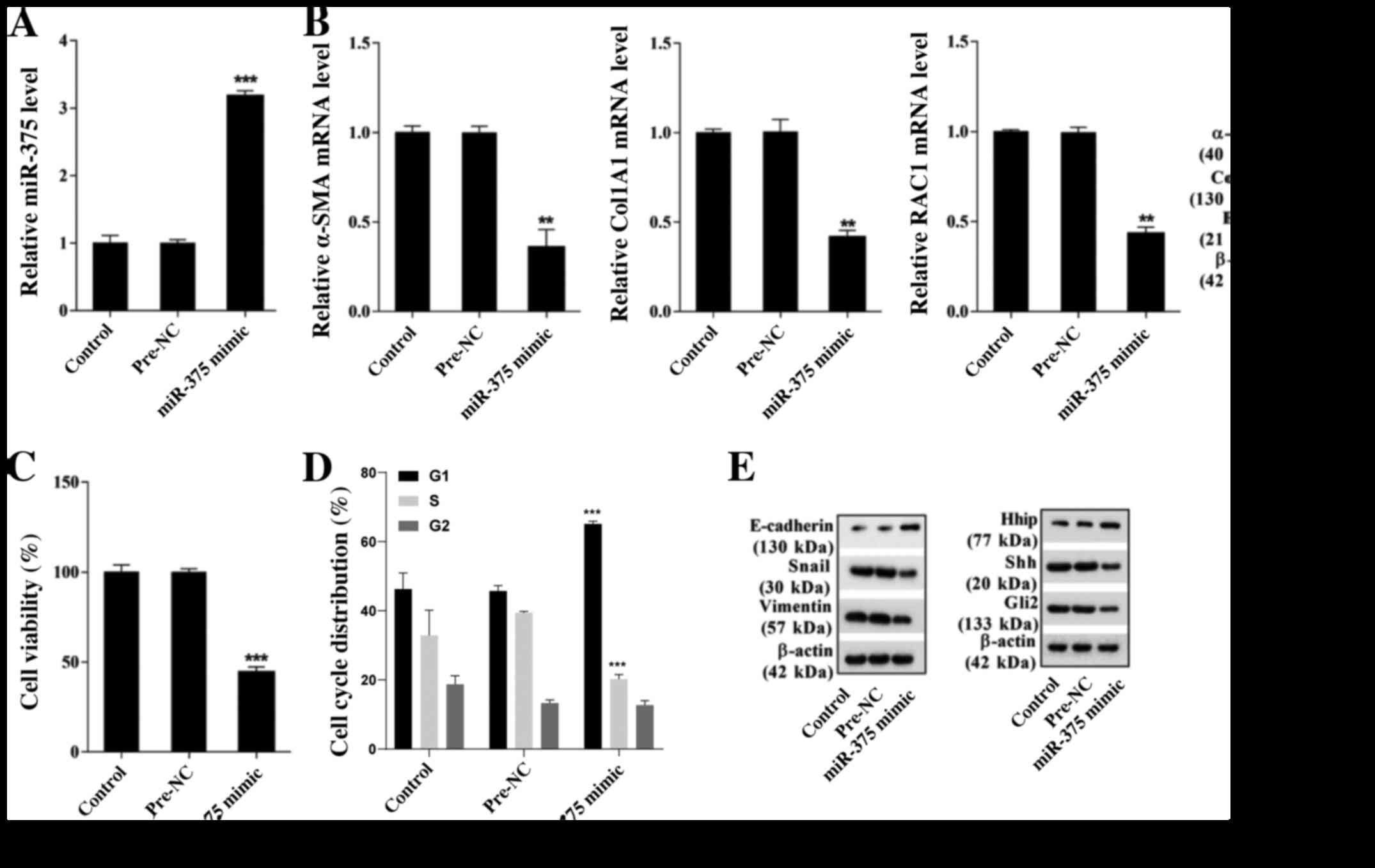 | Figure 2.Effect of miR-375 on mouse HSC
viability, EMT and liver fibrosis. HSCs were isolated from healthy
control mice and cultured in vitro for 1 day to obtain
primary-1-day HSCs. Primary-1-day HSCs were transfected with
miR-375 mimic or pre-NC. (A) miR-375 expression. (B) α-SMA, Col1A1
and RAC1 mRNA and protein expression levels. (C) The MTT assay was
performed to assess HSC viability. (D) Flow cytometry was performed
to assess the cell cycle distribution in HSCs. (E) The protein
expression levels of epithelial marker E-cadherin, mesenchymal
markers Snail and Vimentin, and hedgehog signaling pathway-related
molecules Hhip, Shh and Gli2. **P<0.01 and ***P<0.001 vs.
