Introduction
Breast cancer, which is one of the most prevalent
and aggressive malignancies, is the leading cause of cancer-related
deaths among women worldwide (1,2). Tumor
metastasis is associated with the high mortality rates in patients
with breast cancer (3). Despite
significant advances in the systematic treatment of breast cancer,
which has significantly improved the prognosis rates of patients,
the treatment effect on patients with metastatic breast cancer
remains unsatisfactory (4).
Additionally, chemotherapy resistance is also a major issue in the
treatment of patients with breast cancer (5). Therefore, the molecular mechanisms of
tumorigenesis and development of breast cancer should be further
explored, and new therapeutic targets identified to develop a novel
strategy for breast cancer treatment.
It has been demonstrated that the occurrence and
progression of breast cancer are complex multi-step processes,
which involves genetic alterations and epigenetic modifications
(6). Accumulating evidence has
demonstrated that microRNAs (miRNAs), a class of small non-coding
RNAs, can either promote the degradation of mRNA or inhibit the
translation of the target gene by binding to its 3′untraslated
region (UTR), thereby regulating the expression of the target gene
at the post-transcriptional level (7,8).
Previous studies have revealed that some miRNAs are dysregulated in
various cancers and it has been proposed that they have key roles
in carcinogenesis and tumor progression by regulating the
expression of oncogenes or tumor suppressor genes (9,10).
Recently, several studies have demonstrated that miRNAs play
critical roles in the progression of breast cancer, and may even
function as promising biomarkers or prognostic markers (11–13).
miR-653, located at chromosome 7q21.3, has been previously reported
to be involved in the proliferation and apoptosis of mice
thymocytes by targeting tripartite motif containing 9 (14). Furthermore, Yuan et al
(15) demonstrated that miR-653
regulates cell proliferation, invasion and apoptosis in non-small
cell lung cancer. However, the role of miR-653-5p in the
progression of breast cancer are yet to be elucidated.
In the present study, the biological role of
miR-653-5p in the progression of breast cancer and the underlying
mechanism were investigated. Herein, the data demonstrated that
miR-653-5p mimics inhibited the proliferation, migration and
invasion of breast cancer cells, as well as promoting cell
apoptosis, suggesting the suppressive role of miR-653-5p in breast
cancer. Moreover, investigation into the underlying mechanism
showed that miR-653-5p acted as a tumor suppressor in breast cancer
cells by targeting mitogen-activated protein kinase 6 (MAPK6).
Materials and methods
Cell culture and transfection
Human breast cancer cell lines MCF-7 and MDA-MB-231
were obtained from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences and cultured in RPMI-1640 medium
(HyClone; GE Healthcare Life Sciences) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 U/ml;
Sigma-Aldrich; Merck KGaA) and streptomycin (0.1 mg/ml;
Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2.
miR-653-5p mimics (5′-GUGUUGAAACAAUCUCUACUG-3′) and
miRNA negative control (miR-NC; 5′-UUUGGUAAAAUUCAACCAGCUA-3′) were
synthesized by Guangzhou RiboBio Co., Ltd.. The pcDNA3.1-MAPK6
plasmid was constructed by Guangzhou RiboBio Co., Ltd., the blank
pcDNA3.1 vector was used as control. Cells (2×105
cells/well) were plated into 6-well plates and incubated at 37°C
with 5% CO2 overnight. At 40–60% confluence, MCF-7 and
MDA-MB-231 cells were transfected with miR-653-5p mimics (50 nM),
miR-NC (50 nM), pcDNA3.1-MAPK6 (5 µg) and pcDNA3.1 (5 µg) using
Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc.), following the manufacturer's protocols.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA extracted from transfected MCF-7 and
MDA-MB-231 cells with an Ultrapure RNA kit (CWBio) were reverse
transcribed into cDNA at 42°C for 50 min and 85°C for 5 min with
the miRNA cDNA Synthesis kit (CWBio), following the manufacturer's
manual. RT-qPCR analysis was then performed following the
instructions of the miRNA qPCR Assay kit (CWBio). The following
thermocycling conditions were used for qPCR: Initial denaturation
at 95°C for 10 min; followed by 40 cycles of 95°C for 15 sec and
60°C for 1 min; and final extension at 72°C for 50 sec. miR-653-5p
and MAPK6 expression levels were calculated using the
2−∆∆Cq method (16) and
normalized to the internal reference genes U6 and GAPDH,
respectively. The following primers were used for qPCR: miR-653-5p
forward, 5′-GTGTTGAAACAATCTCTACTG-3′ and reverse,
5′-GAACATGTCTGCGTATCTC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′; MAPK6 forward,
5′-AGCGCTAGAGGAAGCATCAC-3′ and reverse, 5′-GTGGGATGCCTATGGACTCG-3′;
and GAPDH forward, 5′-TATGATGATATCAAGAGGGTAGT-3′ and reverse,
5′-TGTATCCAAACTCATTGTCATAC-3′.
Cell counting kit-8 (CCK-8) assay
Following 24 h of transfection, ~1×103
breast cancer cells were plated into a 96-well plate and cultured
at 37°C. CCK-8 reagent (10 µl; Beijing Solarbio Science &
Technology Co., Ltd.) was added into each well of the plate at 0,
24, 48 and 72 h, according to the manufacturer's instructions.