pre-NC. miR, microRNA; HSC, hepatic stellate cell; EMT,
epithelial-mesenchymal transition; NC, negative control; α-SMA,
α-smooth muscle actin; Col1A1, collagen type I-α1; RAC1, Rac family
small GTPase 1; Snail, snail family transcriptional repressor 1;
Hhip, hedgehog interacting protein; Shh, sonic hedgehog signaling
molecule; Gli2, GLI family zinc finger 2. |
miR-375 inhibits mouse HSC viability
and EMT via the Hh signaling pathway
miR-375-mediated effects on mouse HSC viability and
EMT were further investigated. Primary-1-day HSCs were transfected
with miR-375 mimic and at 48 h post-transfection, cells were
cultured in DMEM supplemented with 0.3 µM SAG (an exogenous Hh
agonist) for 24 h. Compared with pre-NC, miR-375 overexpression
markedly decreased the mRNA and protein expression levels of α-SMA
and Col1A1, which was reversed by treatment with SAG (Fig. 3A). Moreover, the western blotting
results indicated that miR-375 overexpression notably increased the
protein expression levels of E-cadherin and Hhip, but decreased the
expression levels of Snail, Vimentin, Shh and Gli2 compared with
the pre-NC group. miR-375 overexpression-mediated effects on
protein expression were markedly reversed by treatment with SAG