Following incubation at 37°C for 1.5 h, the absorbance at 450 nm
was measured with a microplate reader.
Colony formation assay
Following transfection for 24 h, cells were
collected and digested with 0.25% trypsin. Cells were seeded into
60 mm plates with a density of 5×102 cells/plate and
cultured for 1–2 weeks. After visible colonies were formed, cells
were fixed in 4% paraformaldehyde at room temperature for 30 min
then stained with 0.1% crystal violet stain at room temperature for
30 min. Finally, the number of colonies was counted. Colonies were
defined as the cell population (>50 cells) of the descendants of
>6 generations of a single proliferating cell in
vitro.
Transwell assay
Transwell chambers (EMD Millipore) with or without
Matrigel (BD Biosciences) for 1 h at 37°C in 24-well plates were
used to assess cell migration and invasion in vitro.
Briefly, following transfection for 24 h, cells were suspended in
serum-free medium, then 1×105 cells were added into the
top chamber. RPMI-1640 medium (500 µl) supplemented with 20% FBS
was used as a chemoattractant and added into the bottom chamber.
Following incubation for 24 h, the migrating or invading cells were
fixed in 4% paraformaldehyde for 30 min at room temperature then
stained with 0.1% crystal violet for another 20 min at room
temperature. Stained cells were captured and counted in nine
randomly selected fields of view under a light microscope
(magnification, ×100). The number of migratory or invasive cells
was quantified using ImageJ (version 6.0; National Institutes of
Health).
Flow cytometry
Breast cancer cells transfected with miR-653-5p
mimics or miR-NC for 24 h were collected and cultured in serum-free
medium for another 24 h. After cells were digested with trypsin and
centrifuged at 999 × g for 5 min at 4°C, they were suspended with
buffer solution and the cell density was adjusted to
1–5×106 cells/ml. Cell suspension (100 µl) was stained
with Annexin V/FITC and PI (BioVision, Inc.) at room temperature
for 5 min in the dark. The proportion of apoptotic cells was
analyzed by flow cytometry (BD FACSCanto II; BD Biosciences) and
FlowJo software (version 7.6.5; FlowJo LLC). The proportion of
apoptotic cells was calculated as the sum of early (Annexin
V/FITC+/PI-) and late (Annexin V/FITC+/PI+) apoptotic cells.
Western blot analysis
After 48 h of transfection, total protein was
extracted from the breast cancer cells using RIPA lysis buffer
(CWBio) supplemented with protease inhibitor cocktail (CWBio) and
quantified using a bicinchoninic acid assay kit (Beyotime Institute
of Biotechnology). Total protein (20 µg) for each group were loaded
on a 10% gel, resolved using SDS-PAGE and subsequently transferred
to the PVDF membrane (EMD Millipore). The membranes were blocked
with 5% skimmed milk for 1 h at room temperature and incubated
overnight at 4°C with the following primary antibodies (Cell
Signaling Technology, Inc.): Anti-E-cadherin (1:1,000; cat. no.
14472), anti-N-cadherin (1:1,000; cat. no. 4061), anti-Vimentin
(1:1,000; cat. no. 49636), anti-Bcl-2 (1:1,000; cat. no. 15071),
anti-Bax (1:1,000; cat. no. 2772), anti-cleaved Caspase 3 (1:1,000;
cat. no. 9661), anti-Akt (1:1,000; cat. no. 9272), anti-p-Akt
(1:1,000; Ser473; cat. no. 4060), anti-mTOR (1:1,000; cat. no.
2972), anti-p-mTOR (1:1,000; Ser2448; cat. no. 2971), anti-MAPK6
(1:1,000; cat. no. 4067) and anti-GAPDH (1:1,000; cat. no. 2118).
The blots were incubated with the horseradish peroxidase-conjugated
secondary antibodies (1:3,000; cat. no. SA00001-2; ProteinTech
Group, Inc.) for 1 h at room temperature and visualized using an
ECL reagent (CWBio). Signals were analyzed with Quantity One
software (version 4.6.6; Bio-Rad Laboratories, Inc.).
Dual-luciferase reporter assay
TargetScan (version 7.2; www.targetscan.org/vert_72) and StarBase (version 2.0;
starbase.sysu.edu.cn/starbase2/index.php) were used to
predict the potential target gene of miR-653-5p. The wild-type (wt)
or mutant-type (mut) of the 3′UTR of MAPK6 was cloned into the
pmirGLO vector (Promega Corporation). 293T cells (1×106)
were co-transfected with pmirGLO-MAPK6-wt/-mut plasmid (1 µg) and
50 nM miR-653-5p mimics or miR-NC using Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.).
Following incubation for 48 h, cells were collected and luciferase
activity was measured using the Dual-luciferase Report Assay system
(Promega Corporation). Firefly luciferase activity was normalized
to Renilla luciferase activity.
Statistical analysis
Data are expressed as the mean ± SD of three
independent repeats. An unpaired Student's t-test was used for
comparison between two groups, and one-way ANOVA followed by
Tukey's post hoc test was used for multiple comparisons. All
statistical analyses were performed using GraphPad Prism software
7.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate
a statistically significant difference.
Results
Upregulation of miR-653-5p inhibits
the proliferation of breast cancer cells
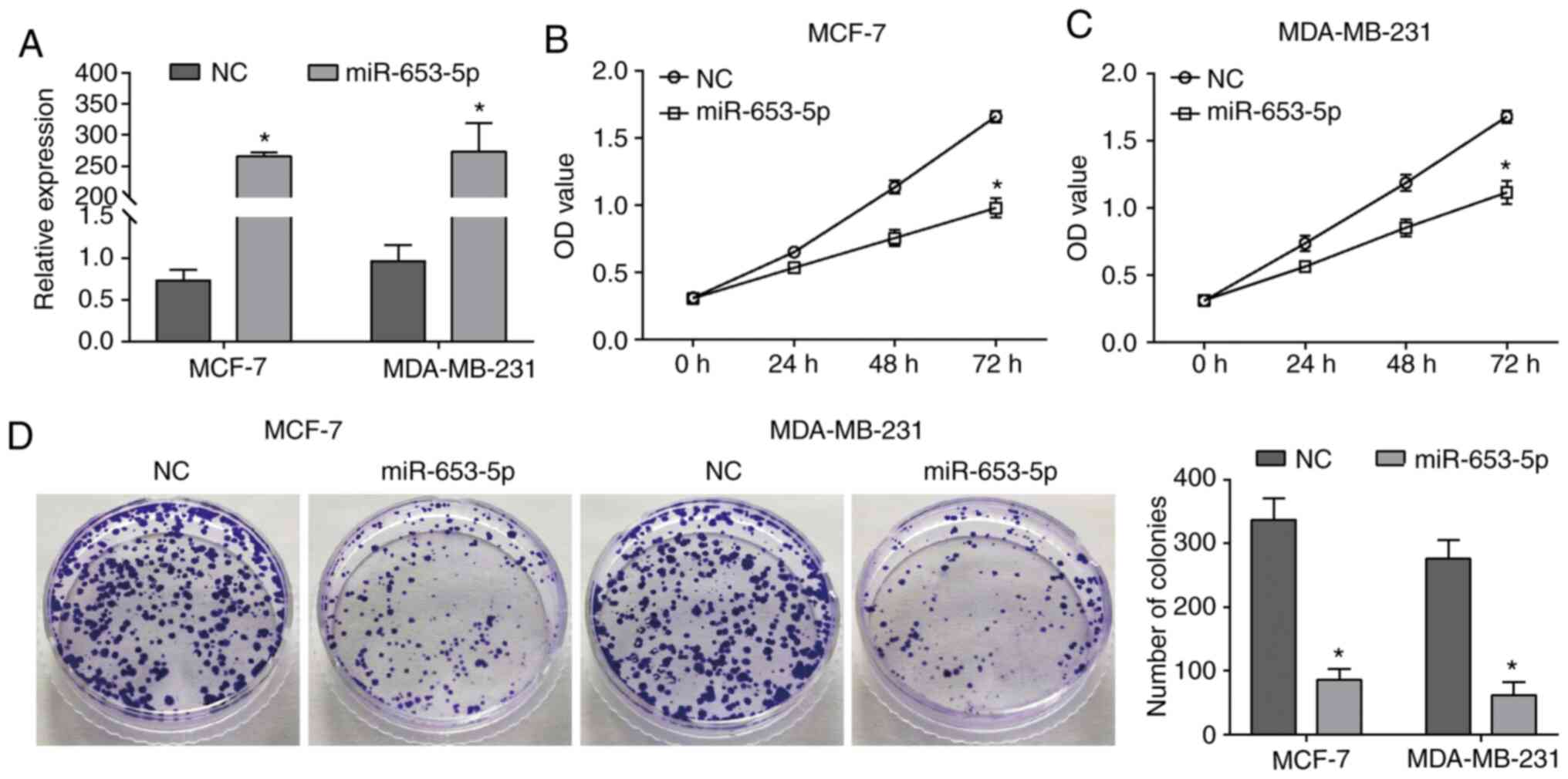
To investigate the function of miR-653-5p in breast
cancer, MCF-7 and MDA-MB-231 cells were transfected with miR-653-5p
mimics to upregulate the expression of miR-653-5p (Fig. 1A). As indicated by the CCK-8 assay,
upregulation of miR-653-5p caused a significant decrease in cell
viability compared with the NC group (Fig. 1B and C). The suppressive effects of
miR-653-5p on cell proliferation was further demonstrated by a
significant decrease in the number of colonies in the miR-653-5p
overexpression group compared with the NC group (Fig. 1D). Therefore, these results
suggested that miR-653-5p functions as a suppressor in the
proliferation of breast cancer cells.