(Fig. 3B). The MTT assay results
demonstrated that compared with the pre-NC group, miR-375
overexpression significantly inhibited mouse HSC viability, which
was significantly reversed by treatment with SAG (Fig. 3C). The cell cycle analysis results
demonstrated that compared with the pre-NC group, miR-375
overexpression significantly inhibited cell cycle progression in
mouse HSCs, which was also significantly reversed by treatment with
SAG (Figs. 3D and S1B). The results further indicated that
miR-375 inhibited mouse HSC viability and EMT via the Hh signaling
pathway.
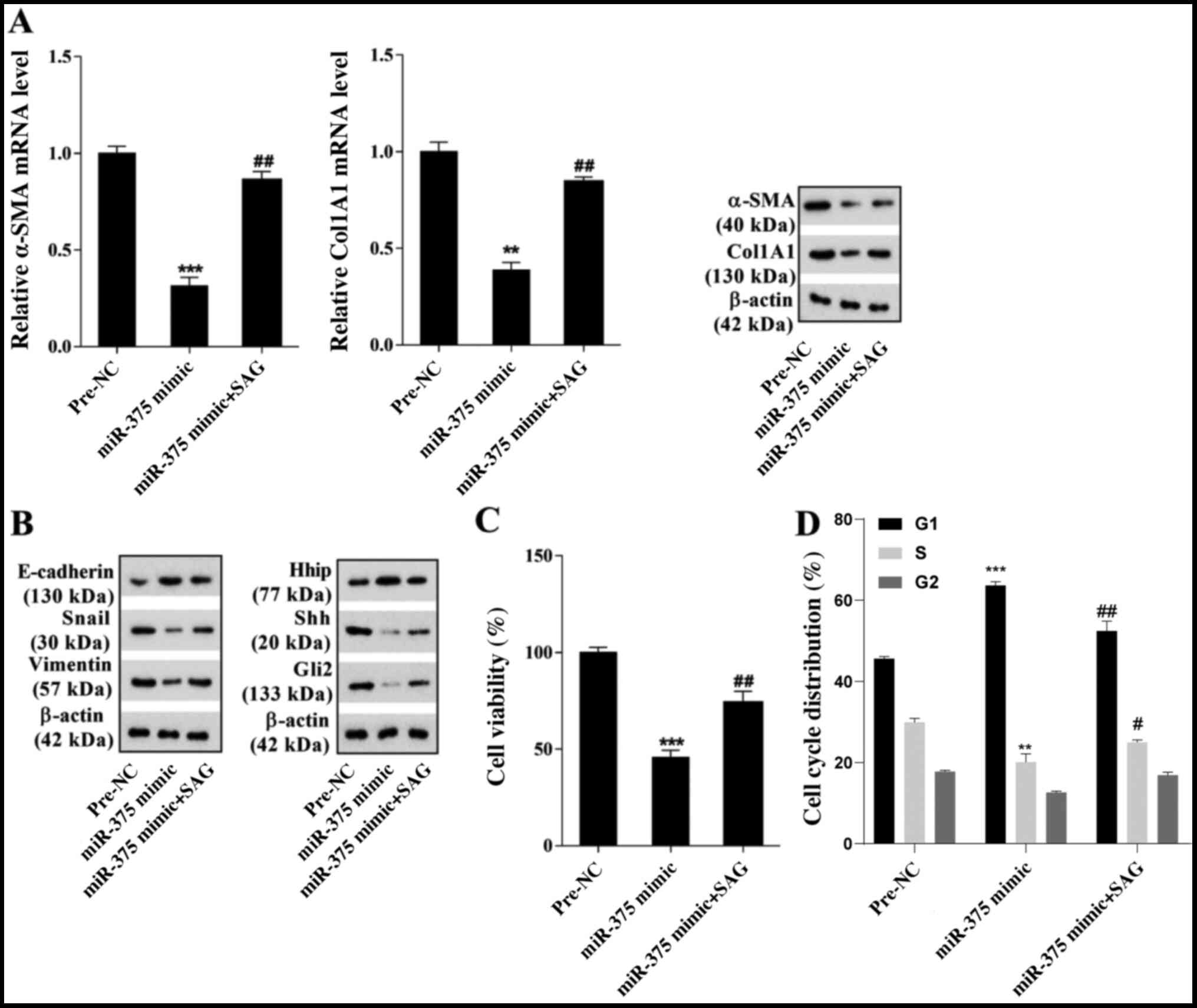 | Figure 3.miR-375 affects mouse HSC viability
and EMT via the Hh signaling pathway. Primary-1-day HSCs were
transfected with miR-375 mimic and at 48 h post-transfection, cells
were cultured in DMEM supplemented with 0.3 µM SAG (an exogenous Hh
agonist) for 24 h. (A) α-SMA and Col1A1 mRNA and protein expression
levels. (B) E-cadherin, Snail, Vimentin, Hhip, Shh and Gli2 protein
expression levels. (C) The MTT assay was performed to assess HSC
viability. (D) Flow cytometry was performed to assess the cell
cycle distribution in HSCs. **P<0.01 and ***P<0.001 vs.
pre-NC; #P<0.05 and ##P<0.01 vs.
miR-375 mimic. miR, microRNA; HSC, hepatic stellate cell; EMT,
epithelial-mesenchymal transition; Hh, hedgehog; SAG, smoothened
agonist; α-SMA, α-smooth muscle actin; Col1A1, collagen type I-α1;
Snail, snail family transcriptional repressor 1; Hhip, hedgehog
interacting protein; Shh, sonic hedgehog signaling molecule; Gli2,
GLI family zinc finger 2; NC, negative control. |
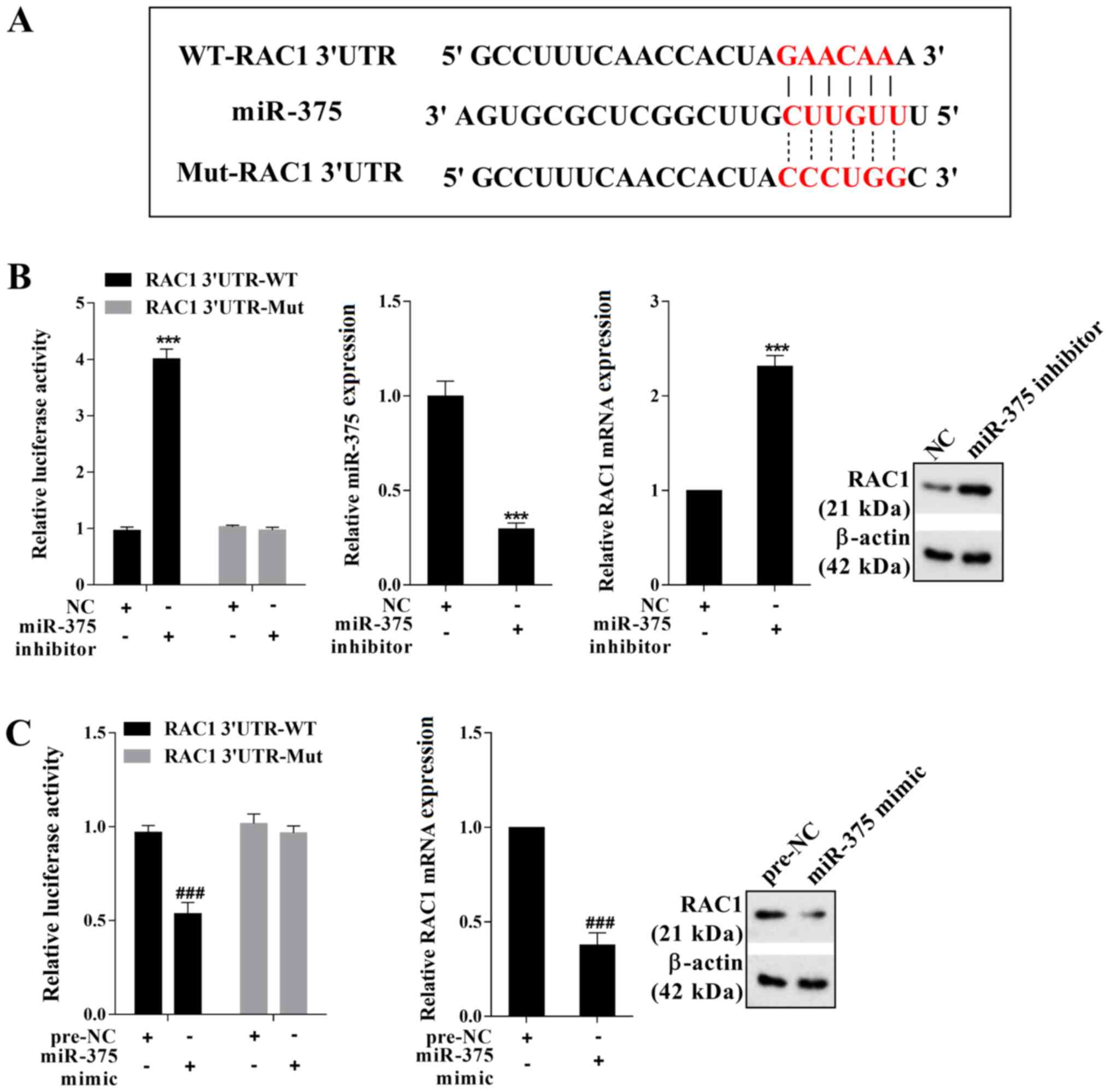
miR-375 negatively regulates RAC1
expression
To determine the mechanism underlying
miR-375-mediated regulation of HSC viability and EMT in mice, the
online bioinformatics software TargetScan was used. The results
indicated that RAC1 was a potential target gene of miR-375
(Fig. 4A). The transfection
efficiency of miR-375 inhibitor was presented in Fig. 4B. The dual-luciferase reporter gene
assay results indicated that miR-375 negatively regulated the
luciferase activity of RAC1 3′UTR-WT (Fig. 4B and C). Moreover, miR-375
negatively regulated the mRNA and protein expression levels of RAC1
(Fig. 4B and C). Therefore, the
aforementioned results indicated that miR-375 negatively regulated
RAC1 expression by binding to the 3′UTR of RAC1.
miR-375 inhibits mouse HSC viability
and EMT by regulating the Hh signaling pathway via RAC1
To further assess whether miR-375 regulated mouse
HSC viability and EMT via targeting RAC1, miR-375 mimic, miR-375
mimic + pcDNA-RAC1 and their corresponding controls were
transfected into mouse primary-1-day HSC.