Upregulation of miR-653-5p reduces
cell migration, invasion and epithelial-mesenchymal transition
(EMT) in breast cancer cells
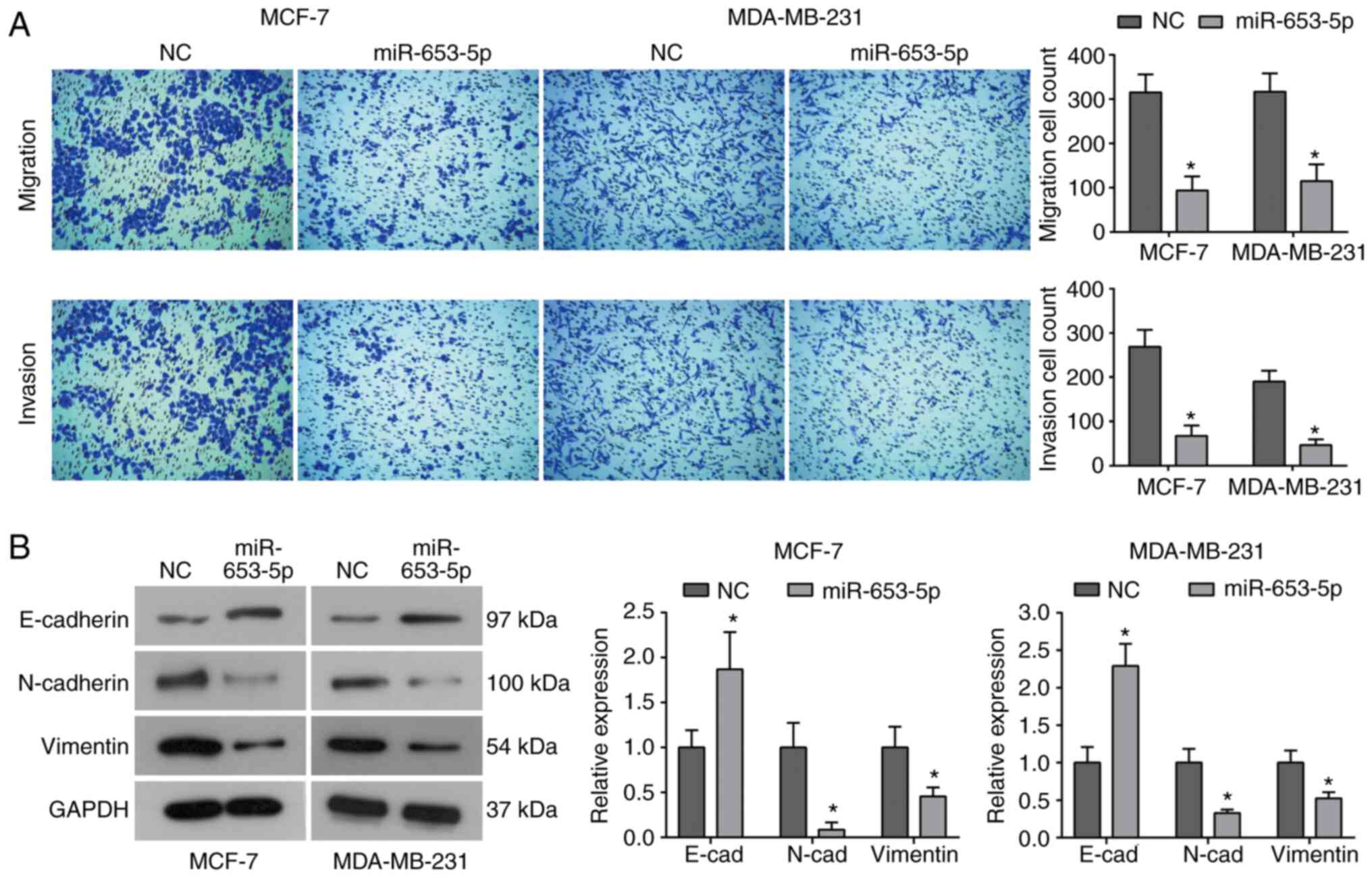
The effect of miR-653-5p on cell migration was
assessed using a Transwell assay. As shown in Fig. 2A, overexpression of miR-653-5p
significantly reduced the migratory activity of both MCF-7 and
MDA-MB-231 cells compared with the control group. Additionally,
cells transfected with miR-653-5p mimics displayed a significant
decrease in the number of cells that invaded through the
Matrigel-coated membrane (Fig. 2A).
As commonly known, EMT is a critical event in the initiation of the
metastatic cascade and contributes to the metastasis of cancer
cells. Therefore, further study was performed to assess the effect
of miR-653-5p on the process of EMT in breast cancer cells. The
data demonstrated that the expression of epithelial marker
E-cadherin was significantly upregulated by miR-653-5p, whereas a
significant decrease in the expression of mesenchymal markers,
N-cadherin and vimentin, was observed in cells transfected with
miR-653-5p mimics (Fig. 2B),
suggesting that miR-653-5p suppresses EMT in breast cancer cells.
Collectively, these data indicated that the inhibitory effect of
miR-653-5p on cell invasion may be related to its ability to
suppress EMT in breast cancer cells.