Compared with the corresponding control groups, RAC1
overexpression significantly increased RAC1 expression, indicating
successful overexpression of RAC1 in mouse primary-1-day HSCs
(Fig. 5A). The RT-qPCR and western
blotting results indicated that compared with the pre-NC group,
miR-375 overexpression markedly decreased the mRNA and protein
expression levels of RAC1, α-SMA and Col1A1, which was reversed by
co-transfection with pcDNA-RAC1 (Fig.
5B-D). Moreover, compared with the pre-NC group, miR-375
overexpression notably increased the protein expression levels of
E-cadherin and Hhip, and decreased the protein expression levels of
Snail, Vimentin, Shh and Gli2. miR-375 overexpression-mediated
effects on protein expression were reversed by co-transfection with
pcDNA-RAC1 (Fig. 5D and E).
Furthermore, compared with the pre-NC group, miR-375 overexpression
significantly inhibited mouse HSC viability, which was
significantly reversed by co-transfection with pcDNA-RAC1 (Fig. 5F). The cell cycle analysis results
suggested that compared with the pre-NC group, miR-375
overexpression significantly restrained cell cycle progression in
mouse HSCs, which was reversed by co-transfection with pcDNA-RAC1
(Figs. 5G and S1C). The results indicated that miR-375
inhibited mouse HSC viability and EMT by regulating the Hh
signaling pathway via RAC1.
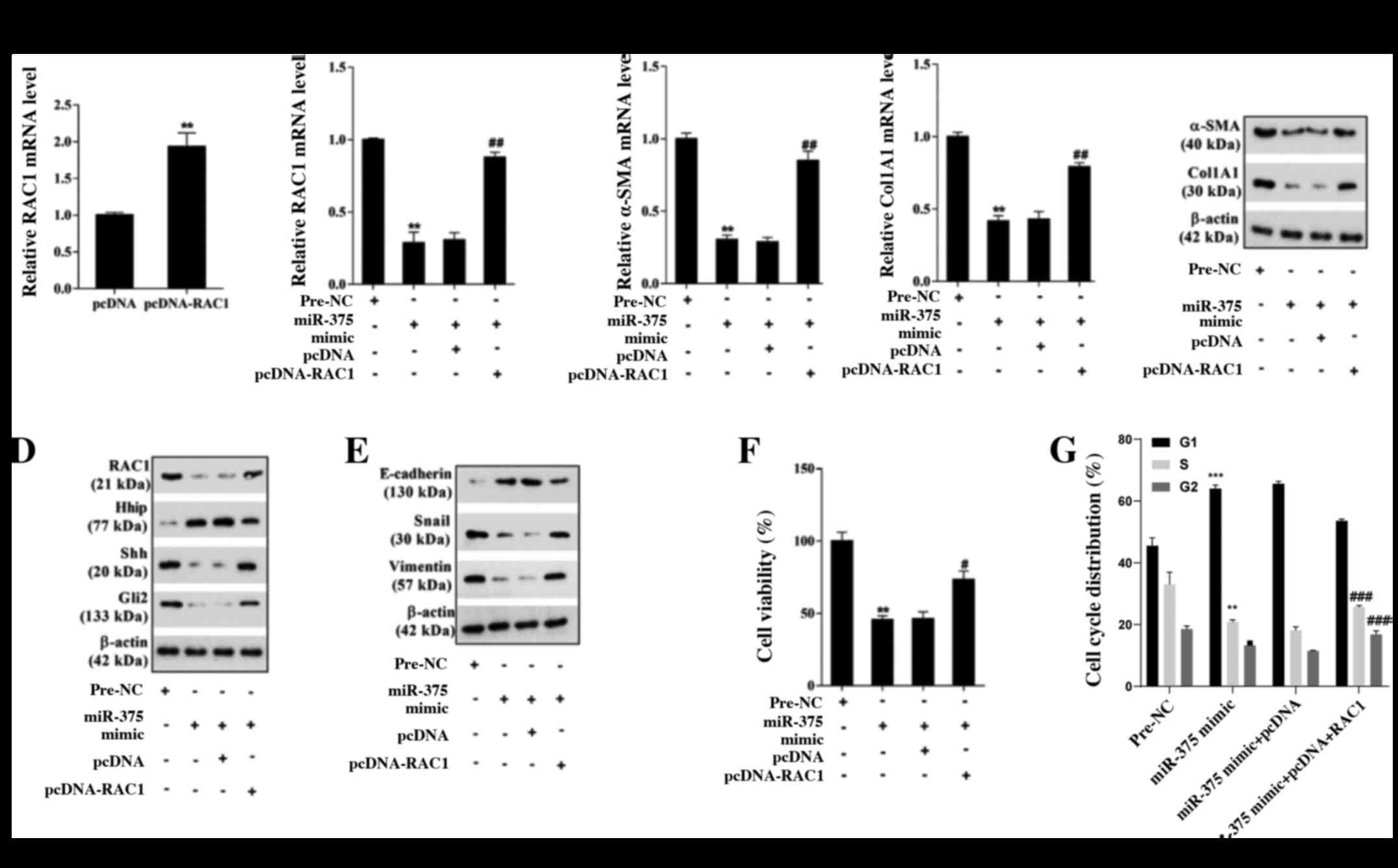 | Figure 5.miR-375 affects mouse HSC viability
and EMT by regulating the hedgehog signaling pathway via RAC1.
Primary-1-day HSCs were transfected with miR-375 mimic, miR-375
mimic + pcDNA-RAC1 or their corresponding controls. Transfection
efficiency of (A) pcDNA-RAC1. (B) RAC1 mRNA expression levels. (C)
α-SMA and Col1A1 mRNA and protein expression levels. (D) RAC1,
Hhip, Shh, Gli2, (E) E-cadherin, Snail and Vimentin protein
expression levels. (F) The MTT assay was performed to assess HSC
viability. (G) Flow cytometry was performed to assess the cell