miR-653-5p induces cell apoptosis and
inhibits the Akt/mammalian target of rapamycin (mTOR) signaling
pathway in breast cancer cells
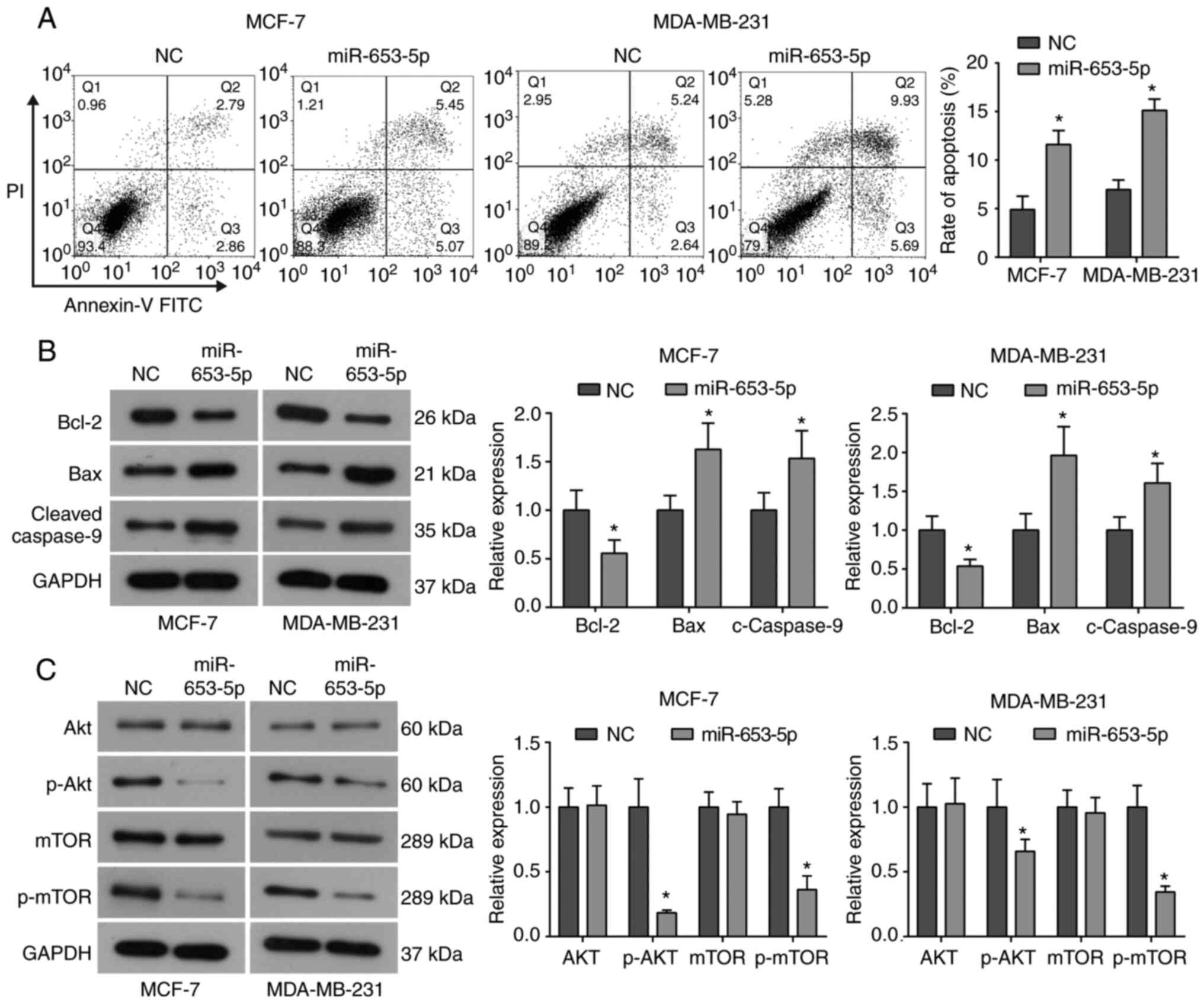
The effect of miR-653-5p on cell apoptosis in breast
cancer cells was investigated using flow cytometry. As indicated in
Fig. 3A, miR-653-5p mimics
increased the apoptotic rate of MCF-7 and MDA-MB-231 cells compared
with the control group. To further investigate the underlying
mechanism of increased apoptosis, western blot analysis was
performed to evaluate the expression of apoptosis-related proteins.
It was demonstrated that, compared with the NC group, miR-653-5p
overexpression downregulated the expression of anti-apoptotic
protein Bcl-2, and upregulated the expression of pro-apoptotic
proteins, Bax and cleaved caspase-9, in both MCF-7 and MDA-MB-231
cells (Fig. 3B). Therefore,
indicating that miR-653-5p promotes apoptosis in breast cancer
cells, possibly by regulating the Bcl-2/Bax axis and activating
caspase-9.
It is commonly known that the Akt/mTOR signaling
pathway plays a critical role in regulating cell proliferation,
differentiation, invasion and survival, as well as tumorigenesis
and progression (17). Thus, the
effect of miR-653-5p on the activation of the Akt/mTOR signaling
pathway was evaluated in breast cancer cells. As determined by
western blotting, it was found that miR-653-5p overexpression did
not affect the total expression of Akt and mTOR, but it did
significantly reduce the phosphorylation of Akt and mTOR in both
MCF-7 and MDA-MB-231 cells (Fig.
3C), suggesting that miR-653-5p inhibits activation of the
Akt/mTOR signaling pathway in breast cancer cells. These results
indicated that the Akt/mTOR signaling pathway may be implicated in
the suppressive role of miR-653-5p in breast cancer cells.
miR-653-5p suppresses the
proliferation and migration of breast cancer cells by targeting
MAPK6
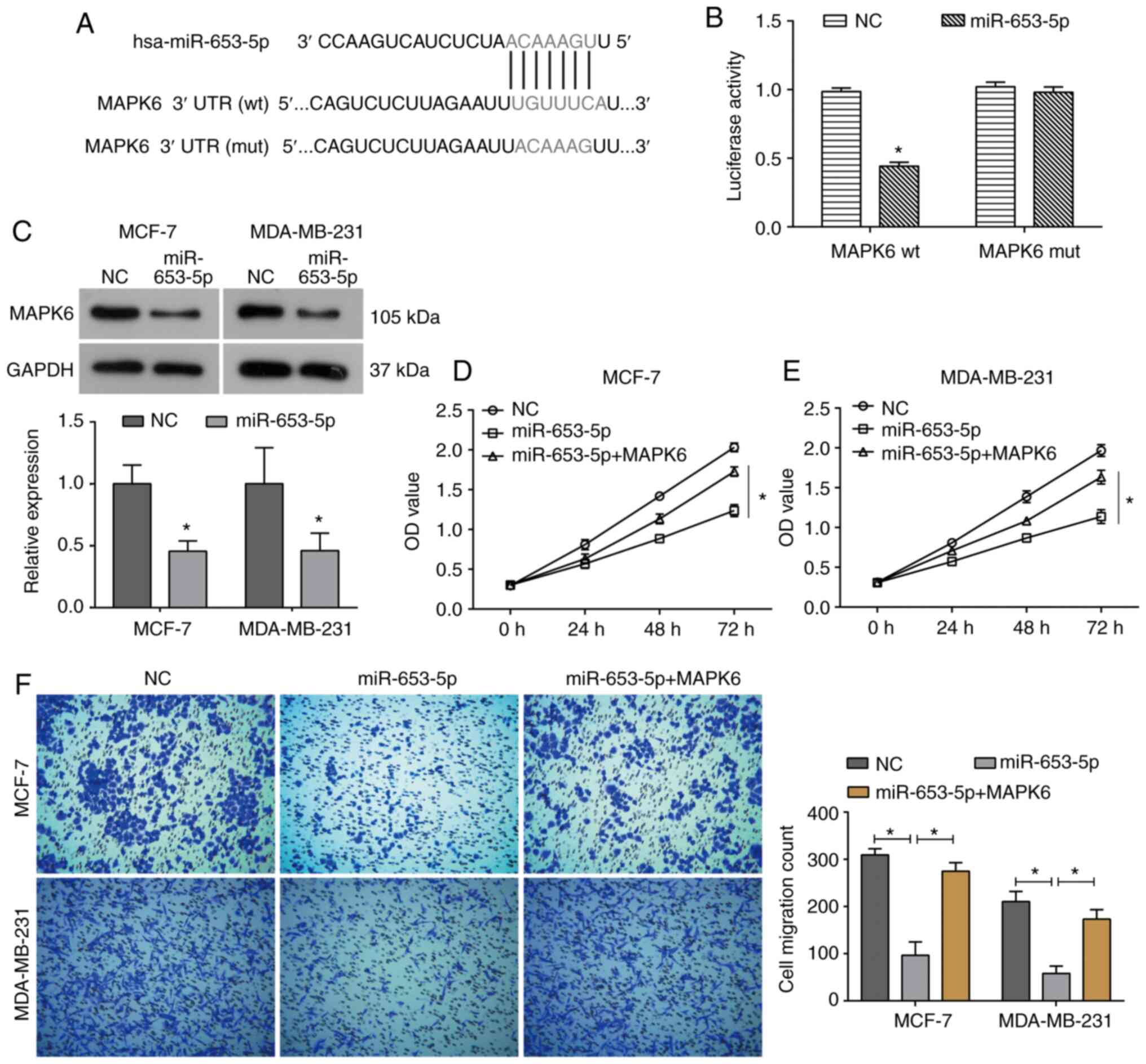
To further elucidate the molecular mechanism by
which miR-653-5p inhibits breast cancer cell proliferation and
migration, the target gene of miR-653-5p was identified by
bioinformatics analysis and a dual-luciferase reporter assay. It
was predicated by bioinformatics analysis (TargetScan and StarBase)
that the 3′UTR of MAPK6 mRNA contained a potential binding site for
miR-653-5p (Fig. 4A). The
dual-luciferase reporter assay demonstrated that miR-653-5p mimics
resulted in a significant decrease in luciferase activity of the wt
3′UTR of MAPK6, but the luciferase activity of the mut 3′UTR of
MAPK6 was not affected by miR-653-5p mimics (Fig. 4B). Moreover, western blotting
further confirmed that miR-653-5p overexpression significantly
decreased the expression of MAPK6 in both MCF-7 and MDA-MB-231
cells (Fig. 4C). MCF-7 and
MDA-MB-231 cells were transfected with pcDNA3.1-MAPK6 to upregulate
its expression (Fig. S1).
Furthermore, upregulation of MPAK6 was able to partially reverse
the miR-653-5p-induced inhibition of cell viability in both MCF-7
and MDA-MB-231 cells (Fig. 4D and
E). Consistently, the decrease in cell migration caused by
miR-653-5p was also reversed by MAPK6 overexpression in breast
cancer cells (Fig. 4F).
Collectively, these data support the hypothesis that miR-653-5p
functions as a tumor suppressor in breast cancer by targeting
MAPK6.
Discussion
It has been reported that miRNAs participate in
regulating the expression of 1/3 of human genes (18,19). A
growing number of studies have revealed that miRNAs are involved in
the occurrence and development of cancers, and function as targets
for cancer diagnosis and treatment (11–13).