cycle distribution in HSCs. **P<0.01 and ***P<0.001 vs. pcDNA
or pre-NC; #P<0.05, ##P<0.01 and
###P<0.001 vs. miR-375 mimic + pcDNA. miR, microRNA;
HSC, hepatic stellate cell; EMT, epithelial-mesenchymal transition;
RAC1, Rac family small GTPase 1; α-SMA, α-smooth muscle actin;
Col1A1, collagen type I-α1; Hhip, hedgehog interacting protein;
Shh, sonic hedgehog signaling molecule; Gli2, GLI family zinc
finger 2; Snail, snail family transcriptional repressor 1; NC,
negative control. |
Discussion
A previous study explored the potential mechanism
underlying miRNAs in liver fibrosis (20). The present study demonstrated that
miR-375 expression was significantly downregulated in liver
fibrosis tissues and cells compared with healthy control tissues
and HCs, respectively. Moreover, the results suggested that miR-375
interacted with RAC1. The further mechanistic studies indicated
that miR-375 regulated the Hh signaling pathway via RAC1 to inhibit
HSC viability and EMT, thus inhibiting liver fibrosis.
A previous study indicated that miRNAs bind to the
3′UTR of their target mRNAs to negatively regulate the expression
of the target mRNA, thus serving key regulatory functions in the
occurrence and development of liver fibrosis (21). For example, miR-146a is
downregulated in liver fibrosis tissues, and miR-146a
overexpression reduces hepatic EMT by targeting SMAD4, thus
inhibiting the progression of liver fibrosis (22). Moreover, miR-375 has been reported
to be downregulated in the liver fibrosis rat model (10). However, the function of miR-375 in
liver fibrosis is not completely understood. The present study
demonstrated that miR-375 expression was significantly
downregulated in mouse liver fibrosis tissues and cells compared
with sham tissues and HCs, respectively. Moreover, compared with
the pre-NC group, miR-375 overexpression inhibited mouse HSC
viability and EMT via the Hh signaling pathway, thus alleviating
liver injury in liver fibrosis model mice.