As a miRNA, miR-653 has been reported to have roles in cell
proliferation, differentiation and apoptosis, which has been
demonstrated in non-small cell lung cancer cells (14,15).
In the present study, it was found that miR-653-5p mimics
significantly inhibited breast cancer cell proliferation, migration
and invasion. Moreover, upregulation of miR-653-5p increased the
expression of E-cadherin and decreased the expression of N-cadherin
and vimentin, suggesting that miR-653-5p suppresses EMT in breast
cancer cells. Additionally, the results indicated that miR-653-5p
promoted apoptosis by regulating Bcl-2/Bax and cleaved caspase-9.
Collectively, these data suggested that miR-653-5p functions as a
tumor suppressor in the growth and metastasis of breast cancer.
However, there are limitations of the present study. For example,
whether miR-653-5p-induced apoptosis is associated with its effect
on cell migration is unknown, which will be further explored in a
future study. Additionally, how miR-653-5p affects the expression
of EMT-related proteins will be further studied.
As a well-known signaling pathway, Akt/mTOR
signaling plays a key role in regulating various cellular
functions, including cancer cell growth, metastasis and survival
(17). It has been revealed that
the Akt/mTOR signaling pathway is frequently overactivated in
cancers (17). As a result,
targeting the Akt/mTOR signaling pathway is considered to be an
effective approach to control the growth and metastasis of tumors
(20,21). The present study found that
miR-653-5p mimics inhibited the activation of the Akt/mTOR
signaling pathway by decreasing the levels of phosphorylated
(p)-Akt and p-mTOR in breast cancer cells. Therefore, miR-653-5p
may be a potential therapeutic target for breast cancer
treatment.
MAPK6, also known as ERK3, is an atypical MAPK. The
MAPK6 pathway has been reported to be involved in inflammatory
responses, and cell growth and differentiation (22). In addition, as an important
downstream factor of MPAK6, p21 protein activated kinase (PAK) has
been identified, and plays a key role in regulating cell adhesion
and migration by regulating actin cytoskeleton dynamics (23). Therefore, MAPK6 plays an important
role in cell migration. Furthermore, previous studies have
demonstrated that MAPK6 is involved in the metastasis of cancer
cells (24–26). MAPK6 has also been revealed to be
involved in cell proliferation and apoptosis in laryngeal carcinoma
as a target of miR-138 (27). MAPK6
knockdown inhibits the proliferation, migration and invasion of
cervical cancer (28). Lv et
al (29), reported that MAPK6
is upregulated in breast cancer and is associated with poor
prognosis of patients. In the present study, MAPK6 was identified
as a target of miR-653-5p that could be negatively regulated by
miR-653-5p in breast cancer cells. Furthermore, upregulation of
MAPK6 was able to partially reverse the inhibitory effects of
miR-653-5p on the proliferation and migration of breast cancer
cells. Taken together, these results indicated that miR-653-5p
inhibits the progression of breast cancer by targeting MAPK6.
Potapova et al (30)
previously reported that MAPK9-induced apoptosis is highly
dependent on p53 in MCF7 and other p53 wt cell lines, except for
MDA-MB-231 cells with a p53 mutation. Previous studies have
revealed that MAPK6, as a target gene of miRNAs, is involved in the
regulation of cell apoptosis (31,32).
At present, we have not found any reports concerning MAPK6 and p53.
We will further explore the specific underlying mechanism of MAPK6
in apoptosis and whether it is related to p53 in the future.
In summary, the present findings support the
hypothesis that miR-653-5p acts as a novel tumor suppressor in the
progression of breast cancer by directly targeting MAPK6.
Consequently, the present study indicates that miR-653-5p may act
as a novel target for the treatment of breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ designed the study and drafted the manuscript.
MZ, HW and XZ performed the experiments. HW and FL performed the
statistical analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Israel BB, Tilghman SL, Parker-Lemieux K
and Payton-Stewart F: Phytochemicals: Current strategies for
treating breast cancer. Oncol Lett. 15:7471–7478. 2018.PubMed/NCBI
|
|
3
|
Calaf GM and Roy D: Metastatic genes
targeted by an antioxidant in an established radiation- and
estrogen-breast cancer model. Int J Oncol. 51:1590–1600. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cristofanilli M and Fortina P: Circulating
tumor DNA to monitor metastatic breast cancer. N Engl J Med.
369:932013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Si W, Shen J, Du C, Chen D, Gu X, Li C,
Yao M, Pan J, Cheng J, Jiang D, et al: A miR-20a/MAPK1/c-Myc
regulatory feedback loop regulates breast carcinogenesis and
chemoresistance. Cell Death Differ. 25:406–420. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen S, Wang Y, Zhang JH, Xia QJ, Sun Q,
Li ZK, Zhang JG, Tang MS and Dong MS: Long non-coding RNA PTENP1
inhibits proliferation and migration of breast cancer cells via AKT
and MAPK signaling pathways. Oncol Lett. 14:4659–4662. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fabbri M: TLRs as miRNA receptors. Cancer
Res. 72:6333–6337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Torres S, García-Palmero I, Bartolomé RA,
Fernandez-Aceñero MJ, Molina E, Calviño E, Segura MF and Casal JI:
Combined miRNA profiling and proteomics demonstrates that different
miRNAs target a common set of proteins to promote colorectal cancer
metastasis. J Pathol. 242:39–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Bi L, Wang Q, Wen M, Li C, Ren Y,
Jiao Q, Mao JH, Wang C, Wei G and Wang Y: miR-1204 targets VDR to
promotes epithelial-mesenchymal transition and metastasis in breast
cancer. Oncogene. 37:3426–3439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
El Helou R, Pinna G, Cabaud O, Wicinski J,
Bhajun R, Guyon L, Rioualen C, Finetti P, Gros A, Mari B, et al:
miR-600 acts as a bimodal switch that regulates breast cancer stem
cell fate through WNT signaling. Cell Rep. 18:2256–2268. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu W, Xu Y, Guan H and Meng H: Clinical
potential of miR-940 as a diagnostic and prognostic biomarker in
breast cancer patients. Cancer Biomark. 22:487–493. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao YL, Dong W, Li YZ and Han W:
MicroRNA-653 inhibits thymocyte proliferation and induces thymocyte
apoptosis in mice with autoimmune myasthenia gravis by
downregulating TRIM9. Neuroimmunomodulation. 26:7–18. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan L, Feng Y, Li L, et al: Effect of
microRNA-653 on biological characteristics of human non-small cell
lung cancer cells by targeting OIP5 gene and regulating mTOR
signaling pathway. J Mod Oncol. 26:831–837. 2018.
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tokunaga E, Oki E, Egashira A, Sadanaga N,
Morita M, Kakeji Y and Maehara Y: Deregulation of the Akt pathway
in human cancer. Curr Cancer Drug Targets. 8:27–36. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Paladini L, Fabris L, Bottai G, Raschioni
C, Calin GA and Santarpia L: Targeting microRNAs as key modulators
of tumor immune response. J Exp Clin Cancer Res. 35:1032016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morgensztern D and McLeod HL:
PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer
Drugs. 16:797–803. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mayer IA and Arteaga CL: The PI3K/AKT
pathway as a target for cancer treatment. Annu Rev Med. 67:11–28.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brand F, Schumacher S, Kant S, Menon MB,
Simon R, Turgeon B, Britsch S, Meloche S, Gaestel M and Kotlyarov
A: The extracellular signal-regulated kinase 3 [mitogen-activated
protein kinase 6 (MAPK6)]-MAPK-activated protein kinase 5 signaling
complex regulates septin function and dendrite morphology. Mol Cell
Biol. 32:2467–2478. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Al-Mahdi R, Babteen N, Thillai K, Holt M,
Johansen B, Wetting HL, Seternes OM and Wells CM: A novel role for
atypical MAPK kinase ERK3 in regulating breast cancer cell
morphology and migration. Cell Adh Migr. 9:483–494. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Evtimova V, Schwirzke M, Tarbé N,
Burtscher H, Jarsch M, Kaul S and Weidle UH: Identification of
breast cancer metastasis-associated genes by chip technology.
Anticancer Res. 21:3799–3806. 2001.PubMed/NCBI
|
|
25
|
Liang B, Wang S, Zhu XG, Yu YX, Cui ZR and
Yu YZ: Increased expression of mitogen-activated protein kinase and
its upstream regulating signal in human gastric cancer. World J
Gastroenterol. 11:623–628. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Long W, Foulds CE, Qin J, Liu J, Ding C,
Lonard DM, Solis LM, Wistuba II, Qin J, Tsai SY, et al: ERK3
signals through SRC-3 coactivator to promote human lung cancer cell
invasion. J Clin Invest. 122:1869–1880. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou YH, Huang YY and Ma M: MicroRNA-138
inhibits proliferation and induces apoptosis of laryngeal carcinoma
via targeting MAPK6. Eur Rev Med Pharmacol Sci. 22:5569–5575.
2018.PubMed/NCBI
|
|
28
|
Wu J, Zhao Y, Li F and Qiao B: MiR-144-3p:
A novel tumor suppressor targeting MAPK6 in cervical cancer. J
Physiol Biochem. 75:143–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lv P, Qiu X, Gu Y, Yang X, Xu X and Yang
Y: Long non-coding RNA SNHG6 enhances cell proliferation, migration
and invasion by regulating miR-26a-5p/MAPK6 in breast cancer.
Biomed Pharmacother. 110:294–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Potapova O, Gorospe M, Dougherty RH, Dean
NM, Gaarde WA and Holbrook NJ: Inhibition of c-Jun N-terminal
kinase 2 expression suppresses growth and induces apoptosis of
human tumor cells in a p53-dependent manner. Mol Cell Biol.
20:1713–1722. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu C, Huang S, Wu F and Ding H: miR-98
inhibits cell proliferation and induces cell apoptosis by targeting
MAPK6 in HUVECs. Exp Ther Med. 15:2755–2760. 2018.PubMed/NCBI
|
|
32
|
Hao W, Zhao ZH, Meng QT, Tie ME, Lei SQ
and Xia ZY: Propofol protects against hepatic ischemia/reperfusion
injury via miR-133a-5p regulating the expression of MAPK6. Cell
Biol Int. 41:495–504. 2017. View Article : Google Scholar : PubMed/NCBI
|