RAC1, an important member of the Rho family of small
GTPase proteins, is associated with the occurrence and development
of various human diseases, including Alzheimer's disease (23,24). A
previous study reported that RAC1 knockdown inhibits the
development of liver fibrosis in mice (25). Choi et al reported that RAC1
activation induces HSC activation and promotes HSC EMT by mediating
the Hh signaling pathway, thus improving liver fibrosis) and
suggesting that RAC1 is involved in the development of liver
fibrosis. Moreover, Venugopal et al demonstrated that RAC1
expression is negatively regulated by miR-194 and serves a role in
liver fibrosis by regulating HSC activation (26). The present study identified binding
sites between RAC1 and miR-375 using TargetScan online
bioinformatics prediction software. Moreover, the results indicated
that RAC1 expression was negatively regulated by miR-375. Further
in-depth studies suggested that miR-375 affected the Hh signaling
pathway by downregulating RAC1 to restrain HSC viability and
EMT.
To conclude, the present study demonstrated that
miR-375 expression was significantly downregulated in liver
fibrosis tissues and cells compared with healthy control tissues
and HCs, respectively. The results indicated that miR-375 regulated
the Hh signaling pathway via RAC1 to inhibit HSC viability and EMT,
thus relieving liver fibrosis. The results of the present study
suggested that the miR-375/RAC1 axis may serve as a novel
therapeutic target for liver fibrosis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the study, wrote the manuscript and
performed the experiments. JL participated in performing the
experiments and writing the manuscript. LZ acquired, analyzed and
interpreted the data. YD substantially contributed to the
conception and design of the study, and critically revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Zhengzhou University
(approval no. 2020-KY-047). Written informed consent was obtained
from all patients before obtaining liver tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Breitkopf-Heinlein K, Meyer C, König C,
Gaitantzi H, Addante A, Thomas M, Wiercinska E, Cai C, Li Q, Wan F,
et al: BMP-9 interferes with liver regeneration and promotes liver
fibrosis. Gut. 66:939–954. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li W, Zhou C, Fu Y, Chen T, Liu X, Zhang Z
and Gong T: Targeted delivery of hyaluronic acid nanomicelles to
hepatic stellate cells in hepatic fibrosis rats. Acta Pharm Sin B.
10:693–710. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu F, Geng W, Dong P, Huang Z and Zheng J:
LncRNA-MEG3 inhibits activation of hepatic stellate cells through
SMO protein and miR-212. Cell Death Dis. 9:10142018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi SS, Syn WK, Karaca GF, Omenetti A,
Moylan CA, Witek RP, Agboola KM, Jung Y, Michelotti GA and Diehl
AM: Leptin promotes the myofibroblastic phenotype in hepatic
stellate cells by activating the hedgehog pathway. J Biol Chem.
285:36551–36560. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu F, Zheng Y, Hong W, Chen B, Dong P and
Zheng J: MicroRNA-200a suppresses epithelial-to-mesenchymal
transition in rat hepatic stellate cells via GLI family zinc finger
2. Mol Med Rep. 12:8121–8128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu F, Lu Z, Chen B, Wu X, Dong P and Zheng
J: Salvianolic acid B-induced microRNA-152 inhibits liver fibrosis
by attenuating DNMT1-mediated Patched1 methylation. J Cell Mol Med.
19:2617–2632. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang C, Luo J, Chen Z, Ye M, Hong Y, Liu
J, Nie J, Zhao Q and Chang Y: MiR-375 impairs the invasive
capabilities of hepatoma cells by targeting HIF1α under hypoxia.
Dig Dis Sci. Mar 25–2020.(Epub ahead of print). doi:
10.1007/s10620-020-06202-9. View Article : Google Scholar
|
|
10
|
Yang YZ, Zhao XJ, Xu HJ, Wang SC, Pan Y,
Wang SJ, Xu Q, Jiao RQ, Gu HM and Kong LD: Magnesium
isoglycyrrhizinate ameliorates high fructose-induced liver fibrosis
in rat by increasing miR-375-3p to suppress JAK2/STAT3 pathway and
TGF-β1/Smad signaling. Acta Pharmacol Sin. 40:879–894. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ungefroren H, Witte D and Lehnert H: The
role of small GTPases of the Rho/Rac family in TGF-β-induced EMT
and cell motility in cancer. Dev Dyn. 247:451–461. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choi SS, Sicklick JK, Ma Q, Yang L, Huang
J, Qi Y, Chen W, Li YX, Goldschmidt-Clermont PJ and Diehl AM:
Sustained activation of Rac1 in hepatic stellate cells promotes
liver injury and fibrosis in mice. Hepatology. 44:1267–1277. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Choi SS, Witek RP, Yang L, Omenetti A, Syn
WK, Moylan CA, Jung Y, Karaca GF, Teaberry VS, Pereira TA, et al:
Activation of Rac1 promotes hedgehog-mediated acquisition of the
myofibroblastic phenotype in rat and human hepatic stellate cells.
Hepatology. 52:278–290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ge B, Wu H, Shao D, Li S and Li F:
Interfering with miR-24 alleviates rotenone-induced dopaminergic
neuron injury via enhancing autophagy by up-regulating DJ-1. Aging
Pathobiol Ther. 1:17–24. 2019. View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu F, Chen B, Dong P and Zheng J: HOTAIR
epigenetically modulates PTEN expression via MicroRNA-29b: A novel
mechanism in regulation of liver fibrosis. Mol Ther. 25:205–217.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou X, Xiong J, Lu S, Luo L, Chen ZL,
Yang F, Jin F, Wang Y, Ma Q, Luo YY, et al: Inhibitory effect of
corilagin on miR-21-regulated hepatic fibrosis signaling pathway.
Am J Chin Med. 47:1541–1569. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu JC, Chen R, Luo X, Li ZH, Luo SZ and Xu
MY: MicroRNA-194 inactivates hepatic stellate cells and alleviates
liver fibrosis by inhibiting AKT2. World J Gastroenterol.
25:4468–4480. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li BB, Li DL, Chen C, Liu BH, Xia CY, Wu
HJ, Wu CQ, Ji GQ, Liu S, Ni W, et al: Potentials of the elevated
circulating miR-185 level as a biomarker for early diagnosis of
HBV-related liver fibrosis. Sci Rep. 6:341572016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei S, Wang Q, Zhou H, Qiu J, Li C, Shi C,
Zhou S, Liu R and Lu L: miR-455-3p alleviates hepatic stellate cell
activation and liver fibrosis by suppressing HSF1 expression. Mol
Ther Nucleic Acids. 16:758–769. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang L, Dong C, Yang J, Yang L, Chang N,
Qi C and Li L: MicroRNA-26b-5p inhibits mouse liver fibrogenesis
and angiogenesis by targeting PDGF receptor-beta. Mol Ther Nucleic
Acids. 16:206–217. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zou Y, Li S, Li Z, Song D, Zhang S and Yao
Q: MiR-146a attenuates liver fibrosis by inhibiting transforming
growth factor-β1 mediated epithelial-mesenchymal transition in
hepatocytes. Cell Signal. 58:1–8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kikuchi M, Sekiya M, Hara N, Miyashita A,
Kuwano R, Ikeuchi T, Iijima KM and Nakaya A: Disruption of a
RAC1-centred network is associated with Alzheimer's disease
pathology and causes age-dependent neurodegeneration. Hum Mol
Genet. 29:817–833. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kai Y, Kon R, Ikarashi N, Chiba Y, Kamei J
and Sakai H: Role of Rac1 in augmented endothelin-1-induced
bronchial contraction in airway hyperresponsive mice. J Pharmacol
Sci. 141:106–110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bopp A, Wartlick F, Henninger C, Kaina B
and Fritz G: Rac1 modulates acute and subacute genotoxin-induced
hepatic stress responses, fibrosis and liver aging. Cell Death Dis.
4:e5582013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Venugopal SK, Jiang J, Kim TH, Li Y, Wang
SS, Torok NJ, Wu J and Zern MA: Liver fibrosis causes
downregulation of miRNA-150 and miRNA-194 in hepatic stellate
cells, and their overexpression causes decreased stellate cell
activation. Am J Physiol Gastrointest Liver Physiol. 298:G101–G106.
2010. View Article : Google Scholar : PubMed/NCBI
